NFE2L2 and STAT3 Converge on Common Targets to Promote Survival of Primary Lymphoma Cells
Abstract
1. Introduction
2. Results
2.1. NFE2L2 and STAT3 Cooperate in Sustaining Cell Survival, MTOR, MAPK3/1 Activation, and Protein Synthesis in PEL Cells
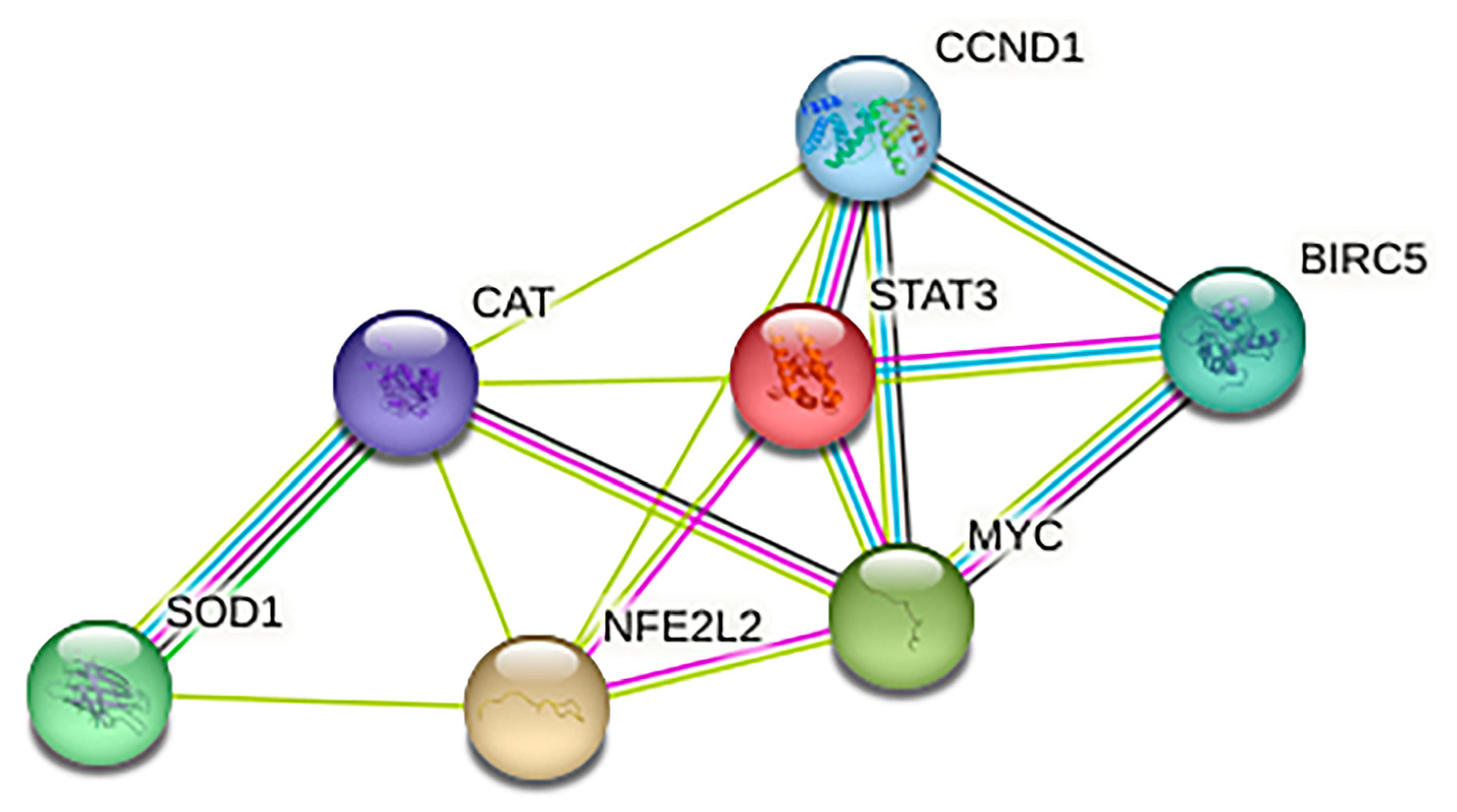
2.2. NFE2L2 and STAT3 Sustain Each Other and Converge in the Regulation of Several Targets
2.3. HSPs and p62/SQSTM1 Are Involved in the NFE2L2 and STAT3 Interplay
2.4. STAT3 and NFE2L2 Concomitant Inhibition More Efficiently Reduces ATM and Molecules Belonging to Both HR and NHEJ DNA Repair Pathways by Downregulating HSPB1 and HSP90A
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Treatments
4.2. MTT Assay
4.3. Cell Assay Viability
4.4. Protein Synthesis Assay—Surface Sensing of Translation (SUnSET)
4.5. Western Blot Analysis
4.6. Knockdown of NFE2L2, STAT3 and p62/SQSTM1 by Small Interfering RNA (siRNA)
4.7. Densitometric Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef]
- Naidu, S.D.; Kostov, R.V.; Dinkova-Kostova, A.T. Transcription factors Hsf1 and Nrf2 engage in crosstalk for cytoprotection. Trends Pharmacol. Sci. 2015, 36, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Ghosh, S.; Mandal, S.; Sau, S.; Pal, M. NRF2 transcriptionally activates the heat shock factor 1 promoter under oxidative stress and affects survival and migration potential of MCF7 cells. J. Biol. Chem. 2018, 293, 19303–19316. [Google Scholar] [CrossRef] [PubMed]
- Jego, G.; Hermetet, F.; Girodon, F.; Garrido, C. Chaperoning STAT3/5 by Heat Shock Proteins: Interest of Their Targeting in Cancer Therapy. Cancers 2019, 12, 21. [Google Scholar] [CrossRef]
- Jego, G.; Hazoume, A.; Seigneuric, R.; Garrido, C. Targeting heat shock proteins in cancer. Cancer Lett. 2013, 332, 275–285. [Google Scholar] [CrossRef]
- Matozaki, M.; Saito, Y.; Yasutake, R.; Munira, S.; Kaibori, Y.; Yukawa, A.; Tada, M.; Nakayama, Y. Involvement of Stat3 phosphorylation in mild heat shock-induced thermotolerance. Exp. Cell Res. 2019, 377, 67–74. [Google Scholar] [CrossRef]
- Gonnella, R.; Arena, A.; Zarrella, R.; Gilardini Montani, M.S.; Santarelli, R.; Cirone, M. HSPs/STAT3 Interplay Sustains DDR and Promotes Cytokine Release by Primary Effusion Lymphoma Cells. Int. J. Mol. Sci. 2023, 24, 3933. [Google Scholar] [CrossRef]
- McHugh, D.; Caduff, N.; Barros, M.H.M.; Ramer, P.C.; Raykova, A.; Murer, A.; Landtwing, V.; Quast, I.; Styles, C.T.; Spohn, M.; et al. Persistent KSHV Infection Increases EBV-Associated Tumor Formation In Vivo via Enhanced EBV Lytic Gene Expression. Cell Host Microbe 2017, 22, 61–73.e67. [Google Scholar] [CrossRef]
- Santarelli, R.; Carillo, V.; Romeo, M.A.; Gaeta, A.; Nazzari, C.; Gonnella, R.; Granato, M.; D’Orazi, G.; Faggioni, A.; Cirone, M. STAT3 phosphorylation affects p53/p21 axis and KSHV lytic cycle activation. Virology 2019, 528, 137–143. [Google Scholar] [CrossRef]
- Santarelli, R.; Gonnella, R.; Di Giovenale, G.; Cuomo, L.; Capobianchi, A.; Granato, M.; Gentile, G.; Faggioni, A.; Cirone, M. STAT3 activation by KSHV correlates with IL-10, IL-6 and IL-23 release and an autophagic block in dendritic cells. Sci. Rep. 2014, 4, 4241. [Google Scholar] [CrossRef]
- Carpenter, R.L.; Lo, H.W. STAT3 Target Genes Relevant to Human Cancers. Cancers 2014, 6, 897–925. [Google Scholar] [CrossRef]
- Arena, A.; Romeo, M.A.; Benedetti, R.; Gilardini Montani, M.S.; Santarelli, R.; Gonnella, R.; D’Orazi, G.; Cirone, M. NRF2 and STAT3: Friends or foes in carcinogenesis? Discov. Oncol. 2023, 14, 37. [Google Scholar] [CrossRef]
- You, L.; Wang, Z.; Li, H.; Shou, J.; Jing, Z.; Xie, J.; Sui, X.; Pan, H.; Han, W. The role of STAT3 in autophagy. Autophagy 2015, 11, 729–739. [Google Scholar] [CrossRef]
- Jiang, T.; Harder, B.; Rojo de la Vega, M.; Wong, P.K.; Chapman, E.; Zhang, D.D. p62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. 2015, 88, 199–204. [Google Scholar] [CrossRef]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. Regulation of the Keap1–Nrf2 pathway by p62/SQSTM1. Curr. Opin. Toxicol. 2016, 1, 54–61. [Google Scholar] [CrossRef]
- Granato, M.; Chiozzi, B.; Filardi, M.R.; Lotti, L.V.; Di Renzo, L.; Faggioni, A.; Cirone, M. Tyrosine kinase inhibitor tyrphostin AG490 triggers both apoptosis and autophagy by reducing HSF1 and Mcl-1 in PEL cells. Cancer Lett. 2015, 366, 191–197. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Sottile, M.L.; Nadin, S.B. Heat shock proteins and DNA repair mechanisms: An updated overview. Cell Stress Chaperones 2018, 23, 303–315. [Google Scholar] [CrossRef]
- Di Veroli, G.Y.; Fornari, C.; Wang, D.; Mollard, S.; Bramhall, J.L.; Richards, F.M.; Jodrell, D.I. Combenefit: An interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016, 32, 2866–2868. [Google Scholar] [CrossRef]
- Yoon, M.S. mTOR as a Key Regulator in Maintaining Skeletal Muscle Mass. Front. Physiol. 2017, 8, 788. [Google Scholar] [CrossRef]
- Wang, L.; Proud, C.G. Ras/Erk signaling is essential for activation of protein synthesis by Gq protein-coupled receptor agonists in adult cardiomyocytes. Circ. Res. 2002, 91, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Cirone, M.; D’Orazi, G. NRF2 in Cancer: Cross-Talk with Oncogenic Pathways and Involvement in Gammaherpesvirus-Driven Carcinogenesis. Int. J. Mol. Sci. 2022, 24, 595. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Feldman, G.M.; Tosato, G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood 2003, 101, 1535–1542. [Google Scholar] [CrossRef] [PubMed]
- Kanda, N.; Seno, H.; Konda, Y.; Marusawa, H.; Kanai, M.; Nakajima, T.; Kawashima, T.; Nanakin, A.; Sawabu, T.; Uenoyama, Y.; et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene 2004, 23, 4921–4929. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Gasperini, P.; Sakakibara, S.; Tosato, G. Contribution of viral and cellular cytokines to Kaposi’s sarcoma-associated herpesvirus pathogenesis. J. Leukoc. Biol. 2008, 84, 994–1000. [Google Scholar] [CrossRef]
- Prahallad, A.; Bernards, R. Opportunities and challenges provided by crosstalk between signalling pathways in cancer. Oncogene 2016, 35, 1073–1079. [Google Scholar] [CrossRef]
- Granato, M.; Rizzello, C.; Gilardini Montani, M.S.; Cuomo, L.; Vitillo, M.; Santarelli, R.; Gonnella, R.; D’Orazi, G.; Faggioni, A.; Cirone, M. Quercetin induces apoptosis and autophagy in primary effusion lymphoma cells by inhibiting PI3K/AKT/mTOR and STAT3 signaling pathways. J. Nutr. Biochem. 2017, 41, 124–136. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, H.; Wang, M.; Wang, R.; Yi, G.; Zhang, M.; Chen, R. Role of STAT3 and NRF2 in Tumors: Potential Targets for Antitumor Therapy. Molecules 2022, 27, 8768. [Google Scholar] [CrossRef]
- Li, D.; Hong, X.; Zhao, F.; Ci, X.; Zhang, S. Targeting Nrf2 may reverse the drug resistance in ovarian cancer. Cancer Cell Int. 2021, 21, 116. [Google Scholar] [CrossRef]
- Garufi, A.; Pistritto, G.; D’Orazi, V.; Cirone, M.; D’Orazi, G. The Impact of NRF2 Inhibition on Drug-Induced Colon Cancer Cell Death and p53 Activity: A Pilot Study. Biomolecules 2022, 12, 461. [Google Scholar] [CrossRef]
- Gjyshi, O.; Roy, A.; Dutta, S.; Veettil, M.V.; Dutta, D.; Chandran, B. Activated Nrf2 Interacts with Kaposi’s Sarcoma-Associated Herpesvirus Latency Protein LANA-1 and Host Protein KAP1 To Mediate Global Lytic Gene Repression. J. Virol. 2015, 89, 7874–7892. [Google Scholar] [CrossRef]
- Granato, M.; Gilardini Montani, M.S.; Angiolillo, C.; D’Orazi, G.; Faggioni, A.; Cirone, M. Cytotoxic Drugs Activate KSHV Lytic Cycle in Latently Infected PEL Cells by Inducing a Moderate ROS Increase Controlled by HSF1, NRF2 and p62/SQSTM1. Viruses 2018, 11, 8. [Google Scholar] [CrossRef]
- Gonnella, R.; Zarrella, R.; Santarelli, R.; Germano, C.A.; Gilardini Montani, M.S.; Cirone, M. Mechanisms of Sensitivity and Resistance of Primary Effusion Lymphoma to Dimethyl Fumarate (DMF). Int. J. Mol. Sci. 2022, 23, 6773. [Google Scholar] [CrossRef]
- Aggarwal, V.; Tuli, H.S.; Varol, A.; Thakral, F.; Yerer, M.B.; Sak, K.; Varol, M.; Jain, A.; Khan, M.A.; Sethi, G. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules 2019, 9, 735. [Google Scholar] [CrossRef]
- Perillo, B.; Tramontano, A.; Pezone, A.; Migliaccio, A. LSD1: More than demethylation of histone lysine residues. Exp. Mol. Med. 2020, 52, 1936–1947. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arena, A.; Di Crosta, M.; Gonnella, R.; Zarrella, R.; Romeo, M.A.; Benedetti, R.; Gilardini Montani, M.S.; Santarelli, R.; D’Orazi, G.; Cirone, M. NFE2L2 and STAT3 Converge on Common Targets to Promote Survival of Primary Lymphoma Cells. Int. J. Mol. Sci. 2023, 24, 11598. https://doi.org/10.3390/ijms241411598
Arena A, Di Crosta M, Gonnella R, Zarrella R, Romeo MA, Benedetti R, Gilardini Montani MS, Santarelli R, D’Orazi G, Cirone M. NFE2L2 and STAT3 Converge on Common Targets to Promote Survival of Primary Lymphoma Cells. International Journal of Molecular Sciences. 2023; 24(14):11598. https://doi.org/10.3390/ijms241411598
Chicago/Turabian StyleArena, Andrea, Michele Di Crosta, Roberta Gonnella, Roberta Zarrella, Maria Anele Romeo, Rossella Benedetti, Maria Saveria Gilardini Montani, Roberta Santarelli, Gabriella D’Orazi, and Mara Cirone. 2023. "NFE2L2 and STAT3 Converge on Common Targets to Promote Survival of Primary Lymphoma Cells" International Journal of Molecular Sciences 24, no. 14: 11598. https://doi.org/10.3390/ijms241411598
APA StyleArena, A., Di Crosta, M., Gonnella, R., Zarrella, R., Romeo, M. A., Benedetti, R., Gilardini Montani, M. S., Santarelli, R., D’Orazi, G., & Cirone, M. (2023). NFE2L2 and STAT3 Converge on Common Targets to Promote Survival of Primary Lymphoma Cells. International Journal of Molecular Sciences, 24(14), 11598. https://doi.org/10.3390/ijms241411598






