Combined Tumor-Based BRCA1/2 and TP53 Mutation Testing in Ovarian Cancer
Abstract
1. Introduction
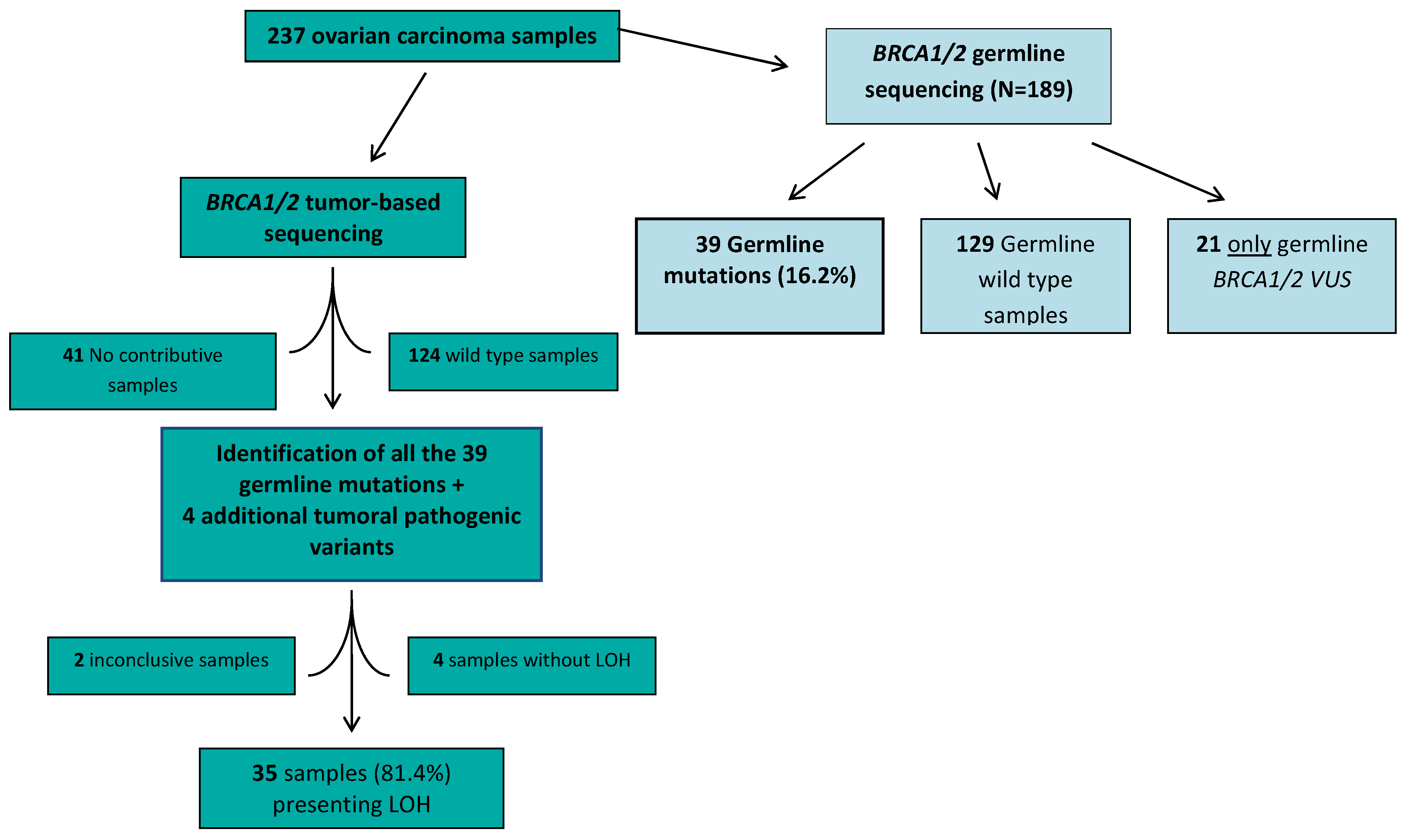
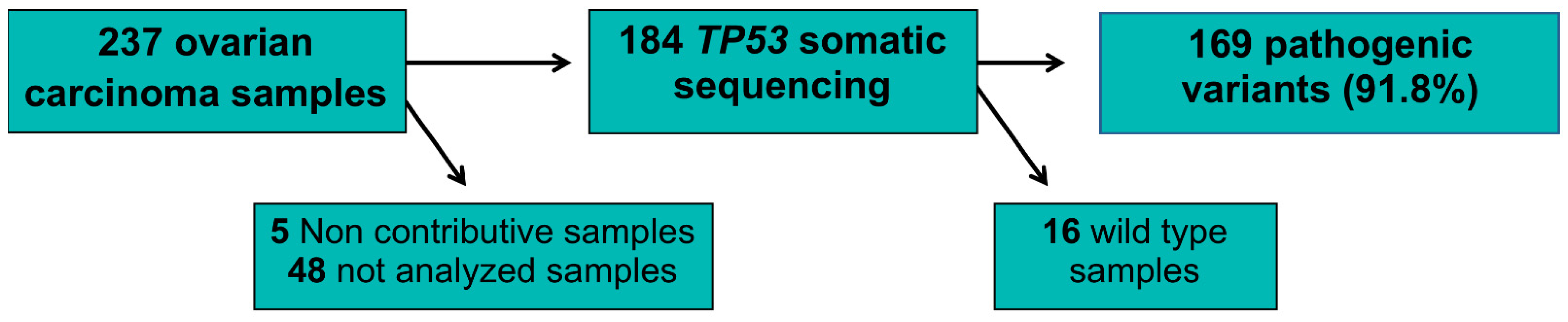
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Helleday, T.; Lo, J.; van Gent, D.C.; Engelward, B.P. DNA Double-Strand Break Repair: From Mechanistic Understanding to Cancer Treatment. DNA Repair 2007, 6, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Harter, P.; Hauke, J.; Heitz, F.; Reuss, A.; Kommoss, S.; Marmé, F.; Heimbach, A.; Prieske, K.; Richters, L.; Burges, A.; et al. Prevalence of Deleterious Germline Variants in Risk Genes Including BRCA1/2 in Consecutive Ovarian Cancer Patients (AGO-TR-1). PLoS ONE 2017, 12, e0186043. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G. The Concept of Synthetic Lethality in the Context of Anticancer Therapy. Nat. Rev. Cancer 2005, 5, 689–698. [Google Scholar] [CrossRef]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib Maintenance Therapy in Platinum-Sensitive Relapsed Ovarian Cancer. N. Engl. J. Med. 2012, 366, 1382–1392. [Google Scholar] [CrossRef]
- Mirza, M.R.; Monk, B.J.; Herrstedt, J.; Oza, A.M.; Mahner, S.; Redondo, A.; Fabbro, M.; Ledermann, J.A.; Lorusso, D.; Vergote, I.; et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016, 375, 2154–2164. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib Maintenance Treatment for Recurrent Ovarian Carcinoma after Response to Platinum Therapy (ARIEL3): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, B.A.; Lai, Z.; Hodgson, D.R.; Orr, M.C.M.; Hawryluk, M.; Sun, J.; Yelensky, R.; Spencer, S.K.; Robertson, J.D.; Ho, T.W.; et al. Biological and Clinical Evidence for Somatic Mutations in BRCA1 and BRCA2 as Predictive Markers for Olaparib Response in High-Grade Serous Ovarian Cancers in the Maintenance Setting. Oncotarget 2017, 8, 43653–43661. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.-G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.N.; Wubbenhorst, B.; Wenz, B.M.; De Sloover, D.; Pluta, J.; Emery, L.; Barrett, A.; Kraya, A.A.; Anastopoulos, I.N.; Yu, S.; et al. BRCA Locus-Specific Loss of Heterozygosity in Germline BRCA1 and BRCA2 Carriers. Nat. Commun. 2017, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- Yost, S.; Ruark, E.; Alexandrov, L.B.; Rahman, N. Insights into BRCA Cancer Predisposition from Integrated Germline and Somatic Analyses in 7632 Cancers. JNCI Cancer Spectr. 2019, 3, pkz028. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, P.; Bandlamudi, C.; Cheng, M.L.; Srinivasan, P.; Chavan, S.S.; Friedman, N.D.; Rosen, E.Y.; Richards, A.L.; Bouvier, N.; Selcuklu, S.D.; et al. Tumour Lineage Shapes BRCA-Mediated Phenotypes. Nature 2019, 571, 576–579. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.N.; Colombo, N.; Scambia, G.; Kim, B.-G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib Following Platinum-Based Chemotherapy in Newly Diagnosed Patients (Pts) with Advanced Ovarian Cancer (OC) and a BRCA1/2 Mutation (BRCAm): Phase III SOLO1 Trial. Ann. Oncol. 2018, 29, viii727. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Del Campo, J.M.; Matulonis, U.A.; Malander, S.; Provencher, D.; Mahner, S.; Follana, P.; Waters, J.; Berek, J.S.; Woie, K.; Oza, A.M.; et al. Niraparib Maintenance Therapy in Patients with Recurrent Ovarian Cancer After a Partial Response to the Last Platinum-Based Chemotherapy in the ENGOT-OV16/NOVA Trial. J. Clin. Oncol. 2019, 37, 2968–2973. [Google Scholar] [CrossRef] [PubMed]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef] [PubMed]
- Pennington, K.P.; Walsh, T.; Harrell, M.I.; Lee, M.K.; Pennil, C.C.; Rendi, M.H.; Thornton, A.; Norquist, B.M.; Casadei, S.; Nord, A.S.; et al. Germline and Somatic Mutations in Homologous Recombination Genes Predict Platinum Response and Survival in Ovarian, Fallopian Tube, and Peritoneal Carcinomas. Clin. Cancer Res. 2014, 20, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Bernards, S.S.; Pennington, K.; Harrell, M.I.; Agnew, K.J.; Norquist, B.M.; Swisher, E.M. Overall Survival in BRCA1 or RAD51C Methylated vs Mutated Ovarian Carcinoma Following Primary Treatment with Platinum Chemotherapy. Gynecol. Oncol. 2017, 145, 24. [Google Scholar] [CrossRef]
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.-Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of All BRCA1 Copies Predicts Response to the PARP Inhibitor Rucaparib in Ovarian Carcinoma. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kondrashova, O.; Nguyen, M.; Shield-Artin, K.; Tinker, A.V.; Teng, N.N.H.; Harrell, M.I.; Kuiper, M.J.; Ho, G.-Y.; Barker, H.; Jasin, M.; et al. Secondary Somatic Mutations Restoring RAD51C and RAD51D Associated with Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2017, 7, 984–998. [Google Scholar] [CrossRef] [PubMed]
| n = 237 Patients | |
|---|---|
| Median age at diagnosis (IQR) * | 62.0 (56.0–68.0) |
| Histological type, n (%) | |
| - High-grade serous carcinoma | 205 (86.5) |
| - Low-grade serous carcinoma | 4 (1.7) |
| - Clear-cell carcinoma | 2 (0.8) |
| - Carcinosarcoma | 4 (1.7) |
| - High-grade endometrioid carcinoma | 15 (6.3) |
| - Undifferentiated carcinoma | 7 (3.0) |
| FIGO stage, n (%) | |
| - I | 8 (3.3) |
| - II | 10 (4.2) |
| - III | 126 (53.2) |
| - IV | 45 (19.0) |
| - NA | 48 (20.3) |
| Type of samples, n (%) | |
| - Biopsy | 93 (39.2) |
| - Surgical samples | 127 (53.6) |
| - NA | 17 (7.2) |
| Sample collection, n (%) | |
| - Primitive | 141 (59.4%) |
| - Primitive post-neoadjuvant treatment | 62 (26.2%) |
| - Relapse | 1 (0.4%) |
| - Relapse post chemotherapy | 27 (11.4%) |
| - NA | 6 (2.5%) |
| Gene | Variant | Protein | Functional Domain | Variant Type | Germline | Allelic Frequency | LOH | TP53 Associated Mutation | Allelic Frequency |
|---|---|---|---|---|---|---|---|---|---|
| BRCA1 | c.134+3A>C | - | - | Splicing | No | 0.43 | Yes | c.375+1G>T | 0.45 |
| BRCA1 | c.1121del | p.Thr374fs | - | Frameshift | Yes | 0.7 | Yes | c.394A>G; p.Lys132Glu | 0.63 |
| BRCA1 | c.212+3A>G | - | Ring finger | Splicing | Yes | 0.85 | Yes | c.673-1G>C | 0.55 |
| BRCA1 | c.1674del | p.Gly559Valfs*13 | - | Nonsense | Yes | 0.67 | Yes | c.742C>T; p.Arg248Trp | 0.57 |
| BRCA1 | c.68_69del | p.Glu23fs | Ring finger + NES1 | Frameshift | Yes | 0.46 | Inconclusive | No | WT |
| BRCA1 | c.190T>C | p.Cys64Arg | Ring finger | Missense | No | 0.19 | Yes | c.578A>T; p.His193Leu | 0.24 |
| BRCA1 | c.5266dup | p.Gln1756_Asp1757fs | Linker | Frameshift | Yes | 0.77 | Yes | c.351del; p.Gly117fs | 0.29 |
| BRCA1 | c.5468-2A>G | - | BRCT2/AD2 | Splicing | Yes | 0.50 | No | c.403T>C; p.Cys135Arg | 0.18 |
| BRCA1 | c.5266dup | p.Gln1756_Asp1757fs | Linker | Frameshift | Yes | 0.74 | Yes | c.840A>C; p.Arg280Ser | 0.62 |
| BRCA1 | c.514C>T | p.Gln172* | - | Nonsense | No | 0.04 | Yes | c.375+5del | 0.05 |
| BRCA1 | c.81-1G>C | - | Ring finger + NES1 | Splicing | No | 0.14 | Yes | c.518T>C; p.Val173Ala | 0.09 |
| BRCA2 | c.2612C>A | p.Ser871* | - | Nonsense | Yes | 0.89 | Yes | NR | NR |
| BRCA1 | c.815_824dup | p.Gly275_Thr276fs | - | Frameshift | No | 0.69 | Yes | c.743G>A; p.Arg248Gln | 0.61 |
| BRCA2 | c.6533_6542del | p.His2178Glnfs*10 | - | Deletion | No | 0.24 | Yes | c.1024C>T; p.Arg342Ter* | 0.29 |
| BRCA1 | c.4183C>T | p.Gln1395* | Coil coiled/AD1 | Nonsense | Yes | 0.62 | Yes | c.527G>T; p.Cys176Phe | 0.44 |
| BRCA1 | c.1789G>T | p.Glu597* | - | Nonsense | No | 0.16 | Yes | c.742C>T, p.Arg248Trp | 0.15 |
| BRCA1 | c.3001G>T | p.Glu1001* | - | Nonsense | Yes | 0.68 | Yes | c.395A>G; p.Lys132Arg | 0.27 |
| BRCA1 | c.5503C>T | p.Arg1835Stop | BRCT2/AD2 | Nonsense | Yes | 0.12 | Yes | c.395A>G; p.Lys132Arg | 0.20 |
| BRCA1 | c.523A>T | p.Lys175* | - | Nonsense | No | 0.39 | No | c.614A>G; p.Tyr205Cys | 0.54 |
| BRCA2 | c.7952del | p.Arg2651fs | Helical domain | Frameshift | No | 0.47 | Yes | c.681_682insT; p.Ser227_Asp228fs | 0.42 |
| BRCA1 | c.4065_4068del | p.Asn1355fs | AD1 | Frameshift | Yes | 0.81 | Yes | c.824G>A; p.Cys275Tyr | 0.62 |
| BRCA1 | c.2389G>T | p.Glu797* | - | Nonsense | No | 0.41 | Yes | c.586C>T; p.Arg196* | 0.26 |
| BRCA2 | c.6591_6592del | p.Thr2197fs | - | Frameshift | Yes | 0.87 | Yes | NR | NR |
| BRCA2 | c.2612C>A | p.Ser871* | - | Nonsense | No | 0.48 | No | c.394A>G; p.Lys132Glu | 0.15 |
| BRCA2 | c.8487+1G>A | - | - | Splicing | No | 1 | Yes | No | WT |
| BRCA2 | c.3785C>G | p.Ser1262* | - | Nonsense | Yes | 0.28 | Yes | c.1025G>A; p.Arg342Gln | 0.35 |
| BRCA2 | c.2612C>A | p.Ser871* | - | Nonsense | Yes | 0.69 | Yes | c.524G>A; p.Arg175His | 0.42 |
| BRCA1 | c.4658del | p.Leu1553fs | AD1 | Frameshift | Inconclusive | 0.87 | Yes | c.376-2A>G | 0.70 |
| BRCA1 | c.3257T>G | p.Leu1086* | - | Nonsense | Yes | 0.55 | No | c.824G>A; p.Cys275Tyr | 0.22 |
| BRCA1 | c.1063A>T | p.Lys355* | - | Nonsense | No | 0.61 | Yes | c.396G>T; p.Lys132Asn | 0.64 |
| BRCA1 | c.3008_3009del | p.Phe1003fs | - | Frameshift | Yes | 0.80 | Yes | c.1010G>T; p.Arg337Leu | 0.52 |
| BRCA1 | c.2679_2682del | p.Lys893fs | - | Frameshift | Yes | 0.63 | Yes | c.818 G>T; p.Arg273Leu | 0.48 |
| BRCA2 | c.2492del | p.Val831fs | - | Frameshift | No | 0.21 | Yes | c.818G>T; p.Arg273Leu | 0.23 |
| BRCA1 | c.4072G>T | p.Glu1358* | AD1 | Nonsense | No | 0.60 | Yes | c.403T>G; p.Cys135Gly | 0.62 |
| BRCA1 | c.3995del | p.Gly1332fs | AD1 | Frameshift | No | 0.28 | Yes | c.783-1_784delinsCTT; p.? | 0.20 |
| BRCA1 | c.4868C>G | p.Ala1623Gly | AD2 | Missense | No | 0.089 | Yes | c.644G>T, p.Ser215Ile | 0.0587 |
| BRCA1 | c.4658del | p.Leu1553fs | AD1 | Frameshift | Yes | 0.87 | Yes | c.376-2A>G | 0.70 |
| BRCA2 | c.5345_5346del | p.Gln1782fs | - | Frameshift | Yes | 0.66 | Yes | c.307_308insGAAAACCT; p.Tyr103_Gln104fs | 0.32 |
| BRCA2 | c.3233_3234insT | p.Val1078_Ser1079fs | - | Frameshift | Yes | 0.76 | Yes | c.262del; p.Ala88fs | 0.68 |
| BRCA2 | c.5682C>G | p.Tyr1894* | - | Nonsense | Yes | 0.91 | Yes | No | - |
| BRCA2 | c.1834G>T | p.Glu612* | - | Nonsense | No | 0.04 | Yes | c.388C>T; p.Leu130Phe | 0.0294 |
| BRCA2 | c.413_417del | - | Frameshift | No | 0.02 | Inconclusive | No | - | |
| BRCA1 | Deletion exon 21 to 24 | p.? | - | Frameshift | Yes | 0.8 * | Yes | c.151del; p.Glu51fs | 0.52 |
| TP53 Variant Type | Frequency |
|---|---|
| Missense | 102 (60%) |
| Frameshift | 34 (20%) |
| Nonsense | 20 (12%) |
| Splicing | 13 (8%) |
| Total | 169 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borcoman, E.; Santana dos Santos, E.; Genestie, C.; Pautier, P.; Lacroix, L.; Caputo, S.M.; Cabaret, O.; Guillaud-Bataille, M.; Michels, J.; Auguste, A.; et al. Combined Tumor-Based BRCA1/2 and TP53 Mutation Testing in Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 11570. https://doi.org/10.3390/ijms241411570
Borcoman E, Santana dos Santos E, Genestie C, Pautier P, Lacroix L, Caputo SM, Cabaret O, Guillaud-Bataille M, Michels J, Auguste A, et al. Combined Tumor-Based BRCA1/2 and TP53 Mutation Testing in Ovarian Cancer. International Journal of Molecular Sciences. 2023; 24(14):11570. https://doi.org/10.3390/ijms241411570
Chicago/Turabian StyleBorcoman, Edith, Elizabeth Santana dos Santos, Catherine Genestie, Patricia Pautier, Ludovic Lacroix, Sandrine M. Caputo, Odile Cabaret, Marine Guillaud-Bataille, Judith Michels, Aurelie Auguste, and et al. 2023. "Combined Tumor-Based BRCA1/2 and TP53 Mutation Testing in Ovarian Cancer" International Journal of Molecular Sciences 24, no. 14: 11570. https://doi.org/10.3390/ijms241411570
APA StyleBorcoman, E., Santana dos Santos, E., Genestie, C., Pautier, P., Lacroix, L., Caputo, S. M., Cabaret, O., Guillaud-Bataille, M., Michels, J., Auguste, A., Leary, A., & Rouleau, E. (2023). Combined Tumor-Based BRCA1/2 and TP53 Mutation Testing in Ovarian Cancer. International Journal of Molecular Sciences, 24(14), 11570. https://doi.org/10.3390/ijms241411570






