The Novel Role of Zfp296 in Mammalian Embryonic Genome Activation as an H3K9me3 Modulator
Abstract
1. Introduction
2. Results
2.1. Zfp296 Is Significantly Expressed in the EGA Stage of Human, Mouse, and Goat, and Has High Species Conservation
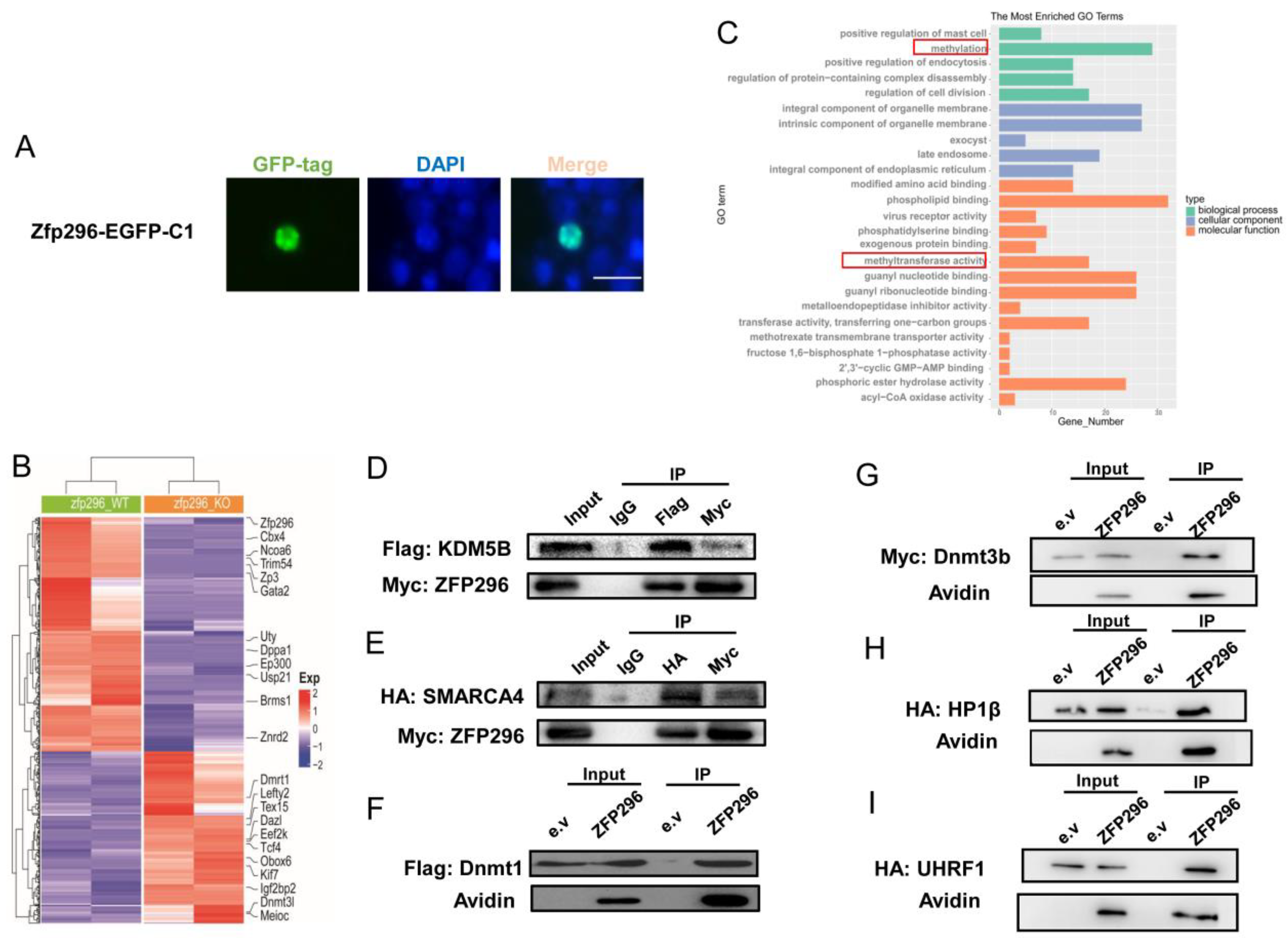
2.2. ZFP296 Localizes to the Nucleus and Interacts with Multiple Proteins Which Were Key to Methylation
2.3. C2H2-TYPE2 and C2H2-TYPE3 of ZFP296 Can Co-Regulate H3K9me3
2.4. Interfering with Zfp296 Expression Arrests Early Embryonic Development of Mice
3. Discussion
3.1. Zfp296 May Play a Similar and Important Role in Early Mammalian Embryonic Development
3.2. Zfp296 Can Affect EGA by Negatively Regulating H3K9me3
4. Materials and Methods
4.1. Data Analysis
4.1.1. Data Access, Analysis, and Visualization of RNA-Seq and ChIP-Seq from Previous Studies
4.1.2. GO Enrichment Analysis
4.1.3. DNA Sequence and Protein Sequence Analysis of Zfp296
4.2. Cell Culture and Transfection
4.3. Construction of Vectors
4.4. Western Blot
4.5. Co-Immunoprecipitation
4.6. Immunofluorescence Staining of Cells
4.7. Collection of Mouse Zygotes
4.8. Immunofluorescence Staining of Embryos
4.9. Microinjection
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EGA | embryonic genome activation |
| ZGA | Zygotic genome activation |
| Zfp296 | zinc finger protein 296 |
| pdaj | adjusted p-value |
| C2H2 | Cys2-His2 |
| KDM5B | lysine demethylase 5B |
| SMARCA4 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| DNMT1 | DNA methyltransferase 1 |
| DNMT3B | DNA methyltransferase 3 beta |
| HP1β | chromobox 1 |
| UHRF1 | ubiquitin like with PHD and ring finger domains 1 |
| H3K9me3 | Trimethylation of lysine 9 on histone H3 protein |
| H3K4me3 | Tri-methylation of lysine 4 on histone H3 |
| DUX | Double homeobox |
| SCNT | Somatic cell nuclear transfer |
| zf | zinc finger |
| mESCs | mouse embryonic stem cells |
| KO | knockout |
| WT | wild type |
| e.v | empty vector |
| GO | gene ontology |
| NC | Negative control |
| KRAB | Krüppel-associated box domain |
| NuRD | The nucleosome-remodeling and deacetylase |
References
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef]
- Vastenhouw, N.L.; Cao, W.X.; Lipshitz, H.D. The maternal-to-zygotic transition revisited. Development 2019, 146, dev161471. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Su, G.; Wang, S.; Yang, L.; Liao, M.; Wei, Z.; Bai, C.; Li, G. Exploring timing activation of functional pathway based on differential co-expression analysis in preimplantation embryogenesis. Oncotarget 2016, 7, 74120–74131. [Google Scholar] [CrossRef] [PubMed]
- Bultman, S.J.; Gebuhr, T.C.; Pan, H.; Svoboda, P.; Schultz, R.M.; Magnuson, T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes. Dev. 2006, 20, 1744–1754. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zhang, C.; Zhang, Y. Epigenetic regulation of mouse preimplantation embryo development. Curr. Opin. Genet. Dev. 2020, 64, 13–20. [Google Scholar] [CrossRef]
- Abe, K.I.; Funaya, S.; Tsukioka, D.; Kawamura, M.; Suzuki, Y.; Suzuki, M.G.; Schultz, R.M.; Aoki, F. Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. USA 2018, 115, E6780–E6788. [Google Scholar] [CrossRef]
- Asami, M.; Lam, B.Y.H.; Ma, M.K.; Rainbow, K.; Braun, S.; VerMilyea, M.D.; Yeo, G.S.H.; Perry, A.C.F. Human embryonic genome activation initiates at the one-cell stage. Cell Stem Cell 2022, 29, 209–216.e4. [Google Scholar] [CrossRef]
- Graf, A.; Krebs, S.; Zakhartchenko, V.; Schwalb, B.; Blum, H.; Wolf, E. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 4139–4144. [Google Scholar] [CrossRef]
- Ling, Y.H.; Zheng, Q.; Li, Y.S.; Sui, M.H.; Wu, H.; Zhang, Y.H.; Chu, M.X.; Ma, Y.H.; Fang, F.G.; Xu, L.N. Identification of lncRNAs by RNA Sequencing Analysis during in vivo Pre-Implantation Developmental Transformation in the Goat. Front. Genet. 2019, 10, 1040. [Google Scholar] [CrossRef]
- Morita, K.; Hatanaka, Y.; Ihashi, S.; Asano, M.; Miyamoto, K.; Matsumoto, K. Symmetrically dimethylated histone H3R2 promotes global transcription during minor zygotic genome activation in mouse pronuclei. Sci. Rep. 2021, 11, 10146. [Google Scholar] [CrossRef]
- Wang, Z.D.; Duan, L.; Zhang, Z.H.; Song, S.H.; Bai, G.Y.; Zhang, N.; Shen, X.H.; Shen, J.L.; Lei, L. Methyl-CpG-Binding Protein 2 Improves the Development of Mouse Somatic Cell Nuclear Transfer Embryos. Cell. Reprogramming 2016, 18, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic Genome Activation in Vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lu, X.; Dean, J. The maternal to zygotic transition in mammals. Mol. Aspects Med. 2013, 34, 919–938. [Google Scholar] [CrossRef]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016, 534, 652–657. [Google Scholar] [CrossRef]
- Liu, G.; Wang, W.; Hu, S.; Wang, X.; Zhang, Y. Inherited DNA methylation primes the establishment of accessible chromatin during genome activation. Genome Res. 2018, 28, 998–1007. [Google Scholar] [CrossRef]
- Xia, W.; Xu, J.; Yu, G.; Yao, G.; Xu, K.; Ma, X.; Zhang, N.; Liu, B.; Li, T.; Lin, Z.; et al. Resetting histone modifications during human parental-to-zygotic transition. Science 2019, 365, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Dahl, J.A.; Jung, I.; Aanes, H.; Greggains, G.D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A.Y.; et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537, 548–552. [Google Scholar] [CrossRef]
- Wang, C.; Liu, X.; Gao, Y.; Yang, L.; Li, C.; Liu, W.; Chen, C.; Kou, X.; Zhao, Y.; Chen, J.; et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 2018, 20, 620–631. [Google Scholar] [CrossRef]
- Sankar, A.; Lerdrup, M.; Manaf, A.; Johansen, J.V.; Gonzalez, J.M.; Borup, R.; Blanshard, R.; Klungland, A.; Hansen, K.; Andersen, C.Y.; et al. KDM4A regulates the maternal-to-zygotic transition by protecting broad H3K4me3 domains from H3K9me3 invasion in oocytes. Nat. Cell Biol. 2020, 22, 380–388. [Google Scholar] [CrossRef]
- Xu, R.; Li, S.; Wu, Q.; Li, C.; Jiang, M.; Guo, L.; Chen, M.; Yang, L.; Dong, X.; Wang, H.; et al. Stage-specific H3K9me3 occupancy ensures retrotransposon silencing in human pre-implantation embryos. Cell Stem Cell 2022, 29, 1051–1066.e8. [Google Scholar] [CrossRef]
- Chitrakar, A.; Noon, M.; Xiao, A.Z. Taming the transposon: H3K9me3 turns foe to friend in human development. Cell Stem Cell 2022, 29, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Liu, Z.; Chen, B.; Wan, Y.; Yang, H.; Zhang, Y.; Cai, Y.; Zhou, J.; Wang, F. Aberrant DNA and histone methylation during zygotic genome activation in goat cloned embryos. Theriogenology 2020, 148, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Gao, Y.; Su, J.; Zhang, J.; Xing, X.; Zhou, C.; Yao, K.; An, Q.; Zhang, Y. H3K9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming. Development 2018, 145, dev158261. [Google Scholar] [CrossRef] [PubMed]
- Schmitges, F.W.; Radovani, E.; Najafabadi, H.S.; Barazandeh, M.; Campitelli, L.F.; Yin, Y.; Jolma, A.; Zhong, G.; Guo, H.; Kanagalingam, T.; et al. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 2016, 26, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- Bian, C.; Chen, Q.; Yu, X. The zinc finger proteins ZNF644 and WIZ regulate the G9a/GLP complex for gene repression. eLife 2015, 4, e05606. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.M.; Parker, J.S.; Liu, F.; Rothbart, S.B.; Ait-Si-Ali, S.; Strahl, B.D.; Jin, J.; Davis, I.J.; Mosley, A.L.; Pattenden, S.G. A Role for Widely Interspaced Zinc Finger (WIZ) in Retention of the G9a Methyltransferase on Chromatin. J. Biol. Chem. 2015, 290, 26088–26102. [Google Scholar] [CrossRef]
- Ueda, J.; Tachibana, M.; Ikura, T.; Shinkai, Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 2006, 281, 20120–20128. [Google Scholar] [CrossRef]
- Dear, T.N. Cloning, structure, expression analysis, and assignment to mouse chromosome 7 of the gene Zfp296 encoding a zinc finger protein. Mamm. Genome 2000, 11, 1037–1039. [Google Scholar] [CrossRef]
- Bedigian, H.G.; Taylor, B.A.; Meier, H. Expression of murine leukemia viruses in the highly lymphomatous BXH-2 recombinant inbred mouse strain. J. Virol. 1981, 39, 632–640. [Google Scholar] [CrossRef]
- Poland, K.S.; Shardy, D.L.; Azim, M.; Naeem, R.; Krance, R.A.; Dreyer, Z.E.; Neeley, E.S.; Zhang, N.; Qiu, Y.H.; Kornblau, S.M.; et al. Overexpression of ZNF342 by juxtaposition with MPO promoter/enhancer in the novel translocation t(17;19)(q23;q13.32) in pediatric acute myeloid leukemia and analysis of ZNF342 expression in leukemia. Genes Chromosomes Cancer 2009, 48, 480–489. [Google Scholar] [CrossRef]
- Fischedick, G.; Klein, D.C.; Wu, G.; Esch, D.; Hoing, S.; Han, D.W.; Reinhardt, P.; Hergarten, K.; Tapia, N.; Scholer, H.R.; et al. Zfp296 is a novel, pluripotent-specific reprogramming factor. PLoS ONE 2012, 7, e34645. [Google Scholar] [CrossRef]
- Kloet, S.L.; Karemaker, I.D.; van Voorthuijsen, L.; Lindeboom, R.G.H.; Baltissen, M.P.; Edupuganti, R.R.; Poramba-Liyanage, D.W.; Jansen, P.; Vermeulen, M. NuRD-interacting protein ZFP296 regulates genome-wide NuRD localization and differentiation of mouse embryonic stem cells. Nat. Commun. 2018, 9, 4588. [Google Scholar] [CrossRef]
- Hackett, J.A.; Huang, Y.; Gunesdogan, U.; Gretarsson, K.A.; Kobayashi, T.; Surani, M.A. Tracing the transitions from pluripotency to germ cell fate with CRISPR screening. Nat. Commun. 2018, 9, 4292. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, J.; Ling, Y.; Ming, H.; Chen, Z.; Fang, F.; Liu, Y.; Cao, H.; Ding, J.; Cao, Z.; et al. Transcription profiles of oocytes during maturation and embryos during preimplantation development in vivo in the goat. Reprod. Fertil. Dev. 2020, 32, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.L.; Zhou, C.X.; Zhang, X.L.; Liu, P.; Jin, Z.; Zhu, G.Y.; Ma, Y.; Li, J.; Yang, Z.X.; Zhang, D. ZP3 is Required for Germinal Vesicle Breakdown in Mouse Oocyte Meiosis. Sci. Rep. 2017, 7, 41272. [Google Scholar] [CrossRef]
- Chen, J.; Melton, C.; Suh, N.; Oh, J.S.; Horner, K.; Xie, F.; Sette, C.; Blelloch, R.; Conti, M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes. Dev. 2011, 25, 755–766. [Google Scholar] [CrossRef]
- Chu, H.P.; Liao, Y.; Novak, J.S.; Hu, Z.; Merkin, J.J.; Shymkiv, Y.; Braeckman, B.P.; Dorovkov, M.V.; Nguyen, A.; Clifford, P.M.; et al. Germline quality control: eEF2K stands guard to eliminate defective oocytes. Dev. Cell 2014, 28, 561–572. [Google Scholar] [CrossRef]
- Liu, H.B.; Muhammad, T.; Guo, Y.; Li, M.J.; Sha, Q.Q.; Zhang, C.X.; Liu, H.; Zhao, S.G.; Zhao, H.; Zhang, H.; et al. RNA-Binding Protein IGF2BP2/IMP2 is a Critical Maternal Activator in Early Zygotic Genome Activation. Adv. Sci. 2019, 6, 1900295. [Google Scholar] [CrossRef]
- Xhabija, B.; Kidder, B.L. KDM5B is a master regulator of the H3K4-methylome in stem cells, development and cancer. Semin. Cancer Biol. 2019, 57, 79–85. [Google Scholar] [CrossRef]
- Jeong, K.W.; Kim, K.; Situ, A.J.; Ulmer, T.S.; An, W.; Stallcup, M.R. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat. Struct. Mol. Biol. 2011, 18, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, Y. Role of Mammalian DNA Methyltransferases in Development. Annu. Rev. Biochem. 2020, 89, 135–158. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Kobayakawa, S.; Yamagata, K.; Abe, K.; Baba, T. Molecular dynamics of heterochromatin protein 1beta, HP1beta, during mouse preimplantation development. J. Reprod. Dev. 2007, 53, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Maenohara, S.; Unoki, M.; Toh, H.; Ohishi, H.; Sharif, J.; Koseki, H.; Sasaki, H. Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genet. 2017, 13, e1007042. [Google Scholar] [CrossRef]
- Matsuura, T.; Miyazaki, S.; Miyazaki, T.; Tashiro, F.; Miyazaki, J.I. Zfp296 negatively regulates H3K9 methylation in embryonic development as a component of heterochromatin. Sci. Rep. 2017, 7, 12462. [Google Scholar] [CrossRef]
- Nicetto, D.; Donahue, G.; Jain, T.; Peng, T.; Sidoli, S.; Sheng, L.; Montavon, T.; Becker, J.S.; Grindheim, J.M.; Blahnik, K.; et al. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 2019, 363, 294–297. [Google Scholar] [CrossRef]
- Smith, C.L.; Lan, Y.; Jain, R.; Epstein, J.A.; Poleshko, A. Global chromatin relabeling accompanies spatial inversion of chromatin in rod photoreceptors. Sci. Adv. 2021, 7, eabj3035. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, P.; Yan, Z.; Yang, M.; Huo, Y.; Nie, Y.; Zhu, X.; Qiao, J.; Yan, L. Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos. Nat. Commun. 2021, 12, 1247. [Google Scholar] [CrossRef]
- De Iaco, A.; Planet, E.; Coluccio, A.; Verp, S.; Duc, J.; Trono, D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017, 49, 941–945. [Google Scholar] [CrossRef]
- Liu, B.; Xu, Q.; Wang, Q.; Feng, S.; Lai, F.; Wang, P.; Zheng, F.; Xiang, Y.; Wu, J.; Nie, J.; et al. The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 2020, 587, 139–144. [Google Scholar] [CrossRef]
- Chen, J.; Litscher, E.S.; Wassarman, P.M. Inactivation of the mouse sperm receptor, mZP3, by site-directed mutagenesis of individual serine residues located at the combining site for sperm. Proc. Natl. Acad. Sci. USA 1998, 95, 6193–6197. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, J.; Diaz, E.S.; Morales, P. Kinases, phosphatases and proteases during sperm capacitation. Cell Tissue Res. 2012, 349, 765–782. [Google Scholar] [CrossRef] [PubMed]
- Florman, H.M.; Wassarman, P.M. O-linked oligosaccharides of mouse egg ZP3 account for its sperm receptor activity. Cell 1985, 41, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Najafabadi, H.S.; Mnaimneh, S.; Schmitges, F.W.; Garton, M.; Lam, K.N.; Yang, A.; Albu, M.; Weirauch, M.T.; Radovani, E.; Kim, P.M.; et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015, 33, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, X.; Blumenthal, R.M.; Cheng, X. A common mode of recognition for methylated CpG. Trends Biochem. Sci. 2013, 38, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Yao, J.; Wang, Y.; Qin, G.; Zhang, Y.; Wang, Y.; Zhao, J. CRISPR/Cas13d-mediated efficient KDM5B mRNA knockdown in porcine somatic cells and parthenogenetic embryos. Reproduction 2021, 162, 149–160. [Google Scholar] [CrossRef]
- Hainer, S.J.; Boskovic, A.; McCannell, K.N.; Rando, O.J.; Fazzio, T.G. Profiling of Pluripotency Factors in Single Cells and Early Embryos. Cell 2019, 177, 1319–1329.e11. [Google Scholar] [CrossRef]
- Pfeiffer, M.J.; Siatkowski, M.; Paudel, Y.; Balbach, S.T.; Baeumer, N.; Crosetto, N.; Drexler, H.C.; Fuellen, G.; Boiani, M. Proteomic analysis of mouse oocytes reveals 28 candidate factors of the “reprogrammome”. J. Proteome Res. 2011, 10, 2140–2153. [Google Scholar] [CrossRef]
- Liu, X.; Gao, Q.; Li, P.; Zhao, Q.; Zhang, J.; Li, J.; Koseki, H.; Wong, J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 2013, 4, 1563. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Chen, J.; Liu, W.; Lai, W.; Liu, B.; Li, X.; Liu, L.; Xu, S.; Dong, Q.; et al. Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature 2018, 564, 136–140. [Google Scholar] [CrossRef]
- Dahlet, T.; Argueso Lleida, A.; Al Adhami, H.; Dumas, M.; Bender, A.; Ngondo, R.P.; Tanguy, M.; Vallet, J.; Auclair, G.; Bardet, A.F.; et al. Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity. Nat. Commun. 2020, 11, 3153. [Google Scholar] [CrossRef]
- Tardat, M.; Albert, M.; Kunzmann, R.; Liu, Z.; Kaustov, L.; Thierry, R.; Duan, S.; Brykczynska, U.; Arrowsmith, C.H.; Peters, A.H. Cbx2 targets PRC1 to constitutive heterochromatin in mouse zygotes in a parent-of-origin-dependent manner. Mol. Cell 2015, 58, 157–171. [Google Scholar] [CrossRef]
- Nielsen, P.R.; Nietlispach, D.; Mott, H.R.; Callaghan, J.; Bannister, A.; Kouzarides, T.; Murzin, A.G.; Murzina, N.V.; Laue, E.D. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 2002, 416, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Liu, H.; Wang, Y.; He, J.; Xu, S.; Chen, Y.; Kuang, J.; Liu, J.; Guo, L.; Li, D.; et al. SETDB1-Mediated Cell Fate Transition between 2C-Like and Pluripotent States. Cell Rep. 2020, 30, 25–36.e6. [Google Scholar] [CrossRef]
- Maksakova, I.A.; Thompson, P.J.; Goyal, P.; Jones, S.J.; Singh, P.B.; Karimi, M.M.; Lorincz, M.C. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin 2013, 6, 15. [Google Scholar] [CrossRef]
- Rowe, H.M.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010, 463, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Puschendorf, M.; Terranova, R.; Boutsma, E.; Mao, X.; Isono, K.; Brykczynska, U.; Kolb, C.; Otte, A.P.; Koseki, H.; Orkin, S.H.; et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 2008, 40, 411–420. [Google Scholar] [CrossRef]
- Burton, A.; Brochard, V.; Galan, C.; Ruiz-Morales, E.R.; Rovira, Q.; Rodriguez-Terrones, D.; Kruse, K.; Le Gras, S.; Udayakumar, V.S.; Chin, H.G.; et al. Heterochromatin establishment during early mammalian development is regulated by pericentromeric RNA and characterized by non-repressive H3K9me3. Nat. Cell Biol. 2020, 22, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Brayer, K.J.; Kulshreshtha, S.; Segal, D.J. The protein-binding potential of C2H2 zinc finger domains. Cell Biochem. Biophys. 2008, 51, 9–19. [Google Scholar] [CrossRef]
- Burdach, J.; O’Connell, M.R.; Mackay, J.P.; Crossley, M. Two-timing zinc finger transcription factors liaising with RNA. Trends Biochem. Sci. 2012, 37, 199–205. [Google Scholar] [CrossRef]
- Yang, H.; Bai, D.; Li, Y.; Yu, Z.; Wang, C.; Sheng, Y.; Liu, W.; Gao, S.; Zhang, Y. Allele-specific H3K9me3 and DNA methylation co-marked CpG-rich regions serve as potential imprinting control regions in pre-implantation embryo. Nat. Cell Biol. 2022, 24, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Zhang, X.; Lian, F.; Liu, D. Comparison of IVF/ICSI outcomes in advanced reproductive age patients with polycystic ovary syndrome and advanced reproductive age normal controls: A retrospective cohort study. BMC Pregnancy Childbirth 2023, 23, 440. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Wang, H.; Fu, L.; Zhou, W.; Li, Y. A method to improve embryo development potential when fertilization is delayed in mice. Syst. Biol. Reprod. Med. 2020, 66, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Blanco, I.; Soto-Heras, S.; Catalá, M.G.; Roura, M.; Izquierdo, D.; Paramio, M.T. Effect of crocetin added to IVM medium for prepubertal goat oocytes on blastocyst outcomes after IVF, intracytoplasmic sperm injection and parthenogenetic activation. Theriogenology 2020, 155, 70–76. [Google Scholar] [CrossRef]
- Van der Weijden, V.A.; Schmidhauser, M.; Kurome, M.; Knubben, J.; Flöter, V.L.; Wolf, E.; Ulbrich, S.E. Transcriptome dynamics in early in vivo developing and in vitro produced porcine embryos. BMC Genom. 2021, 22, 139. [Google Scholar] [CrossRef]
- Skrzyszowska, M.; Samiec, M. Generating Cloned Goats by Somatic Cell Nuclear Transfer-Molecular Determinants and Application to Transgenics and Biomedicine. Int. J. Mol. Sci. 2021, 22, 7490. [Google Scholar] [CrossRef]
- Wakayama, S.; Terashita, Y.; Tanabe, Y.; Hirose, N.; Wakayama, T. Mouse Cloning Using Outbred Oocyte Donors and Nontoxic Reagents. Methods Mol. Biol. 2023, 2647, 151–168. [Google Scholar] [CrossRef]
- Samiec, M.; Skrzyszowska, M. Preimplantation developmental capability of cloned pig embryos derived from different types of nuclear donor somatic cells. Ann. Anim. Sci. 2010, 10, 385–398. [Google Scholar]



| Different Mutants of Zfp296 | Forward | Reverse |
|---|---|---|
| Zfp296 full-length | CCCGAATTCAGATGTCCCGCCGCAAGGC | CGCGGTACCTCAGGCCATCTCTGGGTG |
| Zfp296-Δ2 | GTGTTCCTATGCCTGCGCTCAGAGCAGCAAGCTCAACAG | CTGTTGAGCTTGCTGCTCTGAGCGCAGGCATAGGAACAC |
| Zfp296-Δ3 | TCCCACACTGGTGAGCGACCCTACGGCAGCTGGCACCCG | CGGGTGCCAGCTGCCGTAGGGTCGCTCACCAGTGTGGGA |
| Zfp296-Δ2&Δ3 | TGAGCAGTGCTGCCCGGCGGAGCCCCGGCAGCTGGCACCCGGGA | TCCCGGGTGCCAGCTGCCGGGGCTCCGCCGGGCAGCACTGCTCA |
| Different Vectors | Forward | Reverse |
|---|---|---|
| Zfp296-Bio | CGCGAATTCATGTCCCGCCGCAAGGCC | CCCGGATCCGGCCATCTCTGGGTGCTT |
| Flag-Kdm5b | GGGGTACCATGGAGCCGGCCACCACGCT | GCTCTAGATTACTTTCGGCTTGGTGCGTCCTTC |
| HA-Smarca4 | AAGGAAAAAAGCGGCCGCATGTCTACTCCAGACCCACCCTTGG | AAGGAAAAAAGCGGCCGCTCAGTCTTCCTCACTGCCACTTCC |
| Flag-Dnmt1 | GGGGTACCCAATGCCAGCGCGAACAGCTCCAGCCC | GCTCTAGAGTCCTTGGTAGCAGCCTCCTCTTTT |
| Myc-Dnmt3b | GGAATTCGAATGGGGAAAAAGCAAAACAAGAAGAA | GGGGTACCCTAATTCTTGTCGTCTTTTTTGT |
| HA-Uhrf1 | GGAATTCGAATGTGGATCCAGGTTCGAACTATGG | GGGGTACCTCACCGGCCGCTGCCATAGCCAGG |
| HA-Hp1β | TGAGCAGTGCTGCCCGGCGGAGCCCCGGCAGCTGGCACCCGGGA | TCCCGGGTGCCAGCTGCCGGGGCTCCGCCGGGCAGCACTGCTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Zhang, Z.; Zheng, X.; Wang, F.; Deng, Y.; Zhang, Q.; Wang, G.; Zhang, Y.; Liu, X. The Novel Role of Zfp296 in Mammalian Embryonic Genome Activation as an H3K9me3 Modulator. Int. J. Mol. Sci. 2023, 24, 11377. https://doi.org/10.3390/ijms241411377
Gao L, Zhang Z, Zheng X, Wang F, Deng Y, Zhang Q, Wang G, Zhang Y, Liu X. The Novel Role of Zfp296 in Mammalian Embryonic Genome Activation as an H3K9me3 Modulator. International Journal of Molecular Sciences. 2023; 24(14):11377. https://doi.org/10.3390/ijms241411377
Chicago/Turabian StyleGao, Lu, Zihan Zhang, Xiaoman Zheng, Fan Wang, Yi Deng, Qian Zhang, Guoyan Wang, Yong Zhang, and Xu Liu. 2023. "The Novel Role of Zfp296 in Mammalian Embryonic Genome Activation as an H3K9me3 Modulator" International Journal of Molecular Sciences 24, no. 14: 11377. https://doi.org/10.3390/ijms241411377
APA StyleGao, L., Zhang, Z., Zheng, X., Wang, F., Deng, Y., Zhang, Q., Wang, G., Zhang, Y., & Liu, X. (2023). The Novel Role of Zfp296 in Mammalian Embryonic Genome Activation as an H3K9me3 Modulator. International Journal of Molecular Sciences, 24(14), 11377. https://doi.org/10.3390/ijms241411377





