Abstract
Enterococcus species are known for their ability to form biofilms, which contributes to their survival in extreme environments and involvement in persistent bacterial infections, especially in the case of multi-drug-resistant strains. This review aims to provide a comprehensive understanding of the mechanisms underlying biofilm formation in clinically important species such as Enterococcus faecalis and the less studied but increasingly multi-drug-resistant Enterococcus faecium, and explores potential strategies for their eradication. Biofilm formation in Enterococcus involves a complex interplay of genes and virulence factors, including gelatinase, cytolysin, Secreted antigen A, pili, microbial surface components that recognize adhesive matrix molecules (MSCRAMMs), and DNA release. Quorum sensing, a process of intercellular communication, mediated by peptide pheromones such as Cob, Ccf, and Cpd, plays a crucial role in coordinating biofilm development by targeting gene expression and regulation. Additionally, the regulation of extracellular DNA (eDNA) release has emerged as a fundamental component in biofilm formation. In E. faecalis, the autolysin N-acetylglucosaminidase and proteases such as gelatinase and serin protease are key players in this process, influencing biofilm development and virulence. Targeting eDNA may offer a promising avenue for intervention in biofilm-producing E. faecalis infections. Overall, gaining insights into the intricate mechanisms of biofilm formation in Enterococcus may provide directions for anti-biofilm therapeutic research, with the purpose of reducing the burden of Enterococcus-associated infections.
1. Introduction
As an opportunistic pathogen, the Enterococcus genus is notorious for its ability to form biofilms. Enterococcal species are common in clinical settings, being responsible for 14.7% of healthcare-associated infections in adults [1]. They are frequently resistant to multiple commonly used antibiotics, including vancomycin [1,2]. Enterococcal biofilms have been detected in a variety of infections, such as periodontal [3], wound [4] and urinary tract infections [5], as well as on medical devices such as ureteral stents [6] and intravascular catheters [7]. While not traditionally considered a respiratory pathogen, Enterococcus faecalis has been detected in lower respiratory tract infections [8,9] and has been shown to form biofilms on endotracheal tubes [10,11].
E. faecium is another clinically important species that frequently causes healthcare-associated infections, such as urinary tract infections (UTI) and bacteriemia, with frequent multi-drug resistance, and particularly concerning—and increasing—vancomycin resistance [12].
The Enterococcus genus also includes a wide variety of species besides Enterococcus faecalis and Enterococcus faecium, which are gathered in a group called non-faecalis non-faecium Enterococcus or Other Enterococcus (OE) [13]. The OE group also can be classified into vanC-positive OE (including Enterococcus gallinarum and Enterococcus casseliflavus) and vanC-negative OE (the other species). Although the vanC-positive species have a high clinical relevance due to the presence of an intrinsic resistance to glycopeptides, via the vanC gene, their involvement in biofilm formation has not been extensively studied so far [13]. Also, the vanC-negative OE remains a rare encounter in the clinic setting. However, recent studies have highlighted that, for some species like Enterococcus avium, Enterococcus durans and Enterococcus raffinosus, there might be a site-specific association; however, a definitive conclusion has not yet been made and their involvement in biofilm formation has also not been widely described [14].
The process called biofilm formation comprises four stages: Initial attachment, microcolony formation (in part correlated with quorum sensing), biofilm maturation and dispersal. Each stage is governed by specific genes for each species of Enterococcus, some being very similar to genes across multiple genera [15]. Numerous morphological factors contribute to their pathogenesis via the production of biofilms. Biofilm-related virulence factors can be divided into secreted factors (cytolysin, secreted antigen A and gelatinase) and cell surface factors, which include pili and microbial surface components that recognize adhesive matrix molecules (MSCRAMMs) and aggregation substances (ASs) [16]. Biofilm formation is regulated by a mechanism called quorum sensing. This is a cell-density-dependent mechanism of intercellular communication, mediated by the expression of specific bacterial genes [15].
This paper aims to compile the existing literature concerning the various factors involved in the formation of biofilms by enterococci and to provide a comprehensive insight into this fascinating niche, while also proposing a number of directions for future research into biofilm eradication techniques that may target these factors. Although there are several articles that review the different steps involved in the formation of biofilms by the Enterococcus genus [15,17,18,19,20], the present paper offers a more comprehensive overview of the complexity of factors that govern the process of biofilm formation for this bacterium. It also aims to provide some directions for the future research of anti-biofilm molecules.
We organized the review into four parts: (a) biofilm-related secreted virulence factors; (b) biofilm-related surface proteins; (c) quorum-sensing molecules; and (d) regulators of extracellular DNA release. An overview of the factors discussed is presented in Table 1.

Table 1.
Overview of the factors involved in the production of biofilms by Enterococcus.
2. Biofilm-Related Secreted Virulence Factors
2.1. Gelatinase
Gelatinase is a class 2 metalloproteinase that uses zinc, is synthesized by the gene gelE in E. faecalis, and is composed of 318 amino acids. The main function of this protein is the maturation of the enterocin O16 and the activity of hydrolase against the gelatin, collagen, fibrin, fibrinogen, hemoglobin, complement components C3, C3a, and C5a, endothelin-1, casein and some other small peptides [20,22]. In order for gelatinase to exhibit its protease activity, the 14 C-terminal amino acid needs to be removed. The gelatinase gelE gene, part of the gelE-sprE operon, together with two other loci (ef1097 and fsrABDC), are regulated by the fsr quorum-sensing system. The transcription of gelE occurs because of the two-component system: the first component is a membrane-bound histidine kinase (HK) that detects extracellular signs, hazardous substances or other factors modifying the pH, the osmolarity and redox status of the extracellular environment and, in response, auto phosphorylates itself, and the second component is a response regulator (RR), which regulates DNA transcriptions and, therefore, initiates cellular response [46,47,48].
One of the properties of gelatinase which is most relevant to biofilm formation is its ability to degrade the collagen adhesion protein (Ace), which helps the bacteria adhere to other bacteria and to both biotic and abiotic surfaces [49]. By degrading Ace, gelatinase contributes to dissemination and colonization. Together with a serine protease, gelatinase has an important role in N-acetylglucosaminidase (AtlA) regulation. This enzyme is important in forming the extracellular DNA present in biofilms via the degradation of other bacterial cells. Gelatinase functions as a stimulant for AtlA release, while serine protease inhibits this process. AtlA is the main autolysin (out of three, which are AtlA, AtlB and AtlC) involved in biofilm formation and has three domains. The first one is responsible for peptidoglycan adherence, the second one is responsible for the glycosaminidase activity, while the third one if not yet fully understood [50]. Gelatinase also directly causes cell lysis. It can latch onto serine protease, inhibiting it in response, or onto the peptidoglycan, rupturing the cell wall in the process and releasing AtlA into the extracellular matrix [51,52].
2.2. Cytolysin
Cytolysin is a protein closely related to lantibiotics, a class of bacteriocins that have lanthionine, methyllanthionine, dehydroalanine and dehydrobutyrine as their main amino acids [24], and that is encoded on the pAD1 sex pheromone plasmid or on the same pathogenicity island as the aggregation substance (AS). Cytolysin comprises two polypeptide chains, a small one called CylLS (63 amino acids) and a larger one named CylLL (68 amino acids) [21]. Separately, these subunits have an important role in the quorum-sensing mechanism via the CylR1–CylR2 complex. The operon responsible for cytolysin synthesis consists of eight genes, out of which two are transcribed divergently (the ones responsible for CylR2–CylR2 synthesis) [23]. The role of cytolysin is to destroy other bacteria, targeting especially Gram-negative bacteria, and eukaryotic cells such as red blood cells. Hemolysis has been observed on solid medium but not on liquid cultures. Cell death is necessary for biofilm formation. When cytolysin is present in the medium and other bacteria start to die out, extracellular DNA is formed, which is essential in biofilm synthesis [21,23,24].
After the transcription and translation of the cylLL and cylLS genes, they undergo a series of changes both intracellularly and extracellularly. Firstly, CylM (a 993 amino acid polypeptide), synthesized by the 5th gene in this operon (cylM), induces changes characteristic of lantibiotics [22]. CylM, also named cytolysin synthetase, has two domains, the first of which catalyzes seven reactions involved in the dehydrogenation of serine and threonine residues, turning them into dehydroalanine and dehydrobutyrine, respectively, and the second of which catalyzes three cyclisation reactions, which include the addition of cysteine residues to dehydroamino acids on the large subunit. Thus, CylM generates CylLL* and CylLS*. Both possess a residue with 16 identical amino acids, which helps in CylB recognition. These two intermediates undergo further changes via CylB, the 6th gene in the operon. CylB has two domains [21,23]. The C-terminal domain is an ATP-binding transporter. It takes both subunits and exports them out of the cell. The N-terminal one acts as a proteolytic site, cleaving 24 and 36 amino acids, respectively, from CylLL* and CylLS* outside the cell, thus making CylLL’ and CylLS’, the 2nd intermediate. ATP is needed to transform CylLS*, but not needed to transform CylLL*. CylA, the 7th one in the operon, acts on CylLL’ and CylLS’, cleaving six amino acids (starting from a serine) from the N-terminal end, making CylLL” and CylLS”, two toxic and active subunits. The last gene on the operon, cylI, makes a protein CylI, which acts as a defense mechanism against autolysis by interacting with either subunit via an unknown mechanism [21,23,24].
The CylR1–CylR2 complex is a DNA-binding transmembrane complex that induces the synthesis of the cyl operon. It does not resemble other quorum-sensing mechanisms that use a histidine kinase as a means of activation. CylR1 is theorized to act as the transmembrane subunit, while CylR2 interacts with a promoter region located between the cylR1 and cylR2 (leftward) genes and the rest of the operon (rightward). The mature CylLS’ is capable of interacting with CylR1, leading to the expression of the operon. This mechanism is of utmost importance when discussing cell proximity. When approaching a Gram-negative bacterium or a eukaryotic cell, the large subunit latches onto the cell, while the small subunit acts as an autoinducer [15,21,23,24,53].
2.3. Secreted Antigen A (SagA)
SagA is a gene that encodes a major secreted antigen of Enterococcus faecium, SagA. It facilitates broad-spectrum extracellular matrix binding (ECM) to fibrinogen, fibronectin, laminin, type-I and type-II and type IV collagen [25]. The deletion of SagA decreases biofilm formation. The SagA protein is a peptidoglycan hydrolase that prefers crosslinked Lys-type peptidoglycan fragments. In E. faecium, it is employed in assembling a signature cell wall architecture, which generates smaller muropeptides that more effectively activate nucleotide-binding oligomerization domain-containing protein 2 (NOD2) in mammalian cells [15,54].
3. Biofilm-Related Surface Proteins
3.1. Pili
Pili extend from the surface of bacterial cells and are involved in a variety of functions, such as motility, adhesion, and conjugation. In E. faecalis and E. faecium, pili play an important role in the attachment of the bacteria to host cells and in the formation of biofilms, which can protect the bacteria from the host’s immune system and antibiotics. The specific characteristics and functions of bacterial pili can vary depending on the bacterial species and the type [55].
In E. faecalis, the Ebp (endocarditis and biofilm-associated pili) proteins, namely EbpA, EbpB, and EbpC, are components of Ebp pili; these have been shown to be essential for biofilm formation in vitro with regard to endocarditis in rats [26], as well as urinary tract infections in mice [27].
There are two different types of pili at the surface of a single E. faecium bacterium, pilA and pilB, which both contribute to biofilm formation [28]. They are phenotypically distinct, due to the fact that they are composed of different pilin subunits. Bacteria usually need to be in a high-temperature environment in order to form pili. The standard temperature range in hospitals is 21 °C to 24 °C, which is too low for bacteria to form pili; however, the pilin subunits are shown near the membrane [56,57].
PilB are thicker, more flexible, and they are expressed at the poles of the dividing cells. PilB are often involved in conjugation, and they are expressed in the growth phase. During cell division, the major pilin subunits for pilA and pilB are formed. PilB pilin is translocated around the cross wall (the newly synthesized peptidoglycan layer between the two new cells) and deposited in murein sacculi during its separation. When the cell grows, new murein sacculi arise and the old ones are pushed to the poles, with pilB proteins inside [15,56,57].
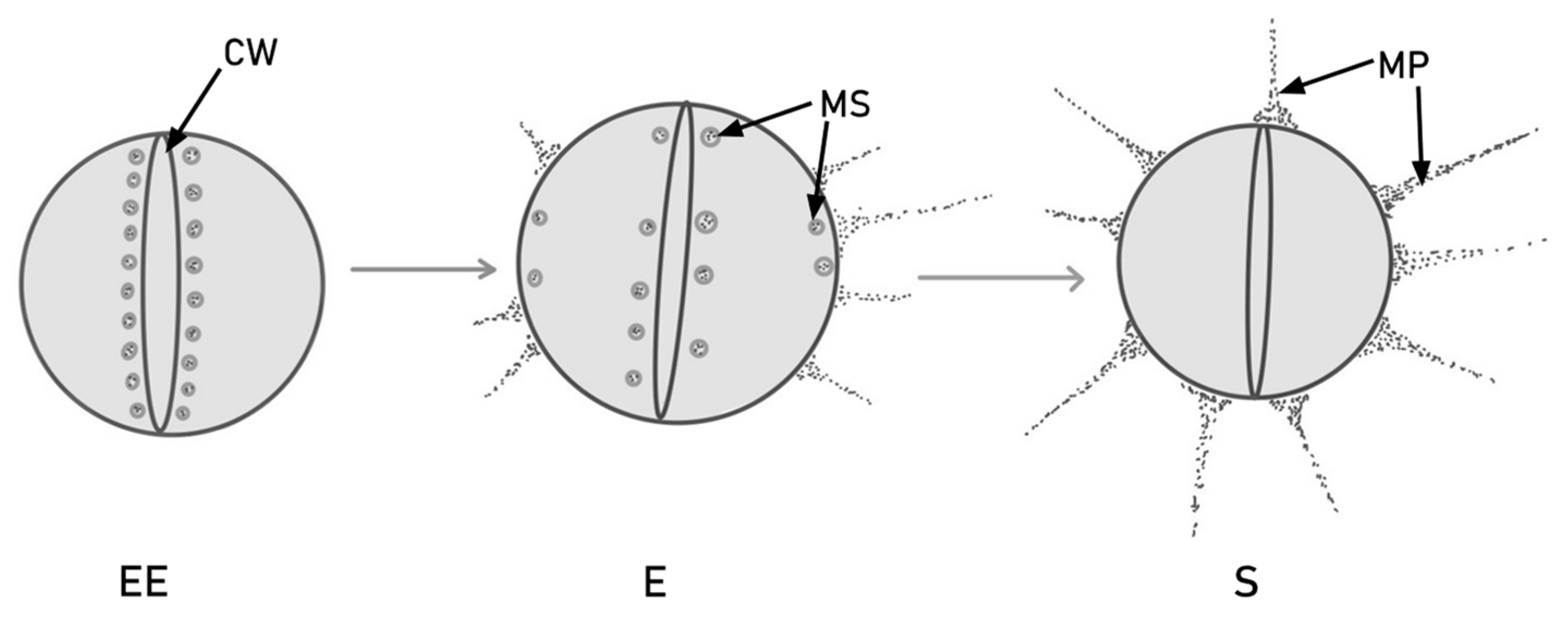
There are three stages of bacterial growth: early exponential (EE), exponential (E) and stationary (S). The expression of the PilB-type pili in dividing cells is different in each of these stages. As the cell grows, so does the level of pilin protein. Later on, in the exponential phase, the murein sacculi approach the plasma membrane and form PilB. The newly formed pilin subunits are deposited in the murein sacculi near the cross wall. PilB pili are thicker than pilA, which indicates the incorporation of more pilin subunits [55]. This process is illustrated in Figure 1.
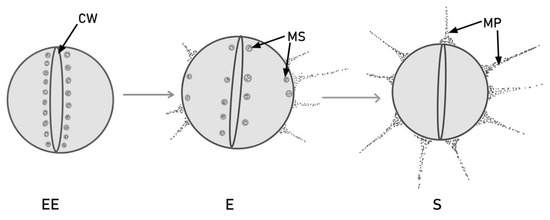
Figure 1.
PilB production and translocation via murein sacculi (MS) during cell division, in different stages of colony growth. In the early exponential (EE) phase, the pilin subunits are stored near the bacterial cross wall (CW). Later on, in the exponential (E) phase, some subunits migrate near the extremities and form pili, but others remain around the cross wall. In the stationary (S) phase, all the pilin subunits are converted into mature PilB-type pili (MP).
The other type of pili in E. faecium is PilA-type pili. These types of pili are absent in cell division. However, the pilA gene and the pilA pilin are present throughout the cell cycle. Their role is to adhere the bacteria to a substrate or to another cell. On liquid media, PilA cannot grow, therefore solid media are used to observe these types of structure [33,58].
Given that pili have been shown to contribute to the formation of biofilms by E. faecalis [26,59] and E. faecium [28], it would be reasonable to assume that the inhibition of pili formation would inhibit enterococcal biofilm formation. This assumption is further sustained by the fact that this phenomenon has been observed in other bacteria such as Pseudomonas aeruginosa [60]. Potential targets for inhibiting biofilm formation could include genes involved in pili formation, such as Cpd and Ccf for E. faecalis, and pilA for E. faecium [28,36]. Another way to eradicate the multi-resistant bacteria that form biofilms could be the disruption of pili-mediated attachment, targeting pilA. In E. faecalis, blocking Ebp using an anti-Ebp vaccine has been shown to inhibit biofilm formation in a mouse CAUTI (catheter-associated urinary tract infection) model [61].
3.2. Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs)
MSCRAMMs are a family of bacterial adhesins capable of binding to the elements of the extracellular matrix, predominantly to collagen. This family comprises three proteins: Ace, Acm and Scm. Ace is found in E. faecalis, while Acm and Scm are found in E. faecium. These surface proteins are usually characterized by an amino acid sequence of leucine, proline, x (any other amino acid) and glycine [30,33].
3.2.1. Ace
The ace gene (adhesion to collagen of E. faecalis) encodes an adhesin that binds to collagen (types I and IV) and laminin. Ace is a CNA-like MSCRAMM that binds to collagen via the multistep ligand-binding mechanism called the collagen hug [29,30]. Most proteins in the CNA-like MSCRAMM sub-family have been shown to act as virulence factors in experimental bacterial infections [62].
Ace expression is dependent on external factors such as the presence of bile salts and serum, as well as internal factors such as the putative Ers box, a nucleotide sequence (AACATTTGTTG) encoding a protein that, in turn, acts as a positive regulator of virulence genes; it also regulates GelE expression, which, in turn, is dependent on a complete fsr system [49,63]. Studies have shown that the administration of monoclonal anti-Ace antibodies (mAb), specifically mAb70, reduces E. faecalis infections in rat models [64].
3.2.2. Acm
In E. faecium, the acm gene (adhesion to collagen of E. faecium) has been identified and found to encode the Acm protein. Acm presents a structure with a N-terminal signal peptide, followed by an A domain, various B sequences (depending on the species) and a C-terminal sequence that enables anchoring to the peptidoglycan layer. In previous studies, considerable homology has been demonstrated with other collagen-binding adhesins present in Staphylococcus aureus and E. faecalis. Like Cna in Staphylococcus aureus (the prototype for the adhesin family), the ligand-binding A domain has three subunits, N1, N2 and N3. The N1N2 subdomains bind with affinity to collagen, through a mechanism called the collagen hug mechanism [31,32].
As has been already demonstrated in samples derived from clinical isolates, there is a correlation between the adherence levels of the pathogen and the amount of Acm protein expressed on the cell surface. The expression of Acm by E. faecium and its adherence to collagen appear to be important parameters for virulent expression in patients with endocarditis, as shown by Nallapareddy et al. [31]. As explained by the collagen hug mechanism and by the reactivity of the serum to the recombinant form, the use of specific antibodies against Acm would facilitate the inhibition of collagen adherence and is therefore a promising therapeutic and prophylactic strategy in patients at risk of E. faecium infections [15,32].
Another important element involved in the pathogenesis of E. faecium producing biofilm infections is certainly autolysin. Autolysin is an endogenous lytic enzyme that is present in all peptidoglycan-containing bacteria. In addition to the breakdown of the peptidoglycan component of cells, it has an important function in cell separation during division and in the release of extracellular DNA (eDNA) into the biofilm matrix, ensuring its stability [50,53]. In E. faecium, the major autolysin identified is AtlAEfm, which appears to be involved in the localization of Acm on the bacterial cell surface, contributing to collagen adherence and the pathogenesis of infections. Therefore, given the fundamental functions expressed in the pathogenic process, as concluded for Acm, AtlAEfm represents a potential target in the treatment of E. faecium producing biofilm infections [31].
3.2.3. Scm
Due to the increase in nosocomial infections caused by E. faecium, more studies have had this bacterium as their main focus. In a recent article, a genomic analysis of the TX0016 strain of E. faecium has shown the importance of Scm (second collagen adhesin of E. faecium, previously known as Fms10 from cell-wall-anchored E. faecium surface protein), a MSCRAMM [33].
In addition to Acm, which binds mostly to type I collagen, the main component of collagen fibers in human tissue, Scm is an adhesin with a much greater affinity for collagen type V, a key component of the cross-linkage between interstitial collagen fibrils and membranous collagen networks. Having two types of collagen adhesins gives E. faecium the ability to fine-tune its adherence phenotype to better suit the given tissue. For collagen type V, which is abundant in the intestinal submucosa, Scm might be the key factor that causes its resistance and persistence in the GI tract [65].
3.3. Aggregation Substance (AS)
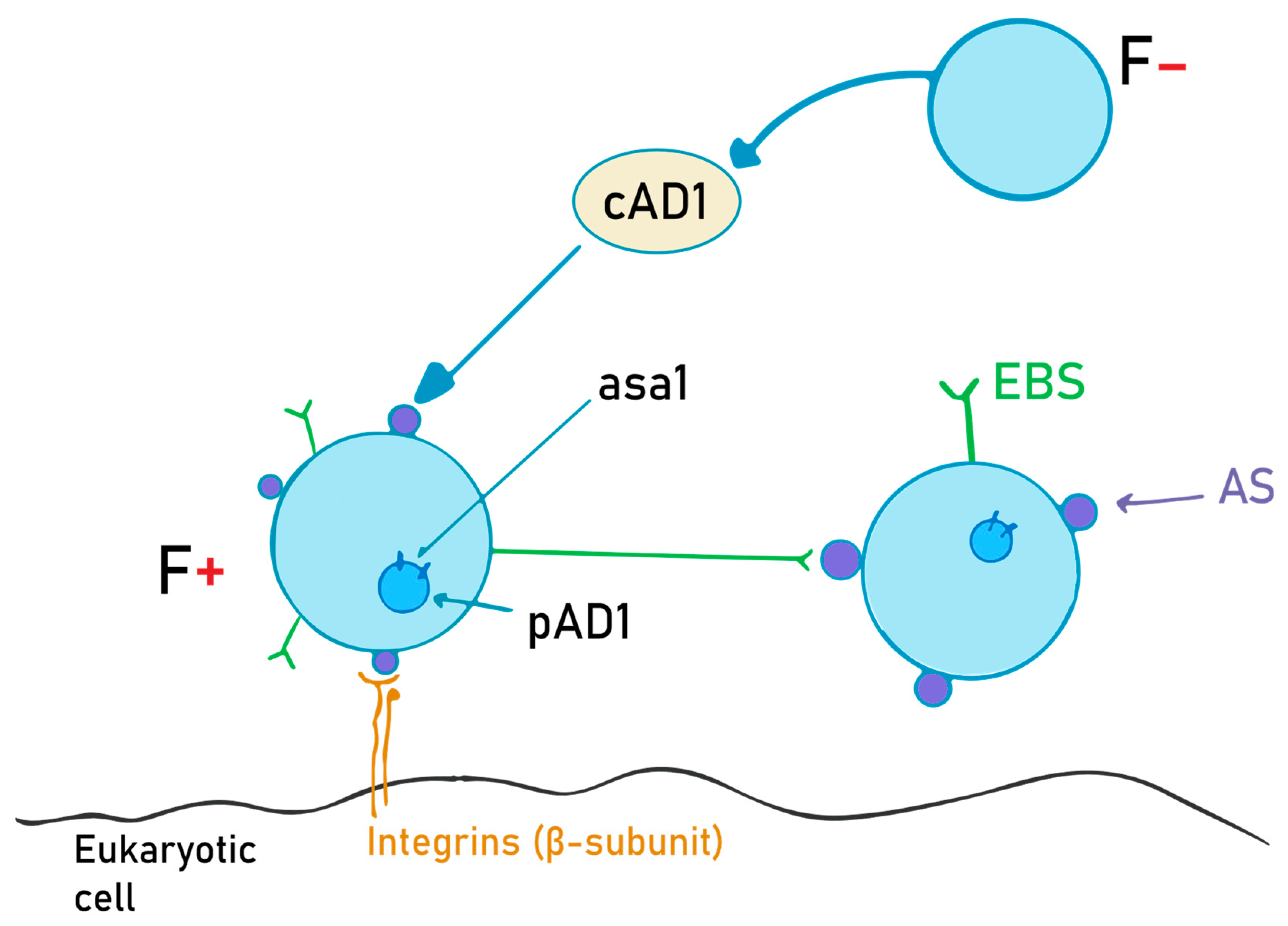
The aggregation substance is a hair-like glycoprotein on the bacterial surface. It is synthesized on “older” structures of the Enterococcus cell wall from the asa1 gene, located on a sex pheromone plasmid, pAD1, when exposed to a sex pheromone, cAD1 [34]. Asc10, also called the aggregation factor, is encoded on the pCF10 plasmid, and it is expressed while within the mammalian bloodstream. Its main role is to help bacteria adhere to other bacteria and eukaryotic cells, and to diminish superoxide function [66,67]. The details of this mechanism are schematically depicted in Figure 2.
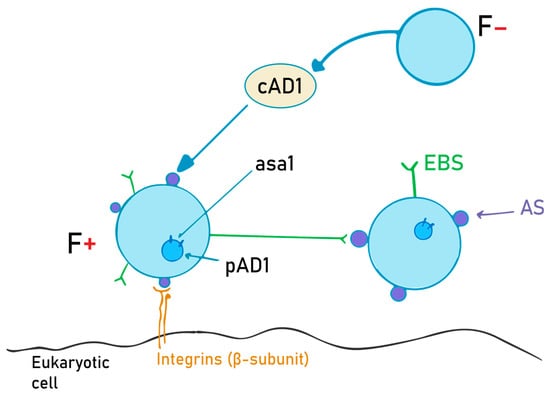
Figure 2.
AS (aggregation substance) production and the role of the cAD1 pheromone. F- bacteria emit cAD1, a sex pheromone, in order to induce AS synthesis in F+ bacteria. Once the AS is expressed on the cell surface, it will interact with the EBS (enterococcal binding substance), and thus the two cells can now exchange the pAD1 plasmid (with the asa1 gene that encodes for AS).
Out of the 1296 amino acids, the main two parts on the AS are the two RGD motifs (rich in arginine, glycine and aspartate). It also has a signal domain comprising 43 amino acids and a C-terminal proline-rich sequence, which is important regarding integration into the bacterial cell wall and membrane. RGD is a sequence comprising three amino acids that is composed of arginine, glycine and aspartic acid, which helps in adhesion. This motif interacts with the β subunit of the integrins of eukaryotic cells, mainly macrophages and epithelial cells, as well as the enterococcus-binding substance (EBS), which is similar in structure to lipoteichoic acid. The N-terminal part of the protein is the one that interacts with both integrins and enterococcal surface proteins, while the C-terminal region is bound to the cell wall. Experimental studies have shown that when N-terminal amino acids are eliminated, biofilm formation decreases significantly compared to the C-terminal mutant counterparts [66,68].
AS also helps with bacterial aggregation. The F- bacterium secretes small peptides called sex pheromones. When the F+ bacterium senses these pheromones, it starts to transcribe the asa1 gene and express AS on the cell surface. When AS interacts with EBS, conjugation occurs, after which both bacteria possess the pAD1 plasmid [68]. The expression of AS leads to bacterial clumping and higher antibiotic resistance, helping in biofilm formation. Because of the aggregation, sonication is needed in order to study a single bacterium. Besides its aggregating properties, AS also helps with regard to integration and resistance inside the macrophages (MF). Lucigenin was used in order to detect superoxide activity inside the MF, and it was shown that CL (a metabolite of lucigenin) had a lower concentration in asa1+ cells in the first 3 h after integration, according to Süßmuth et al. [34]. This indicates that AS lowers superoxide activity and thus helps E. faecalis survive for longer periods of time inside the cell.
The RGD motifs are recognized by the beta subunits of integrins. The main ones are CD18 and CD11b on macrophages, helping with bacterial integration. The same region (RGD) helps in extracellular matrix binding. The presence of AS on the surface of the enterococci increases the adherence to both fibronectin (8-fold) and thrombospondin (4-fold). Monoclonal antibodies against CD18 and CD11b have been shown to reduce bacterial integration, prohibiting bacterial dissemination throughout the organism. RGDS (RGD serine)-containing peptides latch onto the integrins and EBS better than RGD, thus entering into competition with AS. This way, bacteria cannot interact with the surface or with other bacteria, thus reducing the risk of biofilm formation [34,66,68,69].
4. Quorum-Sensing Molecules
The quorum-sensing process works through the synthesis and release of small signal molecules called pheromones. Pheromones are small signal molecules that stimulate an intercellular response among members of the bacterial community. Both Gram-negative and Gram-positive bacteria use this type of cell-to-cell communication [70]. In Gram-positive species, this communication consists of small peptides called autoinducers or quorum-sensing peptides, which are very sensitive to their own receptors. These molecules express an important function in communication between microbes, influencing biofilm stability and development, but are also possibly involved in communication with the host environment [70]. In Enterococcus spp., biofilm formation is regulated by the fsr (fecal streptococci regulator) locus and peptide pheromones. In addition to the communication between microbes, pheromones appear to mediate the transfer of plasmids through the conjugative apparatus. This process facilitates the transfer of genes that promote biofilm formation, such as adhesins involved in the pathogenesis of the infection. Some examples of pheromones in the enterococcal species are Cpd, Cob and Ccf. As demonstrated by Eaton et al. in vitro, these pheromones are chemotactic for human leukocytes, being able to induce the production of superoxides and the secretion of lysosomal enzymes. Therefore, they can be considered virulence factors [71].
These peptide pheromones consist of sequences with seven to nine amino acids and are synthesized by ribosomes. After that, they undergo post-translation modifications and become activated during excretion. Secretion is mediated by a membrane-associated ATP-binding cassette (ABC), which facilitates the achievement of a concentration threshold; this activates a specific receptor through phosphorylation and consequently the transcription of the target gene [70,72]. Details of these mechanisms are explained in Figure 3, Figure 4 and Figure 5.
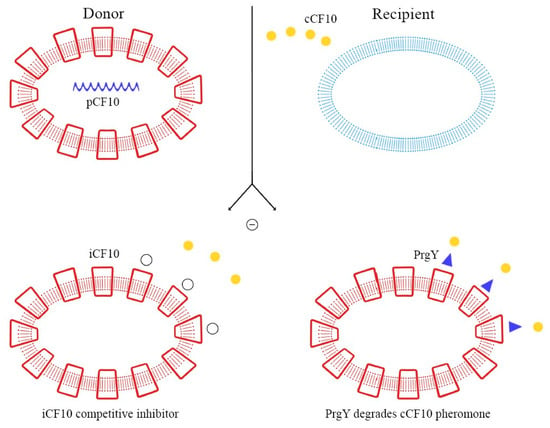
Figure 3.
Autocrine inhibition of pheromone-mediated transfer mechanisms.
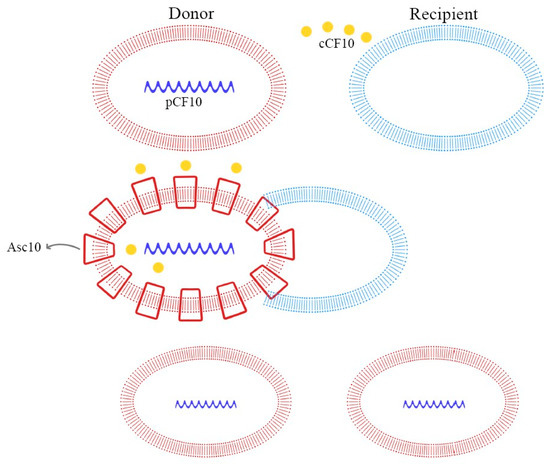
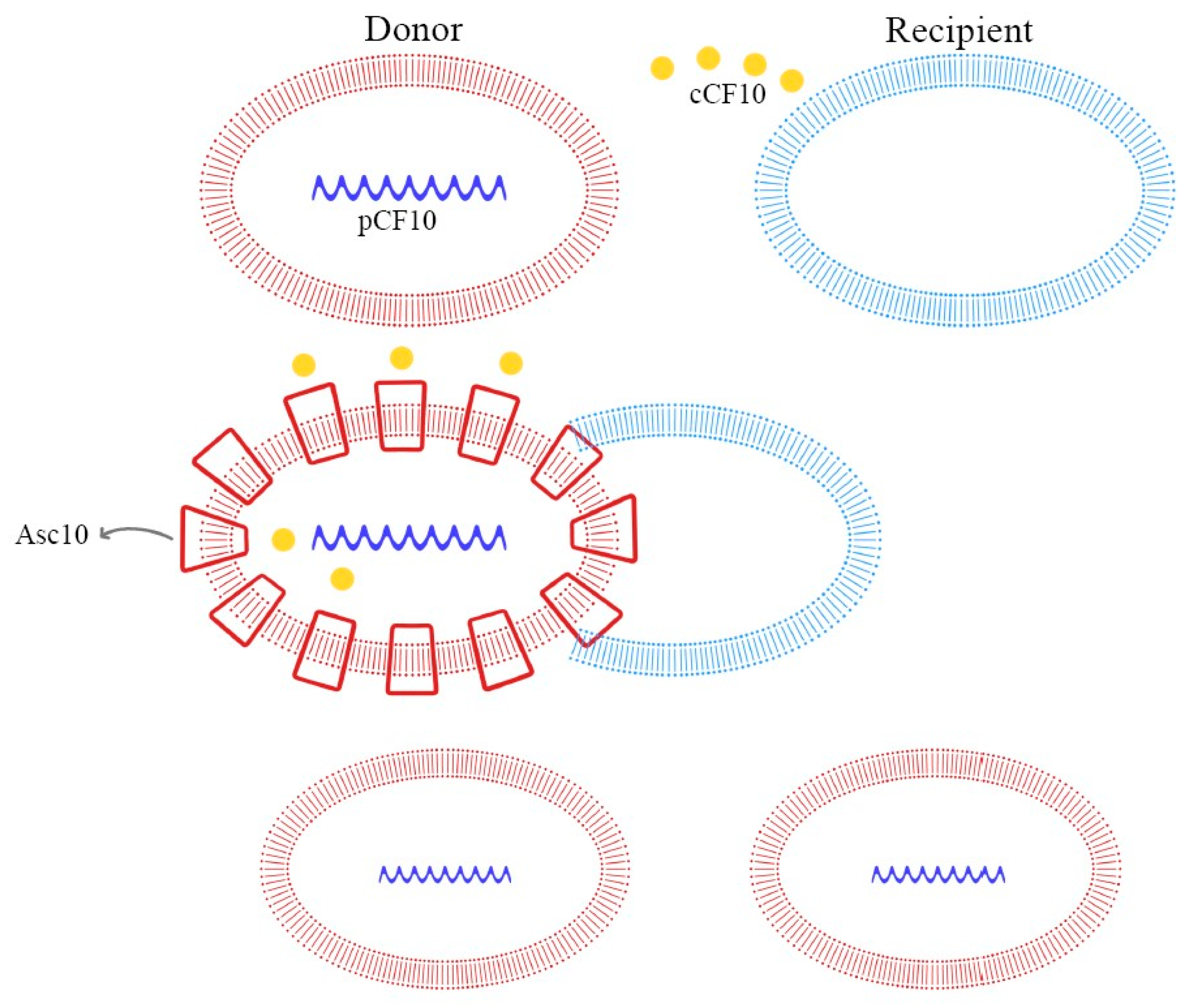
Figure 4.
Pheromone-mediated (pCF10) horizontal plasmid transfer and Asc10 expression.
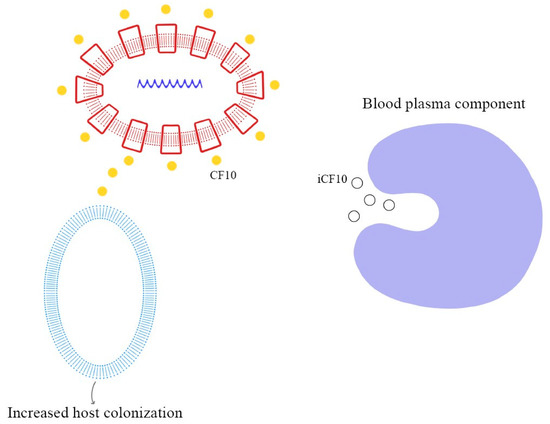
Figure 5.
Degradation of iCF10 and activation of conjugation system resulting in host colonization.
4.1. Cob1
Cob1 is a sex pheromone isolated from E. faecalis that consists of eight hydrophobic amino acids, H–Val–Ala–Val–Leu–Gly–Ala–OH, and induces the conjugal transfer of hemolysin and bacteriocin plasmids (pOB1 and pYI1). Besides these plasmids, Cob1 influences the relationship with the host environment, through the expression of a surface adhesin that mediates the aggregation between recipient and donor cells (aggregation substance), and also the sexual aggregation between donors; it is therefore called a “clumping-inducing agent” (CIA) [35].
In E. faecalis, the transfer of the pCF10 plasmid from donor cells is mediated by the secretion of the cCF10 pheromone from the recipient cells. Donor cells, containing pCF10, internalize the cCF10 pheromone and induce the expression of the aggregation substance (Asc10), which mediates plasmid transfer. This paracrine pheromone signaling pathway is the link between sex pheromone secretion and biofilm formation [66,70,73].
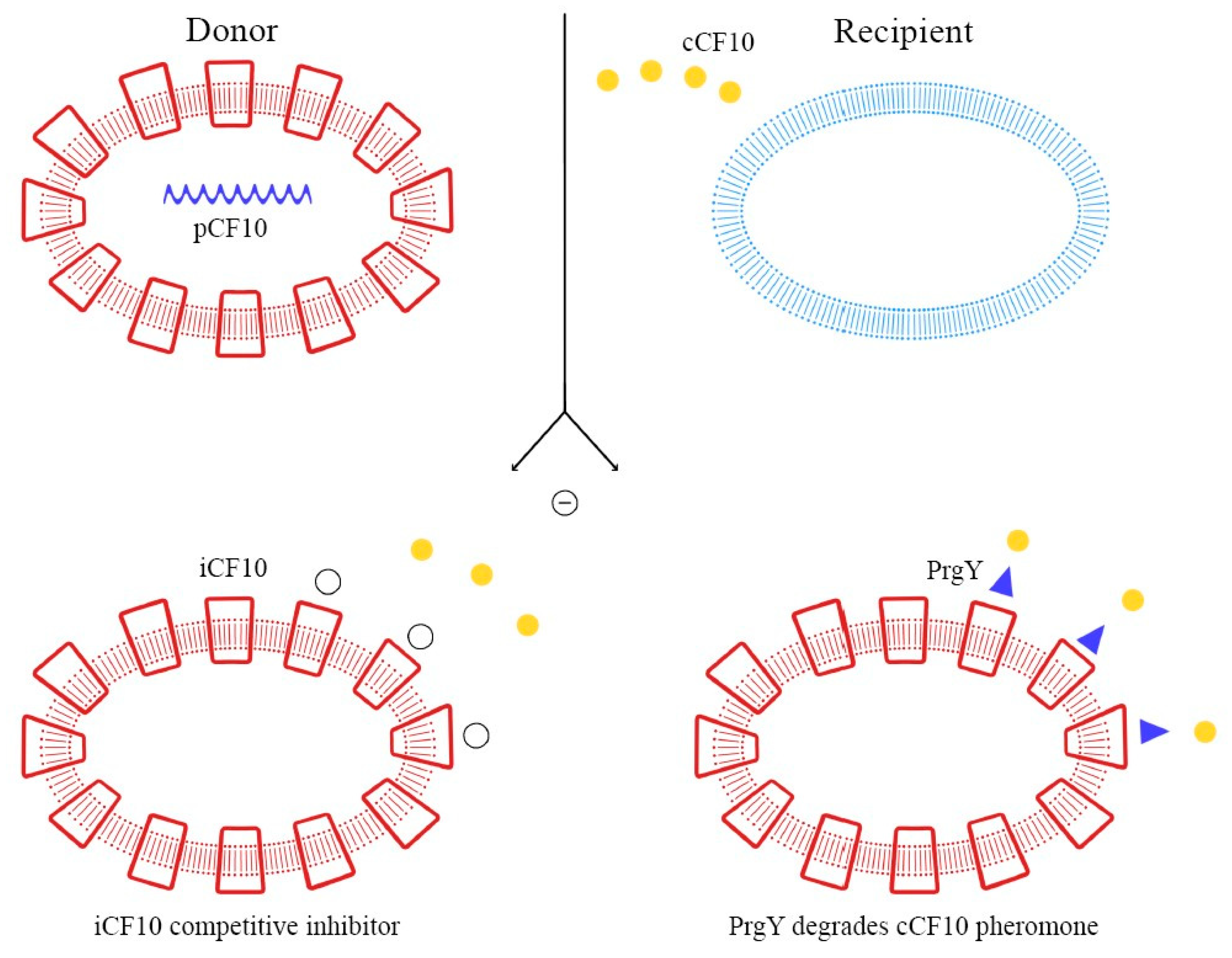
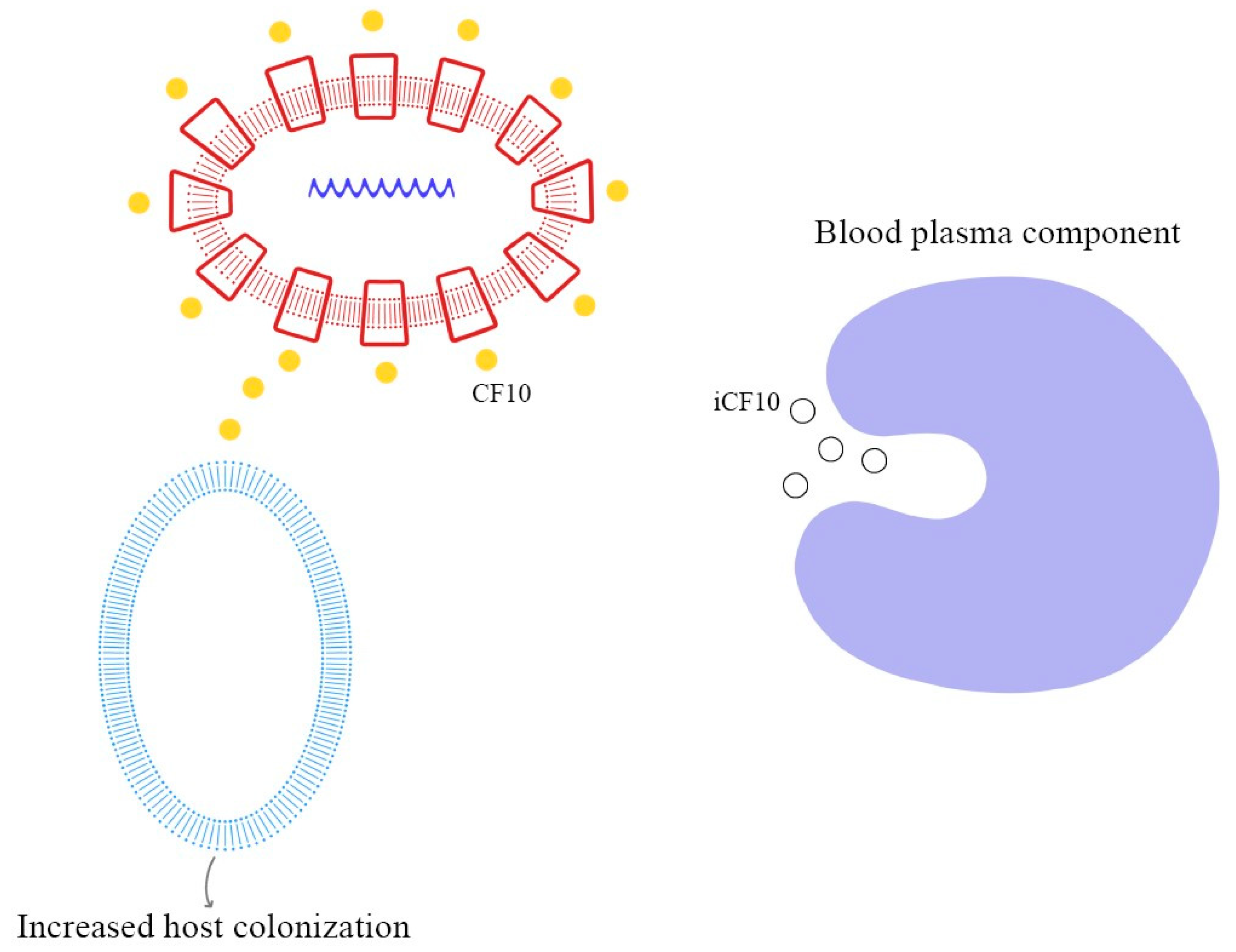
As demonstrated in previous studies, there is a mechanism to prevent the autocrine activation of this process: iCF10 and PrgY. The former is a short protein that acts as a competitive signaling inhibitor on the pheromone, while PrgY directly degrades the CF10 pheromone. The competitive signaling inhibitor iCF10 has been shown to be degraded by a blood plasma component, inducing activation of the conjugation system and thereby increasing the degree of colonization in the host [74,75]. The details of this process are explained in Figure 6. Understanding the synthesis and expression of the pheromones signaling pathway in biofilm-producing bacteria is fundamental in the approach to determining the new therapeutic targets of multi-drug-resistant (MDR) bacteria.
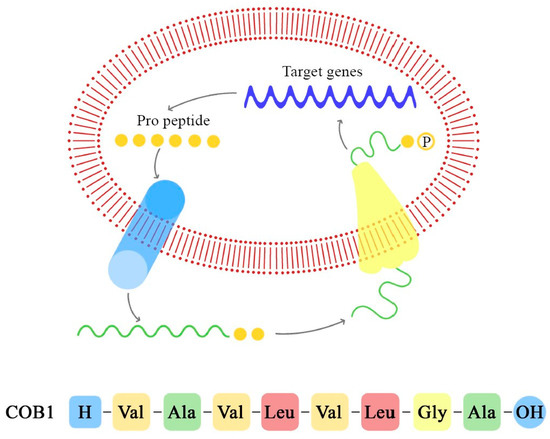
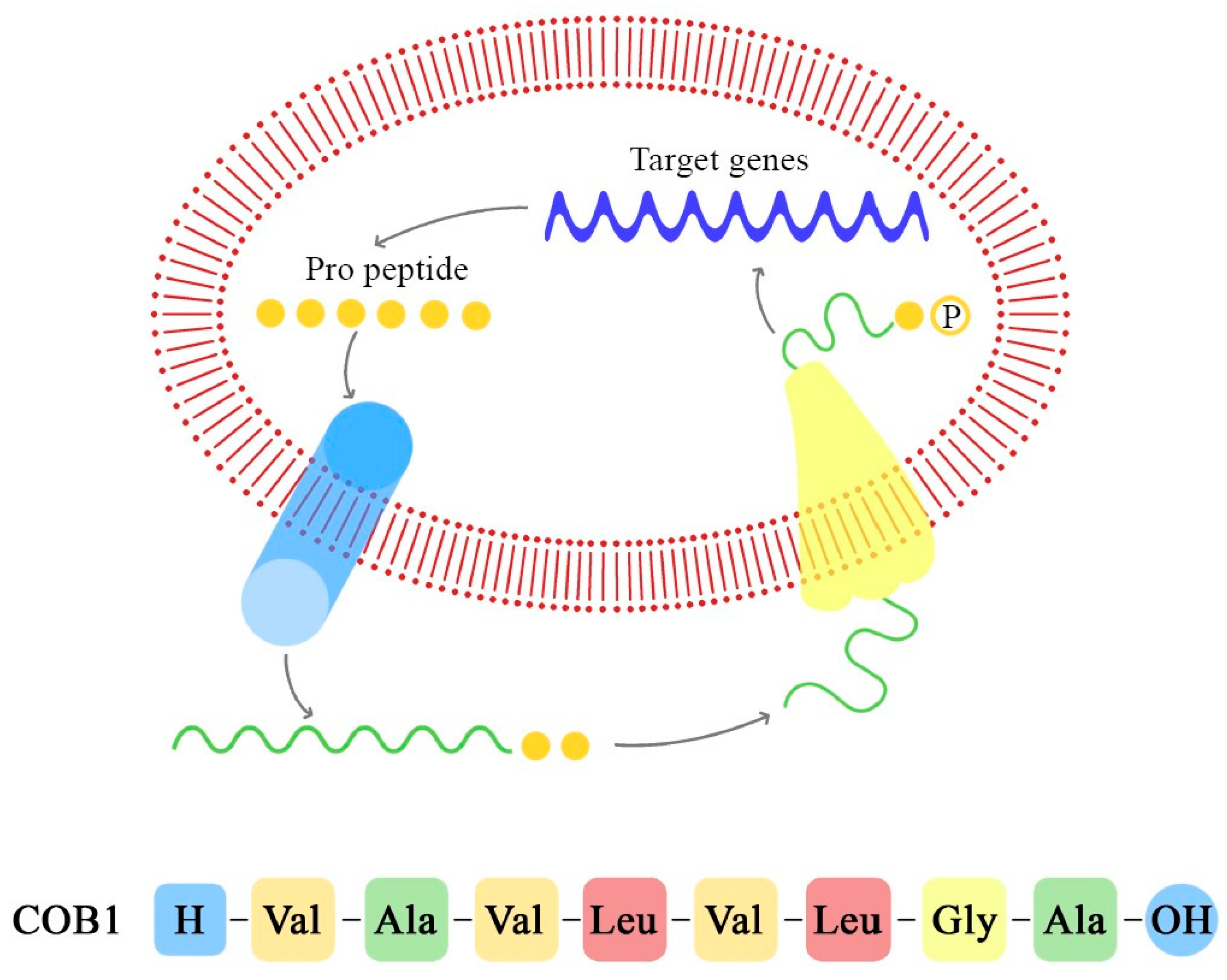
Figure 6.
Pheromone synthesis and target gene activation produced by COB1 pheromone (the amino acids sequence is presented below).
In the case of commensal E. faecalis, cOB1 is encoded by a signaling lipoprotein, EF2496, which possesses the precursor sequence of cOB1. Through a zinc-dependent metalloprotease-mediated cleavage, cOB1 is transported out of the cell by a ATP-binding transporter named the peptide pheromone transporter (PptAB). This proteolytic cleavage of the lipoprotein signal peptide shows similarities to Amyloid β formation in Alzheimer’s disease, and it is a critical step in the production and interaction of cOB1 [76]. As demonstrated by Gour et al., the monomeric form of cOB1 is capable of binding to the specific receptor and of inducing the conjugative transfer of plasmids, while the aggregated form does not bind to the receptor, blocking gene transfer [76]. This inhibition could be a functional therapeutic target against the propagation of virulence factors mediated by peptide pheromones.
As demonstrated by Gilmore et al., when the cOB1 pheromone is released and comes into contact with its receptor on the MDR Enterococcus V583, it modulates the expression of the genes [77]. The major effect of the cOB1 expressed by the commensal species on V583 is to induce transcription of the pTEF2 genes and insertion-sequence-like element accumulation on the V583 chromosome. This interaction results in an incompatibility between V583 and the fecal consortium, causing the killing of V583. Therefore, although little is known about the colonization mechanisms, the destabilization of the native gastrointestinal flora facilitates the colonization of species such as MDRV583 Enterococcus.
4.2. Ccf
The CcfA gene is responsible for the production of the cCF10 pheromone. The CCF10 pheromone has a molecular weight of 789 and has the following amino acid structure: H–Leu–Val–Thr–Leu–Val–Phe–Val–OH [67]. It is necessary for the transfer of the pCF10 tetracycline-resistant plasmid in vivo. The transfer of pCF10 is mediated by two different peptides: one inhibitor, iCF10, and one pheromone, cCF10, both expressed by important genes in E. faecalis. They both interact with the PrgX receptor protein. The transformation of the cCF10 precursor in mature pheromone cCF10 is mediated by another gene called eep, which separates the amino acid terminal end form the rest of the protein [78]. The result is a more hydrophobic amino acid.
The production of the cCF10 pheromone by E. faecalis can be increased by introducing a cloned ccfA gene. It can also make non-pheromone producers, for example, Lactococcus lactis, to produce cCF10 pheromones. Besides plasmid transfer, the cCF10 pheromone can also induce aggregation [36].
4.3. Cpd
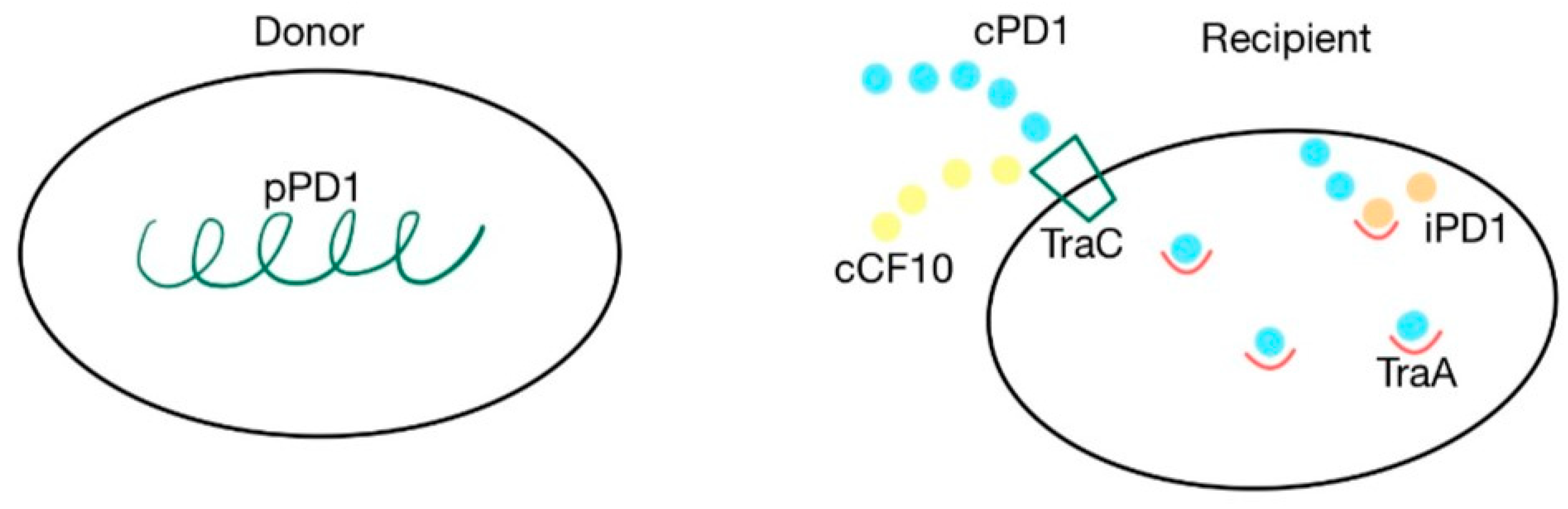
The cpd gene is responsible for the production of the cPD1 pheromone, which mediates the transfer of the pPD1 plasmid. A cell synthesizes the pPD1 plasmid and donates it to another. This plasmid is responsible for synthesizing proteins involved in the mating response to the cPD1 sex pheromone, which binds to the TraA intercellular receptor. This transfer can be blocked by the iPD1 inhibitor, produced by the recipient cells, which is a competitive inhibitor that binds with TraA before the pheromone [37,38,79]. TraC is a pheromone-binding protein, encoded by the pPD1 plasmid, that enables recipient cells to receive cPD1. The cPD1 pheromone has a higher affinity for TraA than for TraC. However, TraC plays an important role in the crossing of the cell wall by the pheromone, acting like a protein carrier. PrgZ, the protein that binds with cCF10, is very similar to TraC. Because of this similarity, cCF10 can also bind with TraC. However, prgZ, encoded by the pCF10 plasmid, has no affinity for cPD1 [35,37,38,79]. The cPD1 pheromone has a molecular weight of 912 and has the following aminoacidic structure: H–Phe–Leu–Val–Met–Phe–Leu–Ser–Gly–OH [79]. The details of this mechanism are presented in Figure 7.
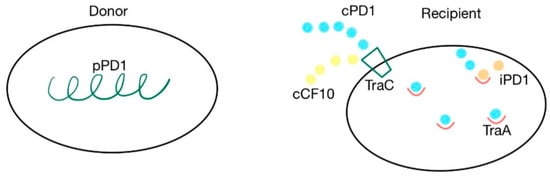
Figure 7.
cPD1 pheromone and pPD1 plasmid interaction via TraA receptor. Pheromone-mediated transfer mechanisms between a donor that contains the pPD1 plasmid and the recipient’s TraC and TraA proteins. cPD1 and cCF10 pheromones can both bind with TraC.
4.4. Eep
A gene known to be involved in quorum sensing is eep. It is involved in the maturation of the peptide pheromones cAD1 and cCF10 through a membrane protease Eep. The protease encoded by eep comprises 422 amino acids and uses Zn2+ as a cofactor. It also facilitates the proteolytic processing of RsiV, the anti-sigma factor for sigV, which is an extracytoplasmic function (ECF) sigma factor, improving stress resistance [39,80].
The innate ability of E. faecalis to resist lysozyme stress is offered by the complete degradation of RsiV, which is a counterpoint to the activation of SigV. In eep deletion, mutant strains were shown to only have a partially degraded RsiV. Eep works together with AhrC and the ArgR, arginine repressors that block the uptake of this amino acid or activate its catabolism, leading to biofilm formation. The deletion of genes that encode these proteins has been shown to lower the burden in UTI and endocarditis [39,80,81].
4.5. The fsr Operon and ef1097 Locus
This system is composed of four genes: fsrA (RR), fsrB, fsrD and fsrC (HK), in order in the fsr operon. When fsrC is transcribed, a membrane-bound HK—fsrC—results, with the role of sensing an 11-amino-acid-long cyclized peptide lactone in the extracellular environment in the form of gelatinase biosynthesis-activating pheromone (GBAP). When the fsrD gene is transcribed, a propeptide fsrD is formed, which undergoes further changes mediated by fsrB. fsrB is a transmembrane protein that belongs to the accessory regulator protein class (AgrB-accessory gene regulator). This protein modifies fsrD, turning it into GBAP, which will further interact with fsrC [44,45,82]. This interaction will lead to fsrA (RR) activation (phosphorylation), a protein belonging to the lytTR family of DNA-binding domains, which will in turn act as a facilitator for the transcription of the ef1097 locus (composed of two genes, ef1097 and ed1097b), fsrB and gelE, facilitating ef1097, ef1097b, the fsr locus, gelE (encoding a gelatinase), and sprE (encoding a serine protease). If the fsr quorum-sensing system is affected, these genes, together with 75 others, are inhibited, including a gene important in biofilm formation, bopD [42].
The role of the ef1097 locus is the synthesis of enterocin O16, an antimicrobial peptide (bacteriocin) composed of 68 C-terminal amino acids [22]. This peptide inhibits the proliferation of Lactobacillus spp., but it does not affect Staphylococcus, Enterococcus or Listeria growth; at higher concentrations, it can also act as an antifungal peptide. This peptide is obtained after EF_1097 (the precursor), a 191-amino-acid-long peptide, is delivered out of the cell via the Sec system, which cuts the peptide between Ala56 and Ser57, and when gelatinase cuts the precursor at the 123rd amino acid, leaving a 68-amino-acid-long peptide residue, previously named enterocin O16 [22]. Unlike in Streptococcus dysgalactiae, the genes associated with bacteriocin resistance (dysI to dysA, the gene that codes for dysgalacticin, in S. dysgalactiae) are not located on the same operon. It has been shown that ef1097b does not confer resistance to enterocin O16 as was expected [22,44,45,82].
5. Regulators of Extracellular DNA Release (eDNA)
The function of eDNA as a component of the biofilm matrix is well established [43,83]. eDNA is known to perform various biological functions in bacteria (adhesion, early biofilm development, stabilization of the biofilm matrix, horizontal gene transfer, phagocytosis prevention, inhibition of inflammation) that vary with the different phases of the biological cycle [43,50]. However, recent studies have remarked upon the lack of a direct correlation between microbial activity and the iDNA/eDNA ratio, as well as the limitations of the current extraction method used for qPCR. The mechanisms that regulate eDNA release from bacterial cells are also less clear. At first, eDNA was only thought to derive from cell lysis, while recently, numerous studies have demonstrated that the release of eDNA in prokaryotic and eukaryotic cells can result from lysis-dependent and lysis-independent mechanisms. This being stated, there is an opportunity for further studies concerning eDNA release in particular species [15,43,83].
In Gram-positive bacteria, endogenous autolysin (AtlA) stimulates eDNA release via QS-dependent lysis. In E. faecalis, eDNA production is QS-dependent, and the lactone peptide (FsrD) triggers the activation of two proteases: gelatinase (GelE) and serine protease (SprE). These two proteases, through different mechanisms, induce the release of eDNA [20,43,83].
E. faecalis produces several autolysins, the most frequently identified being AtlA. This is N-acetylglucosaminidase, an endogenous lytic enzyme that is crucial in the separation of daughter cells during cell division. AtlA sequencing showed a structure composed of three domains: domain I, which hash unknown functions, domain II, which contains the region capable of hydrolyzing peptidoglycans, and domain III, which is composed of six LysM residues recognizing the N-acetylglucosamine residues (GlcNAc) of cell wall peptidoglycans, as is required for AtlA anchoring. Some studies have demonstrated that the inactivation of AtlA results in increased chaining, due to defect septum cleavage, decreased primary attachment, and decreased biofilm production. Consequently, the fundamental role of AtlA can be deduced not only in cell growth and lysis, but also in the initial adhesion and eDNA release in the biofilm composition during the accumulation phase in E. faecalis [20,22,43].
As mentioned previously, QS-dependent eDNA production in E. faecalis is also mediated by the activation of two extracellular proteases. The gene coding for gelatinase (GelE) is located adjacent to fsrC and is co-transcribed with serine protease (SprE). Mutations in the fsr locus, resulting in GelE defects, negatively affect its ability to produce biofilms, indicating an important role in the activation of autolysin and in the regulation of the biological mechanisms of biofilm production. As reported by Thomas et al., there are two possible mechanisms through which these proteases may express the regulatory effect of Atla and influence biofilm development [50,84].
The first mechanism, defined as the autolytic pathway, is that in which GelE localizes itself on the cell wall of producing cells in order to activate autolysis. The levels of SprE present could counteract GelE-induced autolysis but, if insufficient, the cell undergoes autolysis.
The second mechanism, defined as allolytic or fratricidal, involves the diffusion of GelE from a producer cell to a target sister, so as to induce the autolysis of the sister cell. Both types of autolysis, activated by GelE, involve the mediation of AtlA, making these theoretical models similar to the process of programmed cell death in eukaryotic cells. The cytotoxic activity expressed by GelE (the effector) towards a producer cell (autolysis) or sister cells (allolysis) through the mediation of AtlA would result in the release of eDNA, essential in biofilm development. The allolysis (sibling killing sibling) model considers the concept of cell developmental competence, where the inability of a subpopulation of cells to produce SprE (the regulator, immune factor) would result in the death of these cells [43,44,45].
Previous studies have demonstrated the possibility that, following the attachment phase, the fsr quorum-sensing signaling pathway could be activated, resulting in the expression of GelE, SprE and AtlA. When GelE reaches high levels of activity in biofilm production, at later stages of colonization, AtlA could induce cleavage, resulting in small chain cells of E. faecalis. This configuration would increase microbial dissemination and evasion from the host’s defense. In fact, some host immune components, such as LL37, defensin and complement components (C3a and C3b), appear to be able to undergo the proteolytic activity of gelatinase. In addition, gelatinase has been shown to adhere to fibrin, increasing dissemination and providing further evasion of the host immune system. All these data demonstrate how the GelE-mediated autolytic mechanism affects the pathogenic process and the virulence of E. faecalis producing biofilm [43,44,45,52].
Barnes et al., in order to confirm the primary role of eDNA in biofilms, compared the levels of extracellular DNA between samples of E. faecalis producing biofilm and a planktonic form, revealing a significantly higher amount in the biofilm samples [85]. The study also enabled a temporal and morphological analysis of eDNA release in the biofilm ecosystem. In the first 4 h post attachment, there is a significant elevation of eDNA release, which remains stable for the following 24 h. From a morphological point of view, eDNA mainly has two structural forms: a long and intercellular complex (yarn structure), and a globular form (sweater structure), which resembles the appearance of the biofilm matrix. Despite these promising results, the mechanisms that induce DNA export and articulation in biofilm matrix stabilization in E. faecalis remain incompletely understood. Certainly, as reported by Yu et al., extracellular DNA is a potential therapeutic target in the treatment of biofilm-producing E. faecalis infections. In fact, it has been demonstrated that eDNA inhibition decreases its ability to produce biofilms and increases the susceptibility of E. faecalis to NaOCl, even at low concentrations (0.5%) [83].
6. Conclusions
This paper provides an extensive overview of the factors contributing to biofilm formation in Enterococcus species, especially in those involved in human pathology, such as E. faecalis and E. faecium. Some of the factors we identified in this literature review are secreted virulence factors (gelatinase, cytolysin, sagA), some are surface proteins (Acm, Scm, Aggregation substance), some are quorum-sensing molecules (Cob, Ccf, and Cpd, Eep, the fsr operon) and some are molecules regulating eDNA release through cell lysis (AtlA, gelatinase, cytolysin, serin protease). All these factors are involved in biofilm formation, through adhesion, aggregation, matrix formation and stabilization, and some also contribute to gene transfer and bacterial resistance to antibiotics and the immune response.
While some factors are shared between species (aggregation substance), some factors, or, at least, their variants, are species-specific. Even if the overall structure of biofilms, their stages of development and their biological functions are known to be similar across the genus and even multiple genera, there are gaps in the research pertaining to the mechanism of biofilm formation for each particular species. New therapeutic strategies against biofilm-associated infections caused by Enterococcus and other multi-drug resistant bacteria can only be developed by understanding these processes and targeting relevant specific genes or molecules.
Author Contributions
Conceptualization, P.Ș., D.A.T. (Dan Alexandru Toc) and D.A.T. (Doina Adina Todea); methodology P.Ș., D.A.T. (Dan Alexandru Toc) and D.A.T. (Doina Adina Todea); software, G.R., G.B., Ș.-G.G., A.C.G. and A.B.; validation, C.C. and I.A.C.; investigation, G.R., G.B., Ș.-G.G., A.C.G. and A.B.; resources, G.R., G.B., Ș.-G.G., A.C.G. and A.B.; data curation, P.Ș., D.A.T. (Dan Alexandru Toc) and D.A.T. (Doina Adina Todea); writing—original draft preparation, G.R., G.B., Ș.-G.G., A.C.G., A.-G.P., V.S.N. and A.B.; writing—review and editing, P.Ș., D.A.T., V.S.N., A.-G.P., I.A.C., C.C. and D.A.T. (Doina Adina Todea); visualization, G.R., G.B., Ș.-G.G., A.C.G. and A.B.; supervision, P.Ș., D.A.T. (Dan Alexandru Toc) and D.A.T. (Doina Adina Todea). All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
This work was granted by project PDI-PFE-CDI 2021, entitled Increasing the 570 Performance of Scientific Research, Supporting Excellence in Medical Research and Innovation, 571 PROGRES, no. 40PFE/30.12.2021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef]
- Miller, W.R.; Murray, B.E.; Rice, L.B.; Arias, C.A. Vancomycin-Resistant Enterococci: Therapeutic Challenges in the 21st Century. Infect. Dis. Clin. 2016, 30, 415–439. [Google Scholar] [CrossRef]
- Molander, A.; Reit, C.; Dahlén, G.; Kvist, T. Microbiological status of root-filled teeth with apical periodontitis. Int. Endod. J. 1998, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Codelia-Anjum, A.; Lerner, L.B.; Elterman, D.; Zorn, K.C.; Bhojani, N.; Chughtai, B. Enterococcal Urinary Tract Infections: A Review of the Pathogenicity, Epidemiology, and Treatment. Antibiotics 2023, 12, 778. [Google Scholar] [CrossRef] [PubMed]
- Keane, P.; Bonner, M.; Johnston, S.; Zafar, A.; Gorman, S. Characterization of biofilm and encrustation on ureteric stents In Vivo. Br. J. Urol. 1994, 73, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Sandoe, J.A.; Witherden, I.R.; Cove, J.H.; Heritage, J.; Wilcox, M.H. Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J. Med. Microbiol. 2003, 52, 547–550. [Google Scholar] [CrossRef]
- Kelly, B.J.; Imai, I.; Bittinger, K.; Laughlin, A.; Fuchs, B.D.; Bushman, F.D.; Collman, R.G. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome 2016, 4, 7. [Google Scholar] [CrossRef]
- Bonten, M.J.; van Tiel, F.H.; van der Geest, S.; Stobberingh, E.E.; Gaillard, C.A. Enterococcus faecalis pneumonia complicating topical antimicrobial prophylaxis. N. Engl. J. Med. 1993, 328, 209–210. [Google Scholar] [CrossRef]
- van Charante, F.; Wieme, A.; Rigole, P.; De Canck, E.; Ostyn, L.; Grassi, L.; Deforce, D.; Crabbé, A.; Vandamme, P.; Joossens, M.; et al. Microbial diversity and antimicrobial susceptibility in endotracheal tube biofilms recovered from mechanically ventilated COVID-19 patients. Biofilm 2022, 4, 100079. [Google Scholar] [CrossRef]
- Adair, C.; Gorman, S.; Byers, L.; Jones, D.; Feron, B.; Crowe, M.; Webb, H.; McCarthy, G.; Milligan, K. Eradication of endotracheal tube biofilm by nebulised gentamicin. Intensive Care Med. 2002, 28, 426–431. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, R.F.; Leong, K.W.; Cumming, V.; Van Hal, S.J. Vancomycin-resistant Enterococcus faecium and the emergence of new sequence types associated with hospital infection. Res. Microbiol. 2023, 174, 104046. [Google Scholar] [CrossRef] [PubMed]
- Monticelli, J.; Knezevich, A.; Luzzati, R.; Di Bella, S. Clinical management of non-faecium non-faecalis vancomycin-resistant enterococci infection. Focus on Enterococcus gallinarum and Enterococcus casseliflavus/flavescens. J. Infect. Chemother. 2018, 24, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Toc, D.A.; Pandrea, S.L.; Botan, A.; Mihaila, R.M.; Costache, C.A.; Colosi, I.A.; Junie, L.M. Enterococcus raffinosus, Enterococcus durans and Enterococcus avium Isolated from a Tertiary Care Hospital in Romania—Retrospective Study and Brief Review. Biology 2022, 11, 598. [Google Scholar] [CrossRef]
- Ch’ng, J.H.; Chong, K.K.L.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2019, 17, 82–94. [Google Scholar] [CrossRef]
- Nasaj, M.; Mousavi, S.M.; Hosseini, S.M.; Arabestani, M.R. Prevalence of Virulence Factors and Vancomycin-resistant Genes among Enterococcus faecalis and E. faecium Isolated from Clinical Specimens. Iran. J. Public Health 2016, 45, 806–813. [Google Scholar]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef]
- Woźniak-Biel, A.; Bugla-Płoskońska, G.; Burdzy, J.; Korzekwa, K.; Ploch, S.; Wieliczko, A. Antimicrobial resistance and biofilm formation in Enterococcus spp. Isolated from humans and turkeys in Poland. Microb. Drug Resist. 2019, 25, 277–286. [Google Scholar] [CrossRef]
- Stępień-Pyśniak, D.; Hauschild, T.; Kosikowska, U.; Dec, M.; Urban-Chmiel, R. Biofilm formation capacity and presence of virulence factors among commensal Enterococcus spp. from wild birds. Sci. Rep. 2019, 9, 11204. [Google Scholar] [CrossRef]
- Ali, L.; Goraya, M.U.; Arafat, Y.; Ajmal, M.; Chen, J.-L.; Yu, D. Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches. Int. J. Mol. Sci. 2017, 18, 960. [Google Scholar] [CrossRef]
- Shankar, N.; Coburn, P.; Pillar, C.; Haas, W.; Gilmore, M. Enterococcal cytolysin: Activities and association with other virulence traits in a pathogenicity island. Int. J. Med. Microbiol. 2004, 293, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Dundar, H.; Brede, D.A.; La Rosa, S.L.; El-Gendy, A.O.; Diep, D.B.; Nes, I.F. The fsr quorum-sensing system and cognate gelatinase orchestrate the expression and processing of proprotein EF_1097 into the mature antimicrobial peptide enterocin O16. J. Bacteriol. 2015, 197, 2112–2121. [Google Scholar] [CrossRef]
- Rumpel, S.; Razeto, A.; Pillar, C.M.; Vijayan, V.; Taylor, A.; Giller, K.; Gilmore, M.S.; Becker, S.; Zweckstetter, M. Structure and DNA-binding properties of the cytolysin regulator CylR2 from Enterococcus faecalis. EMBO J. 2004, 23, 3632–3642. [Google Scholar] [CrossRef]
- Van Tyne, D.; Martin, M.J.; Gilmore, M.S. Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins 2013, 5, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Kawalec, M.; Weinstock, G.M.; Hryniewicz, W.; Murray, B.E. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect. Immun. 2003, 71, 5033–5041. [Google Scholar] [CrossRef]
- Nallapareddy, S.R.; Singh, K.V.; Sillanpää, J.; Garsin, D.A.; Höök, M.; Erlandsen, S.L.; Murray, B.E. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 2006, 116, 2799. [Google Scholar] [CrossRef]
- Singh, K.V.; Lewis, R.J.; Murray, B.E. Importance of the epa locus of Enterococcus faecalis OGlRF in a mouse model of ascending urinary tract infection. J. Infect. Dis. 2009, 200, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Mohabati Mobarez, A.; Goudarzi, M.; Najar-Peerayeh, S.; Mirzaee, M. Prevalence of biofilm formation and vancomycin-resistant genes among Enterococcus faecium isolated from clinical and environmental specimens in Lorestan hospitals. Iran. J. Microbiol. 2018, 10, 74. [Google Scholar]
- Ross, C.L.; Liang, X.; Liu, Q.; Murray, B.E.; Höök, M.; Ganesh, V.K. Targeted protein engineering provides insights into binding mechanism and affinities of bacterial collagen adhesins. J. Biol. Chem. 2012, 287, 34856–34865. [Google Scholar] [CrossRef]
- Liu, Q.; Ponnuraj, K.; Xu, Y.; Ganesh, V.K.; Sillanpaa, J.; Murray, B.E.; Narayana, S.V.; Hook, M. The Enterococcus faecalis MSCRAMM ace binds its ligand by the collagen hug model. J. Biol. Chem. 2007, 282, 19629–19637. [Google Scholar] [CrossRef]
- Nallapareddy, S.R.; Sillanpää, J.; Ganesh, V.K.; Höök, M.; Murray, B.E. Inhibition of Enterococcus faecium adherence to collagen by antibodies against high-affinity binding subdomains of Acm. Infect. Immun. 2007, 75, 3192–3196. [Google Scholar] [CrossRef] [PubMed]
- Nallapareddy, S.R.; Weinstock, G.M.; Murray, B.E. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 2003, 47, 1733–1747. [Google Scholar] [CrossRef] [PubMed]
- Sillanpää, J.; Nallapareddy, S.R.; Prakash, V.P.; Qin, X.; Hook, M.; Weinstock, G.M.; Murray, B.E. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology 2008, 154, 3199–3211. [Google Scholar] [CrossRef] [PubMed]
- Süßmuth, S.; Muscholl-Silberhorn, A.; Wirth, R.; Susa, M.; Marre, R.; Rozdzinski, E. Aggregation Substance Promotes Adherence, Phagocytosis, and Intracellular Survival of Enterococcus faecalis within Human Macrophages and Suppresses Respiratory Burst. Infect. Immun. 2000, 68, 4900–4906. [Google Scholar] [CrossRef]
- Nakayama, J.; Abe, Y.; Ono, Y.; Isogai, A.; Suzuki, A. Isolation and Structure of the Enterococcus faecalis Sex Pheromone, cOB1, that Induces Conjugal Transfer of the Hemolysin/Bacteriocin Plasmids, pOB1 and pYI1. Biosci. Biotechnol. Biochem. 1995, 59, 703–705. [Google Scholar] [CrossRef]
- Antiporta, M.H.; Dunny, G.M. ccfa the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis, OG1RF. J. Bacteriol. 2002, 184, 1155–1162. [Google Scholar] [CrossRef]
- Folli, C.; Mangiarotti, L.; Folloni, S.; Alfieri, B.; Gobbo, M.; Berni, R.; Rivetti, C. Specificity of the TraA-DNA Interaction in the Regulation of the pPD1-Encoded Sex Pheromone Response in Enterococcus faecalis. J. Mol. Biol. 2008, 380, 932–945. [Google Scholar] [CrossRef]
- Nakayama, J.; Takanami, Y.; Horii, T.; Sakuda, S.; Suzuki, A. Molecular Mechanism of Peptide-Specific Pheromone Signaling in Enterococcus faecalis: Functions of Pheromone Receptor TraA and Pheromone-Binding Protein TraC Encoded by Plasmid pPD1. J. Bacteriol. 1998, 180, 449–456. [Google Scholar] [CrossRef]
- Frank, K.L.; Guiton, P.S.; Barnes, A.M.; Manias, D.A.; Chuang-Smith, O.N.; Kohler, P.L.; Spaulding, A.R.; Hultgren, S.J.; Schlievert, P.M.; Dunny, G.M. AhrC and Eep are biofilm infection-associated virulence factors in Enterococcus faecalis. Infect. Immun. 2013, 81, 1696–1708. [Google Scholar] [CrossRef]
- Frank, K.L.; Barnes, A.M.T.; Grindle, S.M.; Manias, D.A.; Schlievert, P.M.; Dunny, G.M. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense rnas and implicates the protease eep in pathogenesis. Infect. Immun. 2012, 80, 539–549. [Google Scholar] [CrossRef]
- An, F.Y.; Sulavik, M.C.; Clewell, D.B. Identification and Characterization of a Determinant (eep) on the Enterococcus faecalis Chromosome That Is Involved in Production of the Peptide Sex Pheromone cAD1. J. Bacteriol. 1999, 181, 5915–5921. [Google Scholar] [CrossRef] [PubMed]
- Hufnagel, M.; Koch, S.; Creti, R.; Baldassarri, L.; Huebner, J. A Putative Sugar-Binding Transcriptional Regulator in a Novel Gene Locus in Enterococcus faecalis Contributes to Production of Biofilm and Prolonged Bacteremia in Mice. J. Infect. Dis. 2004, 189, 420–430. [Google Scholar] [CrossRef]
- Montanaro, L.; Poggi, A.; Visai, L.; Ravaioli, S.; Campoccia, D.; Speziale, P.; Arciola, C.R. Extracellular DNA in biofilms. Int. J. Artif. Organs. 2011, 34, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Galloway-Peña, J.R.; Bourgogne, A.; Qin, X.; Murray, B.E. Diversity of the fsr-gelE region of the Enterococcus faecalis genome but conservation in strains with partial deletions of the fsr operon. Appl. Environ. Microbiol. 2011, 77, 442–451. [Google Scholar] [CrossRef]
- Hancock, L.E.; Perego, M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 2004, 186, 5629–5639. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Singh, K.V.; Weinstock, G.M.; Murray, B.E. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 2001, 183, 3372–3382. [Google Scholar] [CrossRef]
- Galperin, M.Y. Defining Statement General Principles of Signal Transduction Types of Bacterial Receptors and Signaling Pathways Structural Organization of Signal Transduction Proteins Interaction of Signal Transduction Pathways How E. coli Sees the World? Further Reading. In Sensory Transduction in Bacteria; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Wilke, K.E.; Francis, S.; Carlson, E.E. Activity-Based Probe for Histidine Kinase Signaling. J. Am. Chem. Soc. 2012, 134, 9150–9153. [Google Scholar] [CrossRef]
- Lebreton, F.; Riboulet-Bisson, E.; Serror, P.; Sanguinetti, M.; Posteraro, B.; Torelli, R.; Hartke, A.; Auffray, Y.; Giard, J.C. ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect. Immun. 2009, 77, 2832–2839. [Google Scholar] [CrossRef]
- Thomas, V.C.; Hiromasa, Y.; Harms, N.; Thurlow, L.; Tomich, J.; Hancock, L.E. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 2009, 72, 1022–1036. [Google Scholar] [CrossRef]
- Qin, X.; Singh, K.V.; Weinstock, G.M.; Murray, B.E. Effects of Enterococcus faecalis fsr Genes on Production of Gelatinase and a Serine Protease and Virulence. Infect. Immun. 2000, 68, 2579–2586. [Google Scholar] [CrossRef]
- Kart, D.; Kuştimur, A.S. Investigation of gelatinase gene expression and growth of Enterococcus faecalis clinical isolates in biofilm models. Turk. J. Pharm. Sci. 2019, 16, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Oli, A.K.; Javaregowda, P.K.; Jain, A.; Kelmani, C.R. Mechanism Involved in Biofilm Formation of Enterococcus faecalis. Available online: www.intechopen.com (accessed on 5 May 2023).
- Kim, B.; Wang, Y.C.; Hespen, C.W.; Espinosa, J.; Salje, J.; Rangan, K.J.; Oren, D.A.; Kang, J.Y.; Pedicord, V.A.; Hang, H.C. Enterococcus faecium secreted antigen A generates muropeptides to enhance host immunity and limit bacterial pathogenesis. Elife 2019, 8, e45343. [Google Scholar] [CrossRef]
- Hendrickx, A.P.A.; Bonten, M.J.M.; van Luit-Asbroek, M.; Schapendonk, C.M.E.; Kragten, A.H.M.; Willems, R.J.L. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology 2008, 154, 3212–3223. [Google Scholar] [CrossRef] [PubMed]
- Afonina, I.; Lim, X.N.; Tan, R.; Kline, K.A. Planktonic interference and biofilm alliance between aggregation substance and endocarditis- and biofilm-associated pili in Enterococcus faecalis. J. Bacteriol. 2018, 200, 10–1128. [Google Scholar] [CrossRef]
- La Rosa, S.L.; Montealegre, M.C.; Singh, K.V.; Murray, B.E. Enterococcus faecalis ebp pili are important for cell-cell aggregation and intraspecies gene transfer. Microbiology 2016, 162, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, A.P.A.; Schapendonk, C.M.E.; Van Luit-Asbroek, M.; Bonten, M.J.M.; Van Schaik, W.; Willems, R.J.L. Differential PilA pilus assembly by a hospital-acquired and a community-derived Enterococcus faecium isolate. Microbiology 2010, 156, 2649–2659. [Google Scholar] [CrossRef]
- Singh, K.V.; Nallapareddy, S.R.; Murray, B.E. Importance of the ebp (endocarditis-and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J. Infect. Dis. 2007, 195, 1671–1677. [Google Scholar] [CrossRef]
- Pérez-Martínez, I.; Haas, D. Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 3399–3405. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Pinkner, J.S.; Caparon, M.G.; Hultgren, S.J. EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice. Sci. Transl. Med. 2014, 6, 254ra127. [Google Scholar] [CrossRef]
- Arora, S.; Gordon, J.; Hook, M. Collagen Binding Proteins of Gram-Positive Pathogens. Front. Microbiol. 2021, 12, 628798. [Google Scholar] [CrossRef]
- Cohen, A.L.V.; Roh, J.H.; Nallapareddy, S.R.; Höök, M.; Murray, B.E. Expression of the collagen adhesin ace by Enterococcus faecalis strain OG1RF is not repressed by Ers but requires the Ers box. FEMS Microbiol. Lett. 2013, 344, 18–24. [Google Scholar] [CrossRef]
- Varahan, S.; Iyer, V.S.; Moore, W.T.; Hancock, L.E. Eep Confers Lysozyme Resistance to Enterococcus faecalis via the Activation of the Extracytoplasmic Function Sigma Factor SigV. J. Bacteriol. 2013, 195, 3125–3134. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, M.A.; Iqbal, M.N.; Saddick, S.; Ali, I.; Khan, F.S.; Kanwal, S.; Ahmed, D.; Ibrahim, M.; Afzal, U.; Awais, M. Identification of lead compounds against scm (Fms10) in Enterococcus faecium using computer aided drug designing. Life 2021, 11, 77. [Google Scholar] [CrossRef]
- Chuang, O.N.; Schlievert, P.M.; Wells, C.L.; Manias, D.A.; Tripp, T.J.; Dunny, G.M. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect. Immun. 2009, 77, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Sakagami, Y.; Ishii, Y.; Isogai, A.; Kitada, C.; Fujino, M.; Adsit, J.C.; Dunny, G.M.; Suzuki, A. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 1988, 263, 14574–14578. [Google Scholar] [CrossRef] [PubMed]
- Rozdzinski, E.; Marre, R.; Susa, M.; Wirth, R.; Muscholl-Silberhorn, A. Aggregation substance-mediated adherence of Enterococcus faecalis to immobilized extracellular matrix proteins. Microb. Pathog. 2001, 30, 211–220. [Google Scholar] [CrossRef]
- Muscholl-Silberhorn, A. Analysis of the clumping-mediating domain(s) of sex pheromone plasmid pAD1-encoded aggregation substance. Eur. J. Biochem. 1998, 258, 515–520. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Eaton, T.J.; Gasson, M.J. Molecular Screening of Enterococcus Virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial quorum sensing and microbial community interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef]
- Dang, X.; Wang, G. Spotlight on the Selected New Antimicrobial Innate Immune Peptides Discovered During 2015-2019. Curr. Top. Med. Chem. 2020, 20, 2984–2998. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.R.; Flynn, A.R.; Bryan, E.M.; Dunny, G.M. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J. Bacteriol. 2005, 187, 4830–4843. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.R.; Dunny, G.M. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J. Bacteriol. 2008, 190, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Gour, S.; Kumar, V.; Rana, M.; Yadav, J.K. Pheromone peptide cOB1 from native Enterococcus faecalis forms amyloid-like structures: A new paradigm for peptide pheromones. J. Pept. Sci. 2019, 25, e3178. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Rauch, M.; Ramsey, M.M.; Himes, P.R.; Varahan, S.; Manson, J.M.; Lebreton, F.; Hancock, L.E. Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains. Proc. Natl. Acad. Sci. USA 2015, 112, 7273–7278. [Google Scholar] [CrossRef]
- Hirt, H.; Greenwood-Quaintance, K.E.; Karau, M.J.; Till, L.M.; Kashyap, P.C.; Patel, R.; Dunny, G.M. Enterococcus faecalis Sex Pheromone cCF10 Enhances Conjugative Plasmid Transfer In Vivo. mBio 2018, 9, e00037-18. [Google Scholar] [CrossRef]
- Horii, T.; Nagasawa, H.; Nakayama, J. Functional Analysis of TraA, the Sex Pheromone Receptor Encoded by pPD1, in a Promoter Region Essential for the Mating Response in Enterococcus faecalis. J. Bacteriol. 2002, 184, 6343–6350. [Google Scholar] [CrossRef]
- Frank, K.L.; Vergidis, P.; Brinkman, C.L.; Greenwood Quaintance, K.E.; Barnes, A.M.; Mandrekar, J.N.; Schlievert, P.M.; Dunny, G.M.; Patel, R. Evaluation of the Enterococcus faecalis biofilm-associated virulence factors AhrC and Eep in rat foreign body osteomyelitis and in vitro biofilm-associated antimicrobial resistance. PLoS ONE 2015, 10, e0130187. [Google Scholar] [CrossRef]
- Rouchon, C.N.; Weinstein, A.J.; Hutchison, C.A.; Zubair-Nizami, Z.B.; Kohler, P.L.; Frank, K.L. Disruption of the tagF Orthologue in the epa Locus Variable Region of Enterococcus faecalis Causes Cell Surface Changes and Suppresses an eep -Dependent Lysozyme Resistance Phenotype. J. Bacteriol. 2022, 204, e00247-22. [Google Scholar] [CrossRef]
- Shankar, J.; Walker, R.G.; Ward, D.; Horsburgh, M.J. The Enterococcus faecalis Exoproteome: Identification and Temporal Regulation by Fsr. PLoS ONE 2012, 7, e33450. [Google Scholar] [CrossRef]
- Yu, M.-K.; Kim, M.-A.; Rosa, V.; Hwang, Y.-C.; DEL Fabbro, M.; Sohn, W.-J.; Min, K.-S. Role of extracellular DNA in Enterococcus faecalis biofilm formation and its susceptibility to sodium hypochlorite. J. Appl. Oral Sci. 2019, 27, e20180699. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.C.; Thurlow, L.R.; Boyle, D.; Hancock, L.E. Regulation of Autolysis-Dependent Extracellular DNA Release by Enterococcus faecalis Extracellular Proteases Influences Biofilm Development. J. Bacteriol. 2008, 190, 5690–5698. [Google Scholar] [CrossRef] [PubMed]
- Barnes, A.M.T.; Ballering, K.S.; Leibman, R.S.; Wells, C.L.; Dunny, G.M. Enterococcus faecalis Produces Abundant Extracellular Structures Containing DNA in the Absence of Cell Lysis during Early Biofilm Formation. mBio 2012, 3, e00193-12. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).