Mosaic Genome of a British Cider Yeast
Abstract
:1. Introduction
2. Results
2.1. Genome Sequencing of E1, A British Cider Yeast
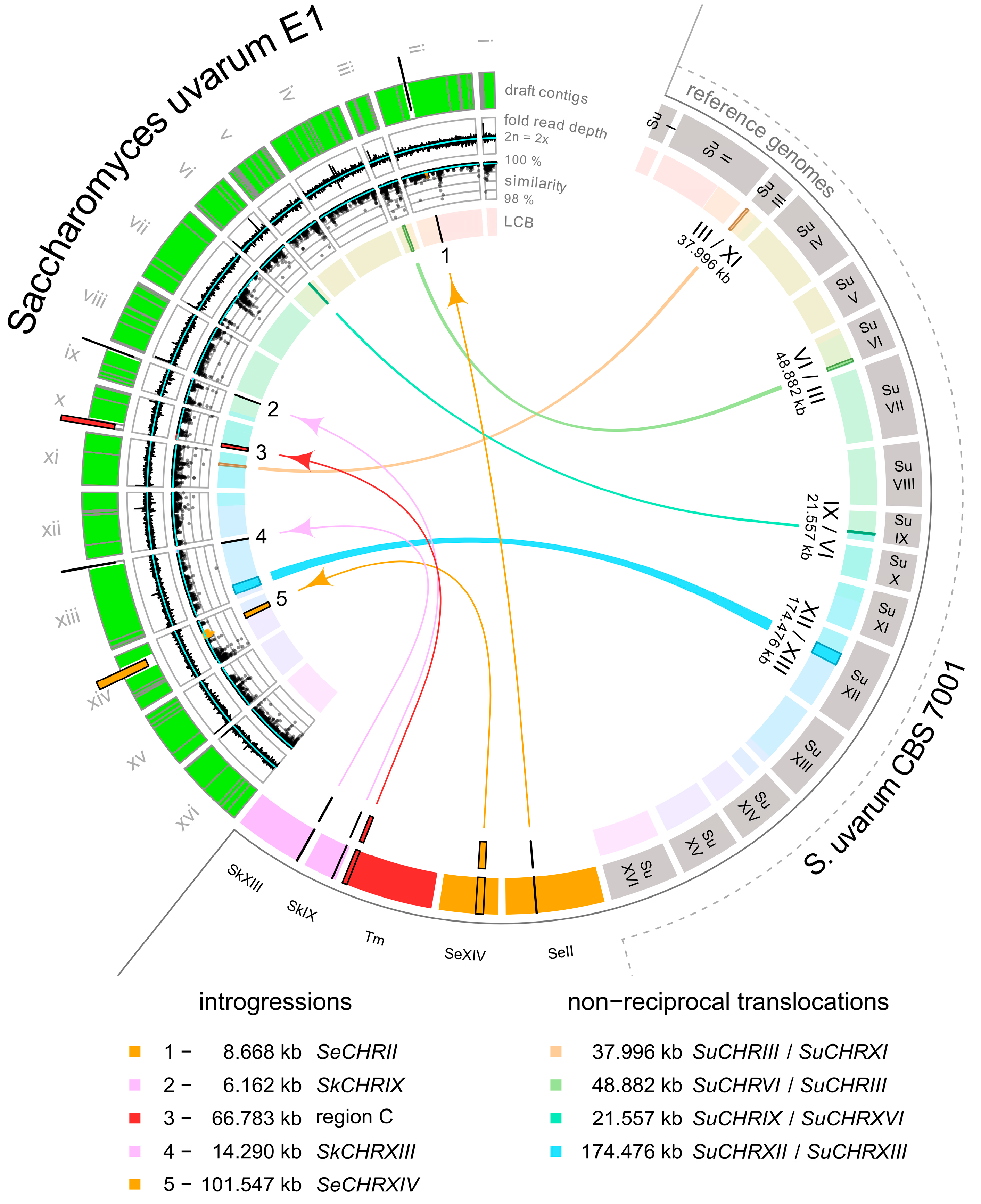
2.2. Analysis of the Introgressed Region Derived from S. kudriavzevii
2.3. Analysis on the Origin of S. eubayanus DNA Introgressed into the E1 Cider Yeast
2.4. Analysis of the Introgressed Region Derived from T. microellipsoides
2.5. Characterization of Growth and Fermentation Performance of E1
2.6. Deletion of FOT Genes in E1 Reduces Nitrogen Uptake
3. Discussion
4. Materials and Methods
4.1. Strains Used and Generated
4.2. Media, Growth and Fermentation Conditions
4.3. FOT-Gene Deletions
4.4. Analytical Methods
4.5. Genome Sequencing and Assembly
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alsammar, H.; Delneri, D. An update on the diversity, ecology and biogeography of the Saccharomyces genus. FEMS Yeast Res. 2020, 20, foaa013. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.V.; Boekhout, T. Characterization of Saccharomyces uvarum (Beijerinck, 1898) and related hybrids: Assessment of molecular markers that predict the parent and hybrid genomes and a proposal to name yeast hybrids. FEMS Yeast Res. 2017, 17, fox014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naumov, G.I.; Masneuf, I.; Naumova, E.S.; Aigle, M.; Dubourdieu, D. Association of Saccharomyces bayanus var. uvarum with some French wines: Genetic analysis of yeast populations. Res. Microbiol. 2000, 151, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Naumov, G.I.; Naumova, E.S.; Antunovics, Z.; Sipiczki, M. Saccharomyces bayanus var. uvarum in Tokaj wine-making of Slovakia and Hungary. Appl. Microbiol. Biotechnol. 2002, 59, 727–730. [Google Scholar] [CrossRef]
- Rementeria, A.; Rodriguez, J.A.; Cadaval, A.; Amenabar, R.; Muguruza, J.R.; Hernando, F.L.; Sevilla, M.J. Yeast associated with spontaneous fermentations of white wines from the “Txakoli de Bizkaia” region (Basque Country, North Spain). Int. J. Food Microbiol. 2003, 86, 201–207. [Google Scholar] [CrossRef]
- McCarthy, G.C.; Morgan, S.C.; Martiniuk, J.T.; Newman, B.L.; McCann, S.E.; Measday, V.; Durall, D.M. An indigenous Saccharomyces uvarum population with high genetic diversity dominates uninoculated Chardonnay fermentations at a Canadian winery. PLoS ONE 2021, 16, e0225615. [Google Scholar] [CrossRef]
- Torriani, S.; Zapparoli, G.; Suzzi, G. Genetic and phenotypic diversity of Saccharomyces sensu stricto strains isolated from Amarone wine. Diversity of Saccharomyces strains from Amarone wine. Antonie Leeuwenhoek 1999, 75, 207–215. [Google Scholar] [CrossRef]
- Rodriguez, M.E.; Perez-Traves, L.; Sangorrin, M.P.; Barrio, E.; Querol, A.; Lopes, C.A. Saccharomyces uvarum is responsible for the traditional fermentation of apple chicha in Patagonia. FEMS Yeast Res. 2017, 17, fow109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-Duarte, R.; Umek, L.; Mendes, I.; Castro, C.C.; Fonseca, N.; Martins, R.; Silva-Ferreira, A.C.; Sampaio, P.; Pais, C.; Schuller, D. New integrative computational approaches unveil the Saccharomyces cerevisiae pheno-metabolomic fermentative profile and allow strain selection for winemaking. Food Chem. 2016, 211, 509–520. [Google Scholar] [CrossRef] [Green Version]
- Saerens, S.M.; Delvaux, F.R.; Verstrepen, K.J.; Thevelein, J.M. Production and biological function of volatile esters in Saccharomyces cerevisiae. Microb. Biotechnol. 2010, 3, 165–177. [Google Scholar] [CrossRef] [Green Version]
- Romano, P.; Braschi, G.; Siesto, G.; Patrignani, F.; Lanciotti, R. Role of Yeasts on the Sensory Component of Wines. Foods 2022, 11, 1921. [Google Scholar] [CrossRef]
- Lyons, S.M.; Morgan, S.C.; McCann, S.; Sanderson, S.; Newman, B.L.; Watson, T.L.; Jiranek, V.; Durall, D.M.; Zandberg, W.F. Unique volatile chemical profiles produced by indigenous and commercial strains of Saccharomyces uvarum and Saccharomyces cerevisiae during laboratory-scale Chardonnay fermentations. OENO One 2021, 55, 101–122. [Google Scholar] [CrossRef]
- Masneuf-Pomarede, I.; Bely, M.; Marullo, P.; Lonvaud-Funel, A.; Dubourdieu, D. Reassessment of phenotypic traits for Saccharomyces bayanus var. uvarum wine yeast strains. Int. J. Food Microbiol. 2010, 139, 79–86. [Google Scholar] [CrossRef]
- Gonzalez Flores, M.; Rodriguez, M.E.; Origone, A.C.; Oteiza, J.M.; Querol, A.; Lopes, C.A. Saccharomyces uvarum isolated from patagonian ciders shows excellent fermentative performance for low temperature cidermaking. Food Res. Int. 2019, 126, 108656. [Google Scholar] [CrossRef]
- Minebois, R.; Perez-Torrado, R.; Querol, A. A time course metabolism comparison among Saccharomyces cerevisiae, S. uvarum and S. kudriavzevii species in wine fermentation. Food Microbiol. 2020, 90, 103484. [Google Scholar] [CrossRef]
- Perez, D.; Denat, M.; Perez-Traves, L.; Heras, J.M.; Guillamon, J.M.; Ferreira, V.; Querol, A. Generation of intra- and interspecific Saccharomyces hybrids with improved oenological and aromatic properties. Microb. Biotechnol. 2022, 15, 2266–2280. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Heras, J.M.; Gamero, A.; Querol, A.; Guillamon, J.M. Impact of Nitrogen Addition on Wine Fermentation by S. cerevisiae Strains with Different Nitrogen Requirements. J. Agric. Food Chem. 2021, 69, 6022–6031. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jia, B.; Sun, X.; Ai, J.; Wang, L.; Wang, C.; Zhao, F.; Zhan, J.; Huang, W. Effect of initial ph on growth characteristics and fermentation properties of Saccharomyces cerevisiae. J. Food Sci. 2015, 80, M800–M808. [Google Scholar] [CrossRef] [PubMed]
- Perez, D.; Jaehde, I.; Guillamon, J.M.; Heras, J.M.; Querol, A. Screening of Saccharomyces strains for the capacity to produce desirable fermentative compounds under the influence of different nitrogen sources in synthetic wine fermentations. Food Microbiol. 2021, 97, 103763. [Google Scholar] [CrossRef] [PubMed]
- Almeida, P.; Goncalves, C.; Teixeira, S.; Libkind, D.; Bontrager, M.; Masneuf-Pomarede, I.; Albertin, W.; Durrens, P.; Sherman, D.J.; Marullo, P.; et al. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 2014, 5, 4044. [Google Scholar] [CrossRef]
- Hall, C.; Brachat, S.; Dietrich, F.S. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1102–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsit, S.; Dequin, S. Diversity and adaptive evolution of Saccharomyces wine yeast: A review. FEMS Yeast Res. 2015, 15, fov067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langdon, Q.K.; Peris, D.; Baker, E.P.; Opulente, D.A.; Nguyen, H.V.; Bond, U.; Goncalves, P.; Sampaio, J.P.; Libkind, D.; Hittinger, C.T. Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 2019, 3, 1576–1586. [Google Scholar] [CrossRef]
- Giannakou, K.; Cotterrell, M.; Delneri, D. Genomic Adaptation of Saccharomyces Species to Industrial Environments. Front. Genet. 2020, 11, 916. [Google Scholar] [CrossRef] [PubMed]
- Lopandic, K.; Pfliegler, W.P.; Tiefenbrunner, W.; Gangl, H.; Sipiczki, M.; Sterflinger, K. Genotypic and phenotypic evolution of yeast interspecies hybrids during high-sugar fermentation. Appl. Microbiol. Biotechnol. 2016, 100, 6331–6343. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Antunovics, Z.; Karanyicz, E.; Sipiczki, M. Diversity and Postzygotic Evolution of the Mitochondrial Genome in Hybrids of Saccharomyces Species Isolated by Double Sterility Barrier. Front. Microbiol. 2020, 11, 838. [Google Scholar] [CrossRef]
- Wendland, J. Lager yeast comes of age. Eukaryot. Cell 2014, 13, 1256–1265. [Google Scholar] [CrossRef] [Green Version]
- Albertin, W.; Chernova, M.; Durrens, P.; Guichoux, E.; Sherman, D.J.; Masneuf-Pomarede, I.; Marullo, P. Many interspecific chromosomal introgressions are highly prevalent in Holarctic Saccharomyces uvarum strains found in human-related fermentations. Yeast 2018, 35, 141–156. [Google Scholar] [CrossRef] [Green Version]
- Novo, M.; Bigey, F.; Beyne, E.; Galeote, V.; Gavory, F.; Mallet, S.; Cambon, B.; Legras, J.L.; Wincker, P.; Casaregola, S.; et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. USA 2009, 106, 16333–16338. [Google Scholar] [CrossRef]
- Galeote, V.; Bigey, F.; Beyne, E.; Novo, M.; Legras, J.L.; Casaregola, S.; Dequin, S. Amplification of a Zygosaccharomyces bailii DNA segment in wine yeast genomes by extrachromosomal circular DNA formation. PLoS ONE 2011, 6, e17872. [Google Scholar] [CrossRef] [Green Version]
- Marsit, S.; Mena, A.; Bigey, F.; Sauvage, F.X.; Couloux, A.; Guy, J.; Legras, J.L.; Barrio, E.; Dequin, S.; Galeote, V. Evolutionary Advantage Conferred by an Eukaryote-to-Eukaryote Gene Transfer Event in Wine Yeasts. Mol. Biol. Evol. 2015, 32, 1695–1707. [Google Scholar] [CrossRef] [Green Version]
- Galeote, V.; Novo, M.; Salema-Oom, M.; Brion, C.; Valerio, E.; Goncalves, P.; Dequin, S. FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high-affinity fructose/H+ symporter. Microbiology 2010, 156 Pt 12, 3754–3761. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Sun, H.; Li, J.; Qin, S.; Yang, W.; Ma, X.; Qiao, X.; Yang, B. Effects of Genetic Background and Altitude on Sugars, Malic Acid and Ascorbic Acid in Fruits of Wild and Cultivated Apples (Malus sp.). Foods 2021, 10, 2950. [Google Scholar] [CrossRef] [PubMed]
- Calugar, P.C.; Coldea, T.E.; Salanță, L.C.; Pop, C.R.; Pasqualone, A.; Burja-Udrea, C.; Zhao, H.; Mudura, E. An Overview of the Factors Influencing Apple Cider Sensory and Microbial Quality from Raw Materials to Emerging Processing Technologies. Processes 2021, 9, 502. [Google Scholar] [CrossRef]
- Morrissey, W.F.; Davenport, B.; Querol, A.; Dobson, A.D. The role of indigenous yeasts in traditional Irish cider fermentations. J. Appl. Microbiol. 2004, 97, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.M.; Swiegers, J.H.; Varela, C.; Pretorius, I.S.; Agosin, E. Influence of wine fermentation temperature on the synthesis of yeast-derived volatile aroma compounds. Appl. Microbiol. Biotechnol. 2007, 77, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Bedrinana, R.P.; Alonso, J.J.M.; Valles, B.S. Evaluation of autochthonous Saccharomyces bayanus strains under stress conditions for making ice ciders. Lwt-Food Sci. Technol. 2017, 81, 217–225. [Google Scholar] [CrossRef]
- Lopez-Malo, M.; Querol, A.; Guillamon, J.M. Metabolomic comparison of Saccharomyces cerevisiae and the cryotolerant species S. bayanus var. uvarum and S. kudriavzevii during wine fermentation at low temperature. PLoS ONE 2013, 8, e60135. [Google Scholar] [CrossRef] [Green Version]
- Peris, D.; Perez-Torrado, R.; Hittinger, C.T.; Barrio, E.; Querol, A. On the origins and industrial applications of Saccharomyces cerevisiae x Saccharomyces kudriavzevii hybrids. Yeast 2018, 35, 51–69. [Google Scholar] [CrossRef] [Green Version]
- Bernardi, B.; Michling, F.; Muno-Bender, J.; Matti, K.; Wendland, J. The genome sequence of the Champagne-Epernay-Geisenheim wine yeast reveals its hybrid nature. FEMS Yeast Res. 2023, in press. [Google Scholar]
- Gangl, H.; Batusic, M.; Tscheik, G.; Tiefenbrunner, W.; Hack, C.; Lopandic, K. Exceptional fermentation characteristics of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. New Biotechnol. 2009, 25, 244–251. [Google Scholar] [CrossRef]
- Gonzalez Flores, M.; Rodriguez, M.E.; Oteiza, J.M.; Barbagelata, R.J.; Lopes, C.A. Physiological characterization of Saccharomyces uvarum and Saccharomyces eubayanus from Patagonia and their potential for cidermaking. Int. J. Food Microbiol. 2017, 249, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, F.; Vidgren, V.; Ruohonen, L.; Gibson, B. Maltose and maltotriose utilisation by group I strains of the hybrid lager yeast Saccharomyces pastorianus. FEMS Yeast Res. 2016, 16, fow053. [Google Scholar] [CrossRef] [Green Version]
- Bergin, S.A.; Allen, S.; Hession, C.; Cinnéide, E.O.; Ryan, A.; Byrne, K.P.; Cróinín, O.C.; Wolfe, K.H.; Butler, G. Identification of European isolates of the lager yeast parent Saccharomyces eubayanus. FEMS Yeast Res. 2022, 22, foac053. [Google Scholar] [CrossRef] [PubMed]
- Scannell, D.R.; Zill, O.A.; Rokas, A.; Payen, C.; Dunham, M.J.; Eisen, M.B.; Rine, J.; Johnston, M.; Hittinger, C.T. The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. G3 2011, 1, 11–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Sinclair, K.; Warner, J.P.; Bonthron, D.T. The ASP1 gene of Saccharomyces cerevisiae, encoding the intracellular isozyme of L-asparaginase. Gene 1994, 144, 37–43. [Google Scholar] [CrossRef]
- Eleuterio Dos Santos, C.M.; Pietrowski Gde, A.; Braga, C.M.; Rossi, M.J.; Ninow, J.; Machado Dos Santos, T.P.; Wosiacki, G.; Jorge, R.M.; Nogueira, A. Apple Aminoacid Profile and Yeast Strains in the Formation of Fusel Alcohols and Esters in Cider Production. J. Food Sci. 2015, 80, C1170–C1177. [Google Scholar] [CrossRef]
- Coral-Medina, A.; Fenton, D.A.; Varela, J.; Baranov, P.V.; Camarasa, C.; Morrissey, J.P. The evolution and role of the periplasmic asparaginase Asp3 in yeast. FEMS Yeast Res. 2022, 22, foac044. [Google Scholar] [CrossRef]
- Gallone, B.; Mertens, S.; Gordon, J.L.; Maere, S.; Verstrepen, K.J.; Steensels, J. Origins, evolution, domestication and diversity of Saccharomyces beer yeasts. Curr. Opin. Biotechnol. 2018, 49, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Rios, E.; Guillamon, J.M. Genomic Adaptations of Saccharomyces Genus to Wine Niche. Microorganisms 2022, 10, 1811. [Google Scholar] [CrossRef] [PubMed]
- Timouma, S.; Balarezo-Cisneros, L.N.; Pinto, J.; De La Cerda, R.; Bond, U.; Schwartz, J.M.; Delneri, D. Transcriptional Profile of the Industrial Hybrid Saccharomyces pastorianus Reveals Temperature-Dependent Allele Expression Bias and Preferential Orthologous Protein Assemblies. Mol. Biol. Evol. 2021, 38, 5437–5452. [Google Scholar] [CrossRef] [PubMed]
- Pontes, A.; Cadez, N.; Goncalves, P.; Sampaio, J.P. A Quasi-Domesticate Relic Hybrid Population of Saccharomyces cerevisiae × S. paradoxus Adapted to Olive Brine. Front. Genet. 2019, 10, 449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becerra-Rodriguez, C.; Marsit, S.; Galeote, V. Diversity of Oligopeptide Transport in Yeast and Its Impact on Adaptation to Winemaking Conditions. Front. Genet. 2020, 11, 602. [Google Scholar] [CrossRef]
- Boeckstaens, M.; Llinares, E.; Van Vooren, P.; Marini, A.M. The TORC1 effector kinase Npr1 fine tunes the inherent activity of the Mep2 ammonium transport protein. Nat. Commun. 2014, 5, 3101. [Google Scholar] [CrossRef] [Green Version]
- Vandenbol, M.; Jauniaux, J.C.; Vissers, S.; Grenson, M. Isolation of the NPR1 gene responsible for the reactivation of ammonia-sensitive amino-acid permeases in Saccharomyces cerevisiae. RNA analysis and gene dosage effects. Eur. J. Biochem. 1987, 164, 607–612. [Google Scholar] [CrossRef]
- Petersen, R.F.; Langkjaer, R.B.; Hvidtfeldt, J.; Gartner, J.; Palmen, W.; Ussery, D.W.; Piskur, J. Inheritance and organisation of the mitochondrial genome differ between two Saccharomyces yeasts. J. Mol. Biol. 2002, 318, 627–636. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Bernardi, B.; Kayacan, Y.; Akan, M.; Wendland, J. Overexpression of RAD51 Enables PCR-Based Gene Targeting in Lager Yeast. Microorganisms 2019, 7, 192. [Google Scholar] [CrossRef] [Green Version]
- Kayacan, Y.; Griffiths, A.; Wendland, J. A script for initiating molecular biology studies with non-conventional yeasts based on Saccharomycopsis schoenii. Microbiol. Res. 2019, 229, 126342. [Google Scholar] [CrossRef]
- Dippel, K.; Matti, K.; Muno-Bender, J.; Michling, F.; Brezina, S.; Semmler, H.; Rauhut, D.; Wendland, J. Co-Fermentations of Kveik with Non-Conventional Yeasts for Targeted Aroma Modulation. Microorganisms 2022, 10, 1922. [Google Scholar] [CrossRef] [PubMed]
- Krause, B.; Löhnertz, O. Saccharopin und Pipecolinsäure: Diagnostische Biomarker in der klassischen Aminosäureanalytik. MTA-Dialog 2017, 18, 316–321. [Google Scholar]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2008, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressivemauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/.
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org.







| Strain | E1 Cider Yeast (ROW 169) |
|---|---|
| Library No. 1 | 2 × 300 bp paired-end, 724,688 read pairs |
| Library No. 2 | 2 × 150 bp paired-end, 1,971,578 read pairs |
| No. of read pairs | 2,696,266 |
| Genome size | 11,987,464 bp |
| No. of scaffolds | 252 |
| Scaffold N50 | 15 |
| Scaffold L50 | 236,996 bp |
| Largest scaffold | 1,000,723 bp |
| GC content | 40% |
| Source | Size [bp] | Position in E1 | DNA Identity | N | Genes | E1 Contigs |
|---|---|---|---|---|---|---|
| S. eubayanus CHRII | 8668 | CHRII | 99.70% ± 0.27% | 17 | 7 | 1 |
| S. eubayanus CHRXIV | 101,547 | CHRXIV | 99.62% ± 0.36% | 202 | 53 | 1 |
| S. kudriavzevii CHRXIII | 6162 | CHRIX | 99.84% ± 0.18% | 10 | 3 | 1 |
| S. kudriavzevii CHRXIII | 14,290 | CHRXIII | 99.67% ± 0.23% | 23 | 9 | 3 |
| T. microellipsoides | 66,783 | CHRX | 99.59% ± 2.32% | 127 | 19 | 2 |
| Strain | Name | E1 | CBS 1503 | CBS 1513 | CBS 1483 | WS 34/70 | UCD646 | UCD650 | FM1318 |
|---|---|---|---|---|---|---|---|---|---|
| S. uvarum | E1 | - | 99.981 | 99.977 | 99.978 | 99.977 | 99.808 | 99.804 | 99.612 |
| Saaz | CBS 1503 | 19 | - | 99.990 | 99.991 | 99.990 | 99.807 | 99.801 | 99.611 |
| Saaz | CBS 1513 | 23 | 10 | - | 99.987 | 99.986 | 99.803 | 99.797 | 99.607 |
| Frohberg | CBS 1483 | 22 | 9 | 13 | - | 99.997 | 99.804 | 99.798 | 99.608 |
| Frohberg | WS 34/70 | 23 | 10 | 14 | 3 | - | 99.803 | 99.797 | 99.607 |
| S. eubayanus | UCD646 | 192 | 193 | 197 | 196 | 197 | - | 99.966 | 99.596 |
| S eubayanus | UCD650 | 196 | 199 | 203 | 202 | 203 | 34 | - | 99.600 |
| S. eubayanus | FM1318 | 400 | 401 | 405 | 404 | 405 | 416 | 412 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardi, B.; Michling, F.; Fröhlich, J.; Wendland, J. Mosaic Genome of a British Cider Yeast. Int. J. Mol. Sci. 2023, 24, 11232. https://doi.org/10.3390/ijms241311232
Bernardi B, Michling F, Fröhlich J, Wendland J. Mosaic Genome of a British Cider Yeast. International Journal of Molecular Sciences. 2023; 24(13):11232. https://doi.org/10.3390/ijms241311232
Chicago/Turabian StyleBernardi, Beatrice, Florian Michling, Jürgen Fröhlich, and Jürgen Wendland. 2023. "Mosaic Genome of a British Cider Yeast" International Journal of Molecular Sciences 24, no. 13: 11232. https://doi.org/10.3390/ijms241311232





