Morphological Observation and Transcriptome Analysis of Ciliogenesis in Urechis unicinctus (Annelida, Echiura)
Abstract
:1. Introduction
2. Results
2.1. Observation of the Body Surface Cilia Distribution on the Embryos and Larvae of U. unicinctus
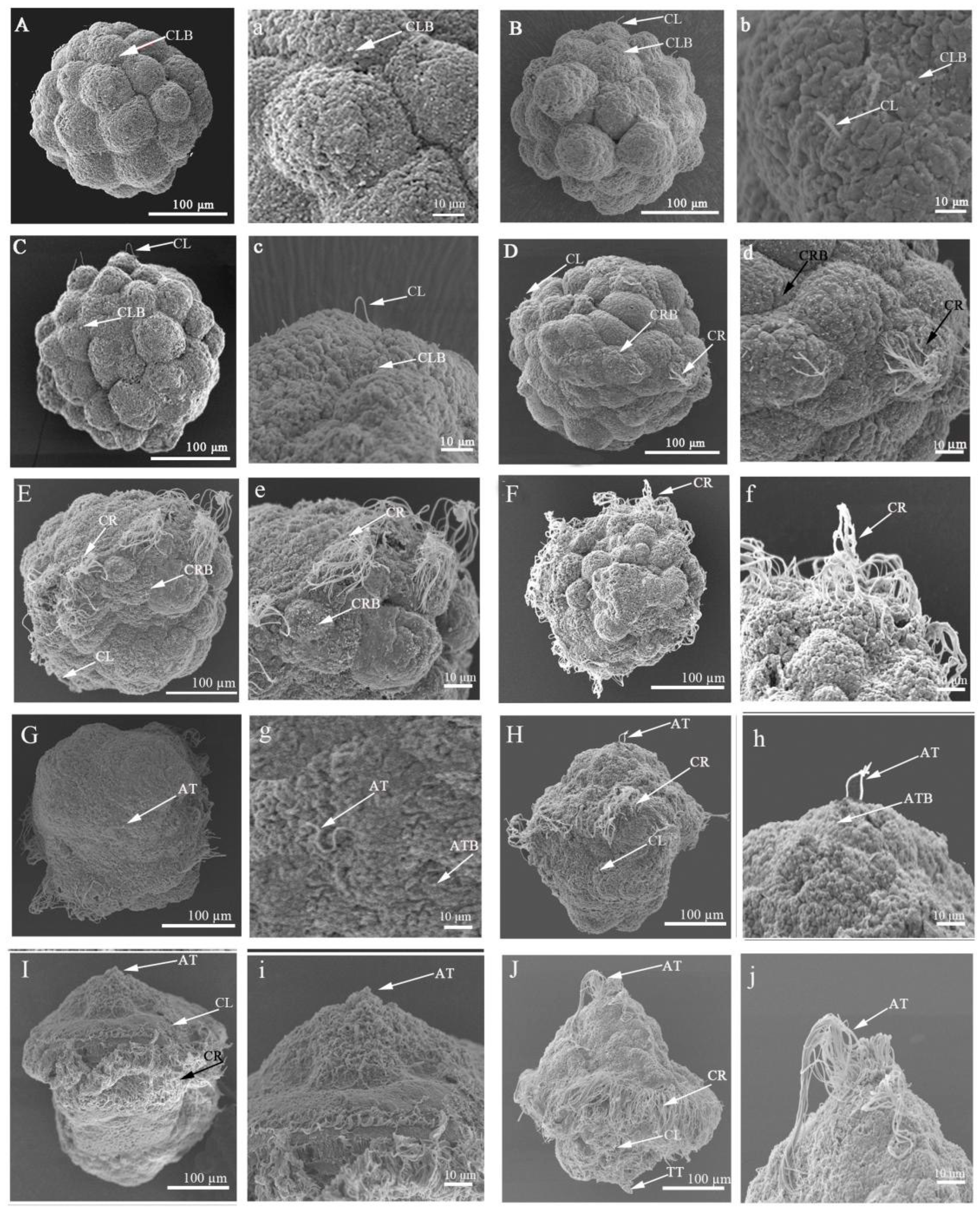
2.2. Scanning Electron Microscopic Observation of Ciliogenesis in Embryos and Larvae of U. unicinctus
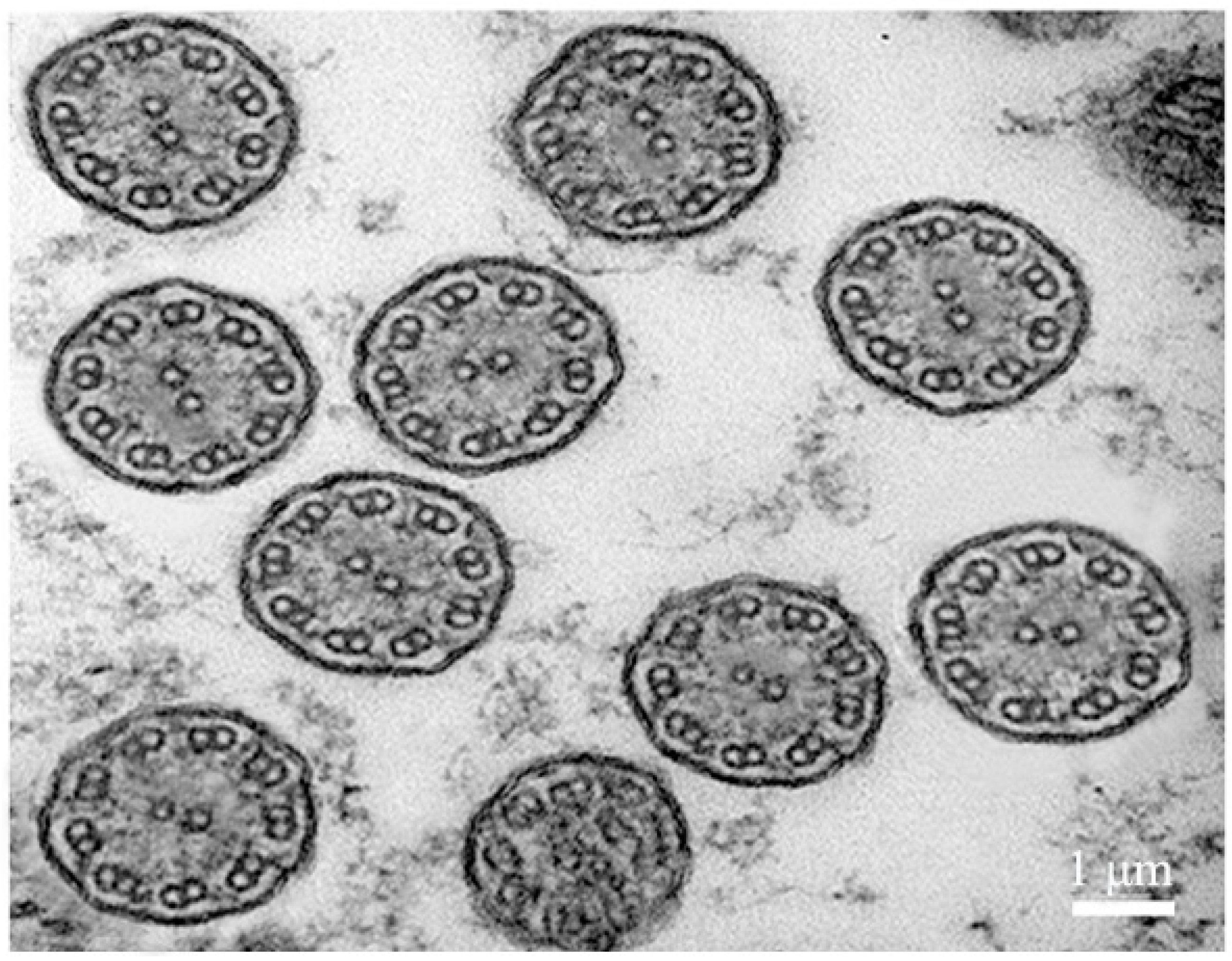
2.3. Transmission Electron Microscope Observation of Body Surface Cilia in U. unicinctus Larva
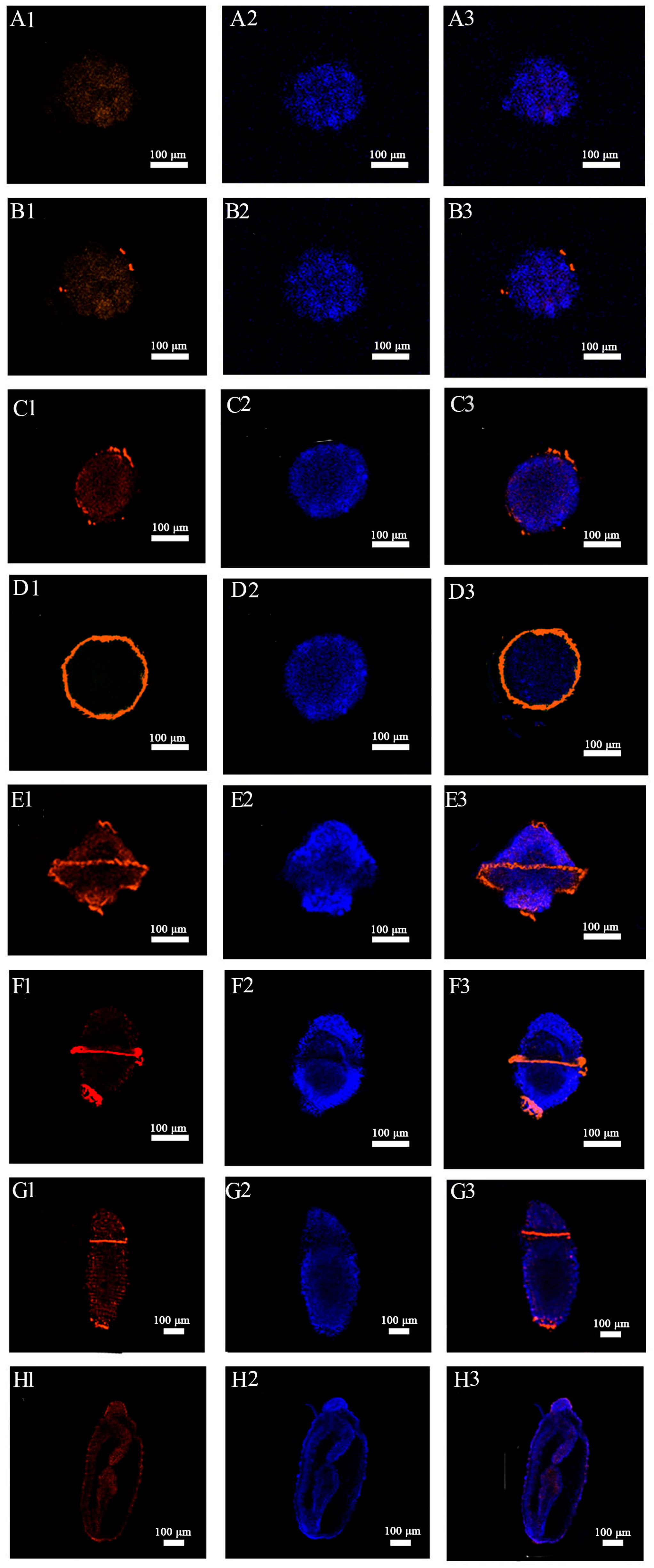
2.4. Immunohistochemical Localization of Acetylated-α-Tubulin in Embryos and Larvae of U. unicinctus
2.5. Illumina Sequencing, De Novo Assembly and Functional Annotation
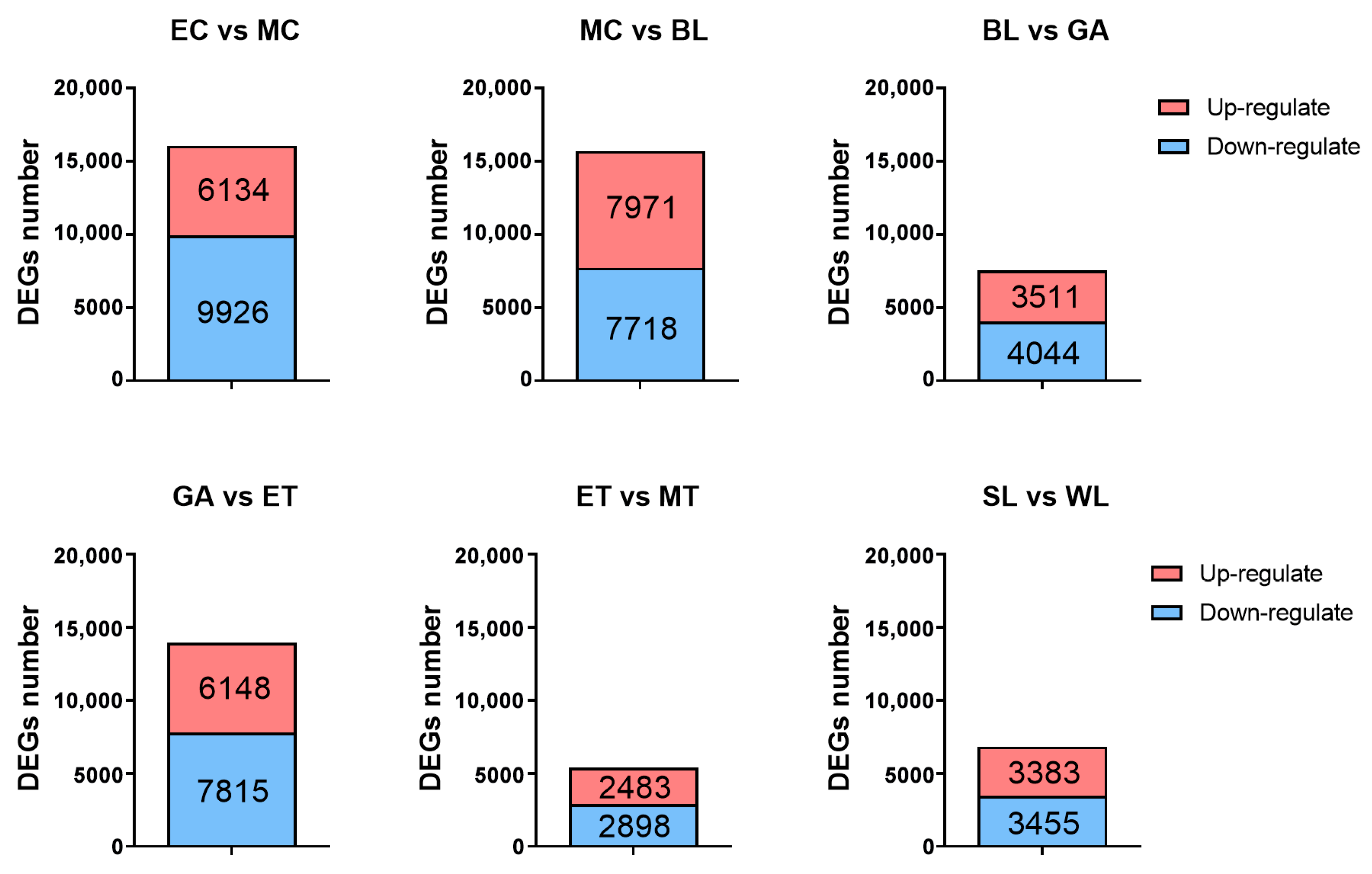
2.6. Identification, Classification and Validation of the Differentially Expressed Genes (DEGs)
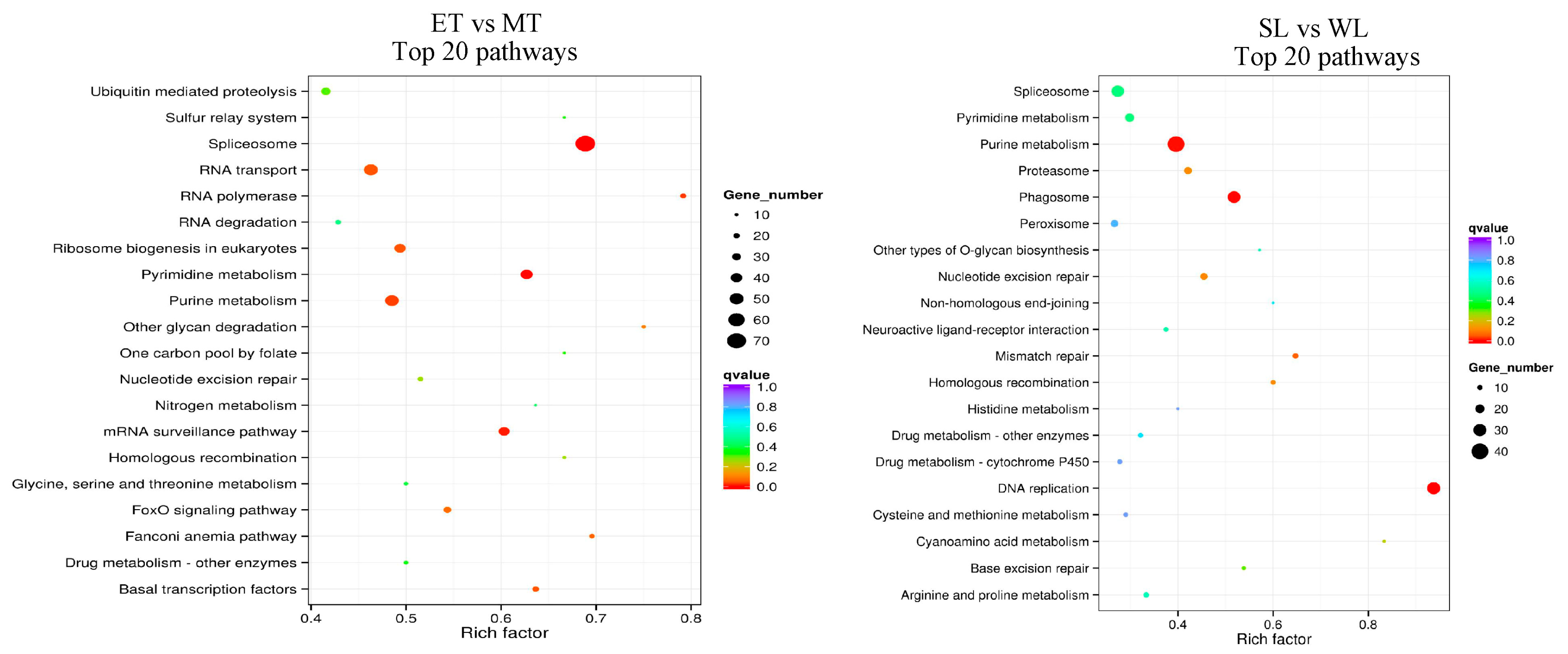
2.7. KEGG Enrichment Analysis Characterizing Possible Pathways Involved in Ciliogenesis
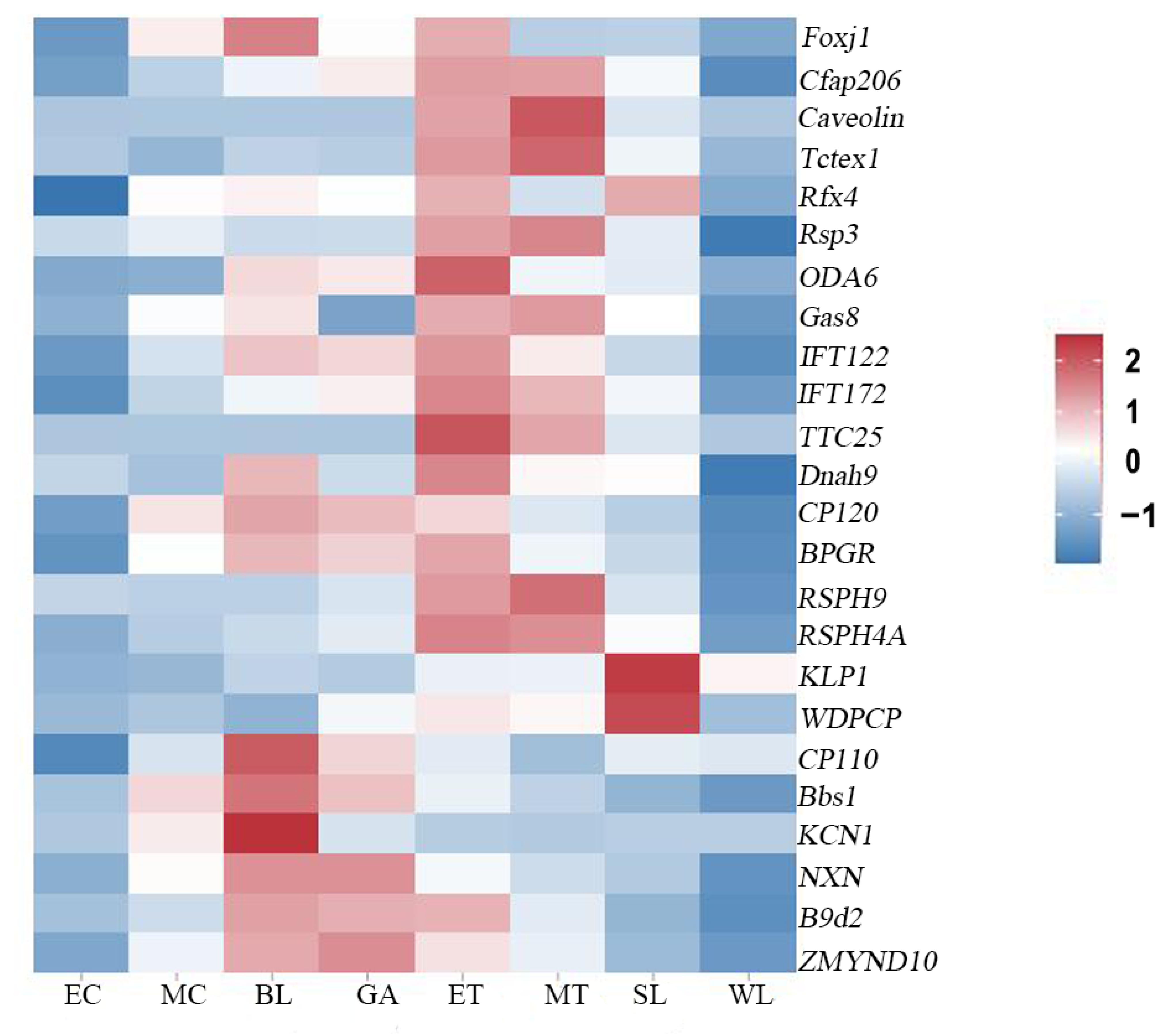
2.8. Expression Profiles of Genes Related to Ciliogenesis
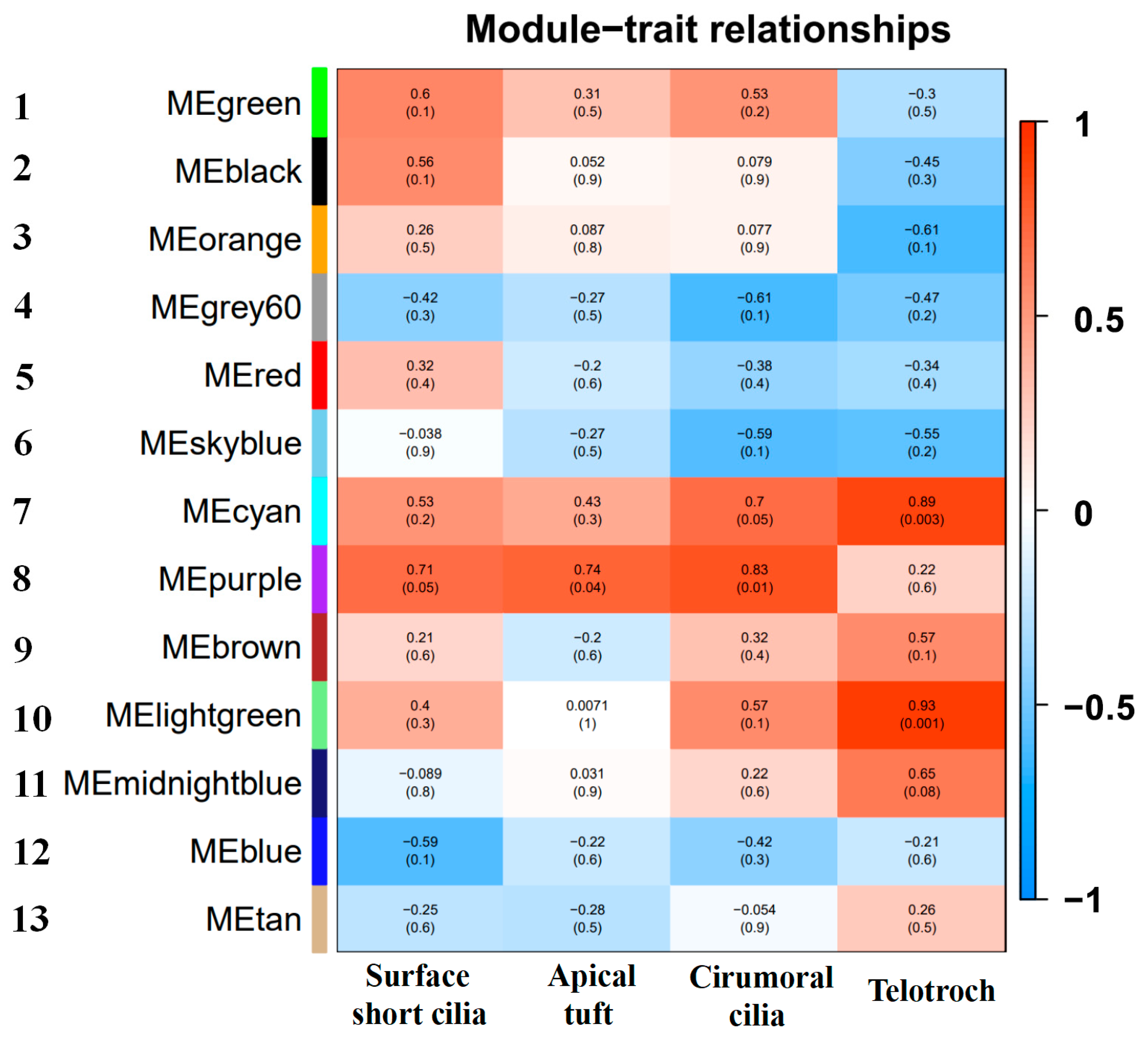
2.9. Weighted Gene Co-Expression Network Analysis Characterizing New Key Genes Involved in Ciliogenesis
3. Discussion
3.1. Similarities and Differences between the Body Surface Cilia of Embryos and Larvae from U. unicinctus and Other Lophotrochoza and Vertebrates
3.2. Possible Key Genes and Pathways for Ciliogenesis on the Body Surface of Embryos and Larvae in U. unicinctus
4. Materials and Methods
4.1. Animals Collection and Larval Culture
4.2. Sample Observation under an Optical Microscope
4.3. Scanning Electron Microscopy
4.4. Transmission Electron Microscope
4.5. Immunohistochemistry (IHC)
4.6. RNA Extraction and Illumina Sequencing
4.7. De Novo Assembly and Function Annotation of Transcriptome
4.8. Enrichment and Analysis of Differentially Expressed Genes (DEGs)
4.9. Construction of Gene Co-Expression Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Orhon, I.; Dupont, N.; Pampliega, O.; Cuervo, A.M.; Codogno, P. Autophagy and regulation of cilia function and assembly. Cell Death Differ. 2015, 22, 389–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grisanti, L.; Revenkova, E.; Gordon, R.E.; Iomini, C. Primary cilia maintain corneal epithelial homeostasis by regulation of the Notch signaling pathway. Development 2016, 143, 2160–2171. [Google Scholar] [PubMed] [Green Version]
- Sugiyama, Y.; Shelley, E.J.; Yoder, B.K.; Kozmik, Z.; May-Simera, H.L.; Beales, P.L.; Lovicu, F.J.; McAvoy, J.W. Non-essential role for cilia in coordinating precise alignment of lens fibres. Mech. Dev. 2016, 139, 10–17. [Google Scholar] [CrossRef] [PubMed]
- May-Simera, H.; Nagel-Wolfrum, K.; Wolfrum, U. Cilia-The sensory antennae in the eye. Prog. Retin. Eye Res. 2017, 60, 144–180. [Google Scholar] [CrossRef]
- Izawa, I.; Goto, H.; Kasahara, K.; Inagaki, M. Current topics of functional links between primary cilia and cell cycle. Cilia 2015, 4, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, T.; Dynlacht, B.D. Regulating the transition from centriole to basal body. J. Cell Biol. 2011, 193, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Langousis, G.; Cavadini, S.; Boegholm, N.; Lorentzen, E.; Kempf, G.; Matthias, P. Structure of the ciliogenesis-associated CPLANE complex. Sci. Adv. 2022, 8, eabn0832. [Google Scholar] [CrossRef]
- Yan, L.; Zheng, Y.F. Cilia and their role in neural tube development and defects. Reprod. Dev. Med. 2022, 6, 67–78. [Google Scholar] [CrossRef]
- Spalluto, C.; Wilson, D.I.; Hearn, T. Nek2 localises to the distal portion of the mother centriole/basal body and is required for timely cilium disassembly at the G2/M transition. Eur. J. Cell Biol. 2012, 91, 675–686. [Google Scholar] [CrossRef]
- Bonnafe, E.; Touka, M.; AitLounis, A.; Baas, D.; Barras, E.; Ucla, C.; Moreau, A.; Flamant, F.; Dubruille, R.; Couble, P.; et al. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol. Cell. Biol. 2004, 24, 4417–4427. [Google Scholar] [CrossRef] [Green Version]
- Stubbs, J.L.; Oishi, I.; Izpisua Belmonte, J.C.; Kintner, C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat. Genet. 2008, 40, 1454–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Ng, C.P.; Habacher, H.; Roy, S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 2008, 40, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Follit, J.A.; Xu, F.; Keady, B.T.; Pazour, G.J. Characterization of mouse IFT complex B. Cell Motil. Cytoskelet. 2009, 66, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Westlake, C.J.; Baye, L.M.; Nachury, M.V.; Wright, K.J.; Ervin, K.E.; Phu, L.; Chalouni, C.; Beck, J.S.; Kirkpatrick, D.S.; Slusarski, D.C.; et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA 2011, 108, 2759–2764. [Google Scholar] [CrossRef]
- Yasunaga, T.; Itoh, K.; Sokol, S.Y. Regulation of basal body and ciliary functions by Diversin. Mech. Dev. 2011, 128, 376–386. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.Z.; Zhao, W.L.; Wang, G.S.; Gu, N.H.; Sun, F. The novel testicular enrichment protein Cfap58 is required for Notch-associated ciliogenesis. Biosci. Rep. 2020, 40, BSR20192666. [Google Scholar] [CrossRef]
- Saternos, H.; Ley, S.; AbouAlaiwi, W. Primary cilia and calcium signaling interactions. Int. J. Mol. Sci. 2020, 21, 7109. [Google Scholar] [CrossRef]
- Williams, E.A.; Jékely, G. Neuronal cell types in the annelid Platynereis dumerilii. Curr. Opin. Neurobiol. 2019, 56, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Marinkovic, M.; Berger, J.; Jekely, G. Neuronal coordination of motile cilia in locomotion and feeding. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2020, 375, 20190165. [Google Scholar] [CrossRef] [Green Version]
- Katow, H.; Katow, T.; Yoshida, H.; Kiyomoto, M. Involvement of huntingtin in development and ciliary beating regulation of larvae of the sea urchin, Hemicentrotus pulcherrimus. Int. J. Mol. Sci. 2021, 22, 5116. [Google Scholar] [CrossRef]
- McDougall, C.; Chen, W.C.; Shimeld, S.M.; Ferrier, D.E. The development of the larval nervous system, musculature and ciliary bands of Pomatoceros lamarckii (Annelida): Heterochrony in polychaetes. Front. Zool. 2006, 3, 16. [Google Scholar] [CrossRef] [Green Version]
- Arenas-Mena, C.; Wong, S.Y.; Arandi-Forosani, N. Ciliary band gene expression patterns in the embryo and trochophore larvae of an indirectly developing polychaete. Gene Expr. Patterns 2007, 7, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Anishchenko, E.; Arnone, M.I.; D’Aniello, S. SoxB2 in sea urchin development: Implications in neurogenesis, ciliogenesis and skeletal patterning. EvoDevo 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huan, P.; Cui, M.; Wang, Q.; Liu, B. CRISPR/Cas9-mediated mutagenesis reveals the roles of calaxin in gastropod larval cilia. Gene 2021, 30, 145640. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, X.; Lian, S.; Li, Y.; Hu, N.; Hu, X.; Bao, Z.; Wang, S. Functional characterization of Cfap206 for bivalve ciliogenesis by RNAi and CRISPR/Cas9 technologies. Front. Mar. Sci. 2022, 9, 392. [Google Scholar] [CrossRef]
- Cruz, C.; Ribes, V.; Kutejova, E.; Cayuso, J.; Lawson, V.; Norris, D.; Stevens, J.; Davey, M.; Blight, K.; Bangs, F.; et al. Foxj1 regulates floor plate cilia architecture and modifies the response of cells to sonic hedgehog signalling. Development 2010, 137, 4271–4282. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, L.; Lian, S.; Qin, Z.; Zhu, X.; Dai, X.; Huang, Z.; Ke, C.; Zhou, Z.; Wei, J.; et al. Evolutionary transcriptomics of metazoan biphasic life cycle supports a single intercalation origin of metazoan larvae. Nat. Ecol. Evol. 2020, 4, 725–736. [Google Scholar] [CrossRef]
- Schmidts, M.; Hou, Y.; Cortés, C.R.; Mans, D.A.; Huber, C.; Boldt, K.; Patel, M.; van Reeuwijk, J.; Plaza, J.-M.; van Beersum, S.E.C.; et al. TCTEX1D2 mutations underlie Jeune asphyxiating thoracic dystrophy with impaired retrograde intraflagellar transport. Nat. Commun. 2015, 6, 7074. [Google Scholar] [CrossRef] [Green Version]
- Ashique, A.M.; Choe, Y.; Karlen, M.; May, S.R.; Phamluong, K.; Solloway, M.J.; Ericson, J.; Peterson, A.S. The Rfx4 transcription factor modulates Shh signaling by regional control of ciliogenesis. Sci. Signal. 2009, 2, ra70. [Google Scholar] [CrossRef]
- Wirschell, M.; Zhao, F.; Yang, C.; Yang, P.; Diener, D.; Gaillard, A.; Rosenbaum, J.L.; Sale, W.S. Building a radial spoke: Flagellar radial spoke protein 3 (RSP3) is a dimer. Cell Motil. Cytoskelet. 2008, 65, 238–248. [Google Scholar] [CrossRef]
- Mitchell, D.R.; Kang, Y. Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J. Cell Biol. 1991, 113, 835–842. [Google Scholar] [CrossRef]
- Colantonio, J.R.; Vermot, J.; Wu, D.; Langenbacher, A.D.; Fraser, S.; Chen, J.N.; Hill, K.L. The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. Nature 2009, 457, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Takahara, M.; Katoh, Y.; Nakamura, K.; Hirano, T.; Sugawa, M.; Tsurumi, Y.; Nakayama, K. Ciliopathy-associated mutations of IFT122 impair ciliary protein trafficking but not ciliogenesis. Hum. Mol. Genet. 2018, 27, 516–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruski, M.; Hu, L.; Yang, C.; Wang, Y.; Zhang, J.B.; Zhang, L.; Huang, Y.; Rajnicek, A.M.; St Clair, D.; McCaig, C.D.; et al. Roles for IFT172 and primary cilia in cell migration, cell division, and neocortex development. Front. Cell Dev. Biol. 2019, 7, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallmeier, J.; Shiratori, H.; Dougherty, G.W.; Edelbusch, C.; Hjeij, R.; Loges, N.T.; Menchen, T.; Olbrich, H.; Pennekamp, P.; Raidt, J.; et al. TTC25 deficiency results in defects of the outer dynein arm docking machinery and primary ciliary dyskinesia with left-right body asymmetry randomization. Am. J. Hum. Genet. 2016, 99, 460–469. [Google Scholar] [CrossRef] [Green Version]
- Fassad, M.R.; Shoemark, A.; Legendre, M.; Hirst, R.A.; Koll, F.; Le Borgne, P.; Louis, B.; Daudvohra, F.; Patel, F.P.; Thomas, L.; et al. Mutations in outer dynein arm heavy chain DNAH9 cause motile cilia defects and situs inversus. Am. J. Hum. Genet. 2018, 103, 984–994. [Google Scholar] [CrossRef] [Green Version]
- Pfalzgraf, S.; Guyot, P.; Laluque, B.; Jouannet-Romaszko, M.; Baud, O.; Romaszko, J.; Pelegrin, S.; Sautou, V. CP-120 Comparison of antibiotic consumption and bacterial resistance in two teaching hospitals: Impact of a multidisciplinary antibiotic management program. BMJ J. 2015, 22, A48. [Google Scholar] [CrossRef]
- Gakovic, M.; Shu, X.; Kasioulis, I.; Carpanini, S.; Moraga, I.; Wright, A.F. The role of RPGR in cilia formation and actin stability. Hum. Mol. Genet. 2011, 20, 4840–4850. [Google Scholar] [CrossRef] [Green Version]
- Vössing, C.; Owczarek-Lipska, M.; Nagel-Wolfrum, K.; Reiff, C.; Jüschke, C.; Neidhardt, J. Translational read-through therapy of RPGR nonsense mutations. Int. J. Mol. Sci. 2020, 21, 8418. [Google Scholar] [CrossRef]
- Castleman, V.H.; Romio, L.; Chodhari, R.; Hirst, R.A.; de Castro, S.C.; Parker, K.A.; Ybot-Gonzalez, P.; Emes, R.D.; Wilson, S.W.; Wallis, C.; et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 2009, 84, 197–209. [Google Scholar] [CrossRef] [Green Version]
- Pidoux, A.L.; LeDizet, M.; Cande, W.Z. Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol. Biol. Cell 1996, 7, 1639–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toriyama, M.; Lee, C.; Taylor, S.P.; Duran, I.; Cohn, D.H.; Bruel, A.L.; Tabler, J.M.; Drew, K.; Kelly, M.R.; Kim, S.; et al. The ciliopathy-associated CPLANE proteins direct basal body recruitment of intraflagellar transport machinery. Nat. Genet. 2016, 48, 648–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spektor, A.; Tsang, W.Y.; Khoo, D.; Dynlacht, B.D. Cep97 and CP110 suppress a cilia assembly program. Cell 2007, 130, 678–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachury, M.V.; Loktev, A.V.; Zhang, Q.; Westlake, C.J.; Peränen, J.; Merdes, A.; Slusarski, D.C.; Scheller, R.H.; Bazan, J.F.; Sheffield, V.C.; et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 2007, 129, 1201–1213. [Google Scholar] [CrossRef] [Green Version]
- Yin, S.; Kaluz, S.; Devi, N.S.; Jabbar, A.A.; de Noronha, R.G.; Mun, J.; Zhang, Z.; Boreddy, P.R.; Wang, W.; Wang, Z.; et al. Arylsulfonamide KCN1 Inhibits in vivo glioma growth and interferes with HIF signaling by disrupting HIF-1α interaction with cofactors p300/CBPAntitumor agent KCN1 disrupts HIF-p300/CBP interactions. Clin. Cancer Res. 2012, 18, 6623–6633. [Google Scholar] [CrossRef] [Green Version]
- Albee, A.J.; Kwan, A.L.; Lin, H.; Granas, D.; Stormo, G.D.; Dutcher, S.K. Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3 Genes Genomes Genet. 2013, 3, 979–991. [Google Scholar] [CrossRef] [Green Version]
- Town, T.; Breunig, J.J.; Sarkisian, M.R.; Spilianakis, C.; Ayoub, A.E.; Liu, X.; Ferrandino, A.F.; Gallagher, A.R.; Li, M.O.; Rakic, P.; et al. The stumpy gene is required for mammalian ciliogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 2853–2858. [Google Scholar] [CrossRef]
- Moore, D.J.; Onoufriadis, A.; Shoemark, A.; Simpson, M.A.; Zur Lage, P.I.; De Castro, S.C.; Bartoloni, L.; Gallone, G.; Petridi, S.; Woollard, W.J.; et al. Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013, 93, 346–356. [Google Scholar] [CrossRef]
- Hadfield, M.G.; Meleshkevitch, E.A.; Boudko, D.Y. The apical sensory organ of a gastropod veliger is a receptor for settlement cues. Biol. Bull. 2000, 198, 67–76. [Google Scholar] [CrossRef]
- Conzelmann, M.; Offenburger, S.L.; Asadulina, A.; Keller, T.; Münch, T.A.; Jékely, G. Neuropeptides regulate swimming depth of Platynereis larvae. Proc. Natl. Acad. Sci. USA 2011, 108, 1174–1183. [Google Scholar] [CrossRef]
- Nielsen, C. Trochophora larvae: Cell-lineages, ciliary bands and body regions. 2. Other groups and general discussion. J. Exp. Zool. Part B Mol. Dev. Evol. 2005, 304B, 401–447. [Google Scholar] [CrossRef]
- Seaver, E.C.; Kaneshige, L.M. Expression of ‘segmentation’ genes during larval and juvenile development in the polychaetes Capitella sp. I and H. elegans. Dev. Biol. 2006, 289, 179–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouse, G.W. Trochophore concepts: Ciliary bands and the evolution of larvae in spiralian metazoan. Biol. J. Linn. Soc. 1999, 66, 411–464. [Google Scholar] [CrossRef]
- Nielsen, C. Trochophora larvae: Cell-lineages, ciliary bands, and body regions. 1. Annelida and Mollusca. J. Exp. Zool. Part B Mol. Dev. Evol. 2004, 302, 35–68. [Google Scholar] [CrossRef]
- Conzelmann, M.; Williams, E.A.; Tunaru, S.; Randel, N.; Shahidi, R.; Asadulina, A.; Berger, J.; Offermanns, S.; Jékely, G. Conserved MIP receptor–ligand pair regulates Platynereis larval settlement. Proc. Natl. Acad. Sci. USA 2013, 110, 8224–8229. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C. Animal Evolution: Interrelationships of the Living Phyla; Oxford University Press on Demand: Oxford, UK, 2012. [Google Scholar]
- Carrillo-Baltodano, A.M.; Seudre, O.; Guynes, K.; Martín-Durán, J.M. Early embryogenesis and organogenesis in the annelid Owenia fusiformis. EvoDevo 2021, 12, 5. [Google Scholar] [CrossRef]
- Gerdes, J.M.; Davis, E.E.; Katsanis, N. The vertebrate primary cilium in development, homeostasis, and disease. Cell 2009, 137, 32–45. [Google Scholar] [CrossRef] [Green Version]
- Giuliani, F.; Giuliani, G.; Bauer, R.; Rabouille, C. Innexin3, a new gene required for dorsal closure in Drosophila embryo. PLoS ONE 2013, 8, e69212. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Li, M.; Zhang, Y.; Pang, Z.; Xiao, W.; Yang, Y.; Luo, K. A role for Innexin2 and Innexin3 proteins from Spodoptera litura in apoptosis. PLoS ONE 2013, 8, e70456. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.P.; Chen, F.Y.; Dong, L.X.; Zhang, Y.-Q.; Chen, H.-Y.; Qiao, K.; Wang, K.-J. A novel Innexin2 forming membrane hemichannel exhibits immune responses and cell apoptosis in Scylla paramamosain. Fish Shellfish. Immunol. 2015, 47, 485–499. [Google Scholar] [CrossRef]
- Richard, M.; Bauer, R.; Tavosanis, G.; Hoch, M. The gap junction protein Innexin3 is required for eye disc growth in Drosophila. Dev. Biol. 2017, 425, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, X.; Hou, X.; Wang, J.; Yue, W.; Huang, S.; Xu, G.; Yan, J.; Lu, G.; Hofreiter, M.; et al. “Omics” data unveil early molecular response underlying limb regeneration in the Chinese mitten crab, Eriocheir sinensis. Sci. Adv. 2022, 8, eabl4642. [Google Scholar] [CrossRef] [PubMed]
- Miao, G.; Godt, D.; Montell, D.J. Integration of migratory cells into a new site in vivo requires channel-independent functions of innexins on microtubules. Dev. Cell 2020, 54, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Chen, K.H.; Chen, Y.Y.; Tsao, L.-H.; Yeh, T.-H.; Chen, Y.-C.; Wu, P.-Y.; Wang, T.-W.; Yu, J.-Y. βPS-Integrin acts downstream of Innexin2 in modulating stretched cell morphogenesis in the Drosophila ovary. G3 Genes Genomes Genet. 2021, 11, jkab215. [Google Scholar] [CrossRef]
- Bauer, R.; Lehmann, C.; Fuss, B.; Eckardt, F.; Hoch, M. The Drosophila gap junction channel gene Innexin2 controls foregut development in response to Wingless signalling. J. Cell Sci. 2002, 115 Pt 9, 1859–1867. [Google Scholar] [CrossRef]
- Bauer, R.; Löer, B.; Ostrowski, K.; Martini, J.; Weimbs, A.; Lechner, H.; Hoch, M. Intercellular communication: The Drosophila innexin multiprotein family of gap junction proteins. Chem. Biol. 2005, 12, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.; Qin, Z.; Wei, M.; Fu, Z.; Liu, R.; Lu, L.; Bai, S.; Ma, Y.; Zhang, Z. Identification of the neuropeptide precursor genes potentially involved in the larval settlement in the echiuran worm Urechis unicinctus. BMC Genom. 2020, 21, 892. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene ID | Gene Name | KME | Gene Function |
|---|---|---|---|
| evm.model.Hic_asm_10.155 | FAM181B (family with sequence similarity 181 member B) | 0.995 | Involved in embryonic development, meiosis, cell differentiation, nervous system and other processes. |
| evm.model.Hic_asm_6.1358 | TRP5 (Tetratricopeptide repeat protein 5) | 0.993 | Actin regulation, autophagy, and DNA repair. |
| evm.model.Hic_asm_14.687 | Innexin | 0.992 | Development of embryos and organs. |
| evm.model.Hic_asm_2.868 | P450c17 (steroid 17 alpha-hydroxylase/17,20 lyase) | 0.989 | Involved in hormone synthesis. |
| evm.model.Hic_asm_10.436 | E3 ubiquitin-protein ligase RBX1 | 0.989 | Involved in embryonic development and cell survival. |
| evm.model.Hic_asm_8.163 | PCSK6 (Proprotein convertase subtilisin/kexin type 6) | 0.988 | Regulates cell proliferation and inflammatory response. |
| evm.model.Hic_asm_8.1080 | U2AF1L4 (Splicing factor U2AF 26 kDa subunit) | 0.988 | Acts as a pre-mRNA splicing factor. |
| evm.model.Hic_asm_14.236.3 | UreD (Urease accessory protein D) | 0.987 | Involved in activating urease apolipoprotein. |
| evm.model.Hic_asm_9.1210 | Capsl | 0.987 | Development of embryos and organs. |
| evm.model.Hic_asm_10.721.1 | Cnbd2 (Cyclic nucleotide-binding domain-containing protein 2) | 0.987 | Controls the development of spermatoflagellar curvature |
| Gene ID | Gene Name | KME | Gene Function |
|---|---|---|---|
| evm.model.Hic_asm_12.772 | NACHRA10 (Neuronal acetylcholine receptor protein subunit alpha 10) | 0.995 | Involved in the regulation of auditory stimuli |
| evm.model.Hic_asm_13.362 | AP2S1 (AP-2 complex subunit sigma) | 0.994 | Involved in clathrin-dependent endocytosis. |
| evm.model.Hic_asm_13.51 | Mpv17-like protein | 0.993 | Supports mitochondrial function. |
| evm.model.Hic_asm_0.559 | Innexin | 0.993 | Development of embryos and organs. |
| evm.model.Hic_asm_3.848.1 | GSTs (Glutathione S-transferase) | 0.993 | One of the most important metabolic enzymes in biotransformation. |
| evm.model.Hic_asm_10.1040 | SPATA18 (Mitochondria-eating protein) | 0.991 | Degrade damaged mitochondria and mitochondrial proteins. |
| evm.model.Hic_asm_4.311 | CHL1 (Neural cell adhesion molecule L1-like protein) | 0.989 | Regulates neuron migration, axon growth, and dendrite projection. |
| evm.model.Hic_asm_3.845 | SULT1B1(Sulfotransferase Family 1B Member 1) | 0.988 | Ontogeny and hormone regulation. |
| evm.model.Hic_asm_1.737 | MDH1B (Malate Dehydrogenase 1B) | 0.988 | Involved in the catalytic dehydrogenation of malate in the tricarboxylic acid cycle. |
| evm.model.Hic_asm_8.1588 | Chst3 (carbohydrate sulfotransferase 3) | 0.988 | Catalyzed proteoglycan sulfation. |
| Gene ID | Gene Name | KME | Gene Function |
|---|---|---|---|
| evm.model.Hic_asm_13.963 | LRRC74A (Leucine-rich repeat-containing protein 74A) | 0.997 | The initiator of Met-1 or Met-27. |
| evm.model.Hic_asm_13.966 | TMIE (Transmembrane inner ear expressed protein) | 0.996 | Play role in a cellular membrane location. |
| evm.model.Hic_asm_14.88 | FGL1 (Fibrinogen-like protein 1) | 0.992 | Immune-suppressive molecule. |
| evm.model.Hic_asm_0.423 | RAB8B (Ras-related protein Rab-8B) | 0.991 | The small GTPases Rab are key regulators of intracellular membrane trafficking, from the formation of transport vesicles to their fusion with membranes. |
| evm.model.Hic_asm_13.1164 | Mcfd2 (Multiple coagulation factor deficiency protein 2 homolog) | 0.990 | The MCFD2-LMAN1 complex forms a specific cargo receptor for the ER-to-Golgi transport of selected proteins. |
| evm.model.Hic_asm_6.554 | Adcy8 (Adenylate cyclase type 2) | 0.989 | Converting ATP into cAMP triggers a cellular signaling response. |
| evm.model.Hic_asm_6.414 | VNN1 (Pantetheinase) | 0.988 | Amidohydrolase that hydrolyzes specifically one of the carboamide linkages in D-pantetheine, thus recycling pantothenic acid (vitamin B5) and releasing cysteamine. |
| evm.model.Hic_asm_14.1693 | Chrna3 (Neuronal acetylcholine receptor subunit alpha-3) | 0.987 | After binding acetylcholine, the AChR responds by an extensive change in conformation that affects all subunits and leads to opening of an ion-conducting channel across the plasma membrane. |
| evm.model.Hic_asm_15.505.1 | qvr (Protein quiver) | 0.987 | Important regulator of the Sh K+ channel. |
| evm.model.Hic_asm_11.797 | DYDC2 (DPY30 domain-containing protein 2) | 0.987 | Functions as part of axonemal radial spoke complexes that play an important part in the motility of sperm and cilia (by similarity). Plays a crucial role during acrosome biogenesis. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, D.; Wei, M.; Liu, D.; Zhang, Z.; Ma, Y.; Zhang, Z. Morphological Observation and Transcriptome Analysis of Ciliogenesis in Urechis unicinctus (Annelida, Echiura). Int. J. Mol. Sci. 2023, 24, 11537. https://doi.org/10.3390/ijms241411537
Kong D, Wei M, Liu D, Zhang Z, Ma Y, Zhang Z. Morphological Observation and Transcriptome Analysis of Ciliogenesis in Urechis unicinctus (Annelida, Echiura). International Journal of Molecular Sciences. 2023; 24(14):11537. https://doi.org/10.3390/ijms241411537
Chicago/Turabian StyleKong, Dexu, Maokai Wei, Danwen Liu, Zhengrui Zhang, Yubin Ma, and Zhifeng Zhang. 2023. "Morphological Observation and Transcriptome Analysis of Ciliogenesis in Urechis unicinctus (Annelida, Echiura)" International Journal of Molecular Sciences 24, no. 14: 11537. https://doi.org/10.3390/ijms241411537





