T Cells in Colorectal Cancer: Unravelling the Function of Different T Cell Subsets in the Tumor Microenvironment
Abstract
:1. Introduction
1.1. Immune Cell Infiltration into the Consensus Molecular CRC Subtypes
- (I)
- Consensus Molecular Subtype (CMS) 1 is characterized by MSI and a hypermutated profile of mismatch repair genes and BRAF genes. Most CRCs with microsatellite instability are in the CMS1 subtype [9]. Due to microsatellite instability, CMS1 tumors have a high amount of neo-antigens that are not expressed in healthy tissues. CMS1 patients have an immune infiltrate consisting of T helper 1 (Th1) cells, natural killer (NK) cells, CD8+ cytotoxic T lymphocytes (CTL) and dendritic cells (DCs). About 14% of CRCs belong to the CMS1 subtype.
- (II)
- The CMS2 subtype includes CRCs with higher chromosomal instability (CIN) and microsatellite-stable tumors. CMS2 tumors lack DCs in the tumor microenvironment, indicating that the CMS2 subtype is only poorly immunogenic, with few tumor-infiltrating immune cells. CMS2 patients are characterized by the infiltration of a few naïve CD4+ T cells and resting NK cells. The WNT signaling pathway correlates with T cell exclusion in CRCs [10]. Approximately 37% of CRCs belong to CMS2.
- (III)
- The CMS3 subtype is the metabolic subtype, characterized by chromosomal instability and a higher level of KRAS mutations compared with other CMS phenotypes. Like CMS2, CMS3 shows only poor immune infiltration and no immune regulatory cytokines. Tumor-infiltrating lymphocytes (TILs) consist of naïve T cells and T helper 17 (Th17) cells. About 13% belong to the CMS3 subtype.
- (IV)
- The CMS4 subtype shows a high expression of genes specific for immunosuppressive TGF-β signaling and of genes specific to regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs) [11]. CMS4 is also characterized by an upregulated expression of genes that encode chemokines that attract myeloid cells and T cells that produce interleukin 17 (IL-17), which are known to enhance carcinogenesis [12]. The CMS4 phenotype shows high levels of infiltrating macrophages and stromal cells. Approximately 23% of CRC tumors fall into CMS4. Although CMS4 patients show high levels of leukocyte infiltration, patients with CMS4 tumors have the worst prognosis of the four subtypes.

1.2. Differences between Cold, Altered-Excluded, Altered-Immunosuppressed and Hot CRCs
- (1)
- The optimal immunity with a high immunoscore and hot, “T cell inflamed” tumors.
- (2)
- The altered immunity with an intermediate immunoscore and an immunosuppressed phenotype.
- (3)
- The exclusion phenotype with an intermediate immunoscore and a high density of T cells only at the margin of the tumor.
- (4)
- The phenotype with a low immunoscore and cold, “non-T cell inflamed” tumors in which other immune cells, such as myeloid cells, can be observed.
1.3. T Cell Subsets in CRC
1.3.1. Conventional αβ CD4-Positive T Cells
1.3.2. Conventional αβ CD8-Positive T Cells
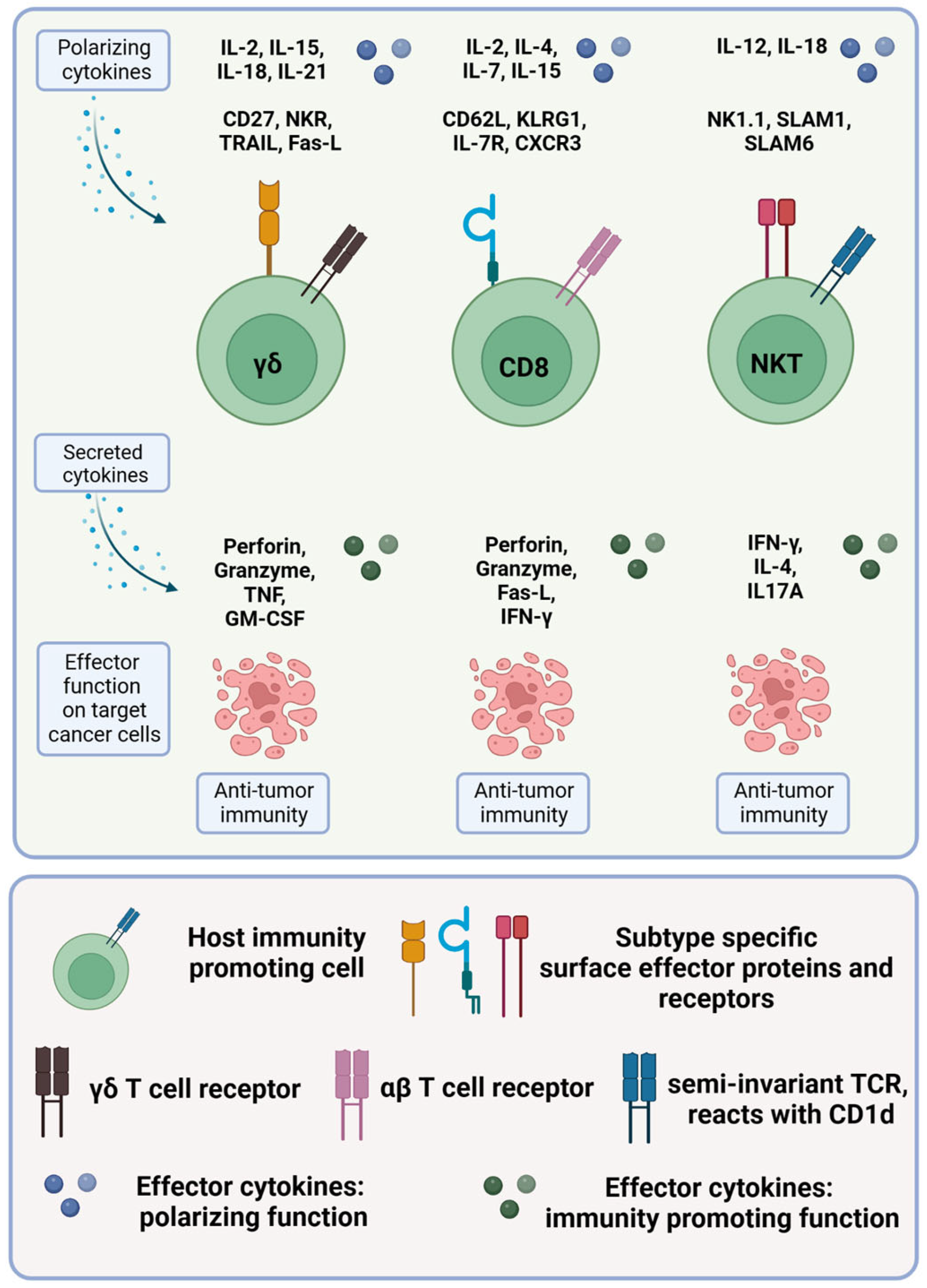
1.3.3. γδ T Cells
1.3.4. NKT Cells
| Name | Surface Marker | Polarizing Cytokines | Function | Secreted Cytokines | Reference |
|---|---|---|---|---|---|
| CTL | CD8+ CCR4+ CXCR3+ IFNγR+ αβ TCR | IL-2, IL-7, IL-4, IL-15 | Destruction of tumor cells by cytotoxic molecules, e.g., perforin, granzymes, granulysin and Fas ligand. | IFN-γ, TNF-α, IL-2 | [142,143] |
| iNKT | CXCR3+, NK1.1+ Vα24-Jα18 TCR | IL-12, IL-18 | Anti-tumoral functions mainly mediated by IFN-γ, TNF-α and downstream activation of NK cells and CTLs. Destruction of tumor cells by cytotoxic molecules. | IFN-γ, TNF-α, IL-2, IL-4, IL-5, IL-3, IL-13, IL-10, IL-17, IL-21, IL-22 | [132,136] |
| Type II NKT | αβ TCR | Pro-tumoral functions mediated by IL-4 and IL-13. | IL-4, IL-13 | [132,136] | |
| γδ T1 | γδ TCR CD27+ CD122+ CD45RB+ Fas-L+ | IL-2, IL-15 | Anti-tumoral functions mainly mediated by IFN-γ, TNF-α and downstream activation of NK cells and CTLs. Destruction of tumor cells by cytotoxic molecules. | IFN-γ, TNF-α, IL-2 | [119,144] |
| γδ T17 | γδ TCR CCR6+ SCART-2+ | IL-23, IL-1β, IL-6, TGF-β | Regulator of epithelial barrier integrity. | IL-17 | [145,146] |
| γδ T2 | γδ TCR | IL-4 | Maintain tolerance to self-antigens, prevent the induction of auto-antibodies. Suppression of effector T cell-mediated immune responses in colorectal cancer. | IL-4, TGF-β | [147] |
2. Tumor Initiation: Recognition of Colon Cancer Cells by T Cells
3. Tumor Progression: When Cancer Cells Escape T Cell Recognition
3.1. T Cell Exclusion
3.2. Dysfunctional T Cells
3.3. Tolerance, Anergy and Ignorance
3.4. Exhaustion
4. Immunotherapeutic Interventions
4.1. Immunotherapies with Immune Checkpoint Inhibitors
- (i)
- Ipilimumab, an antibody that binds CTLA-4, for melanoma treatment [191].
- (ii)
- Nivolumab, an antibody that binds PD-1, for melanoma treatment [192].
- (iii)
- Avelumab, an antibody against PD-L1 targeting the PD-L1/PD-1 pathway, for metastatic Merkel cell carcinoma [193].
- (iv)
- Atezolizumab, an engineered humanized immunoglobulin G1 antibody against PD-L1, for metastatic urothelial carcinoma [194].
- (v)
- Urelumab, an anti-CD137 antibody [195], for the treatment of different solid cancers, such as bladder cancer, renal cell cancer, colorectal cancer, gliosarcoma, etc.
- (vi)
- Relatlimab, an anti-lymphocyte activation gene 3 (LAG-3) antibody in combination with nivolumab, for the treatment of resectable melanoma [196].
- (vii)
- Lirilumab, an antibody that blocks the killer immunoglobulin-like receptor (KIR)/human leukocyte antigen-C (HLA-C) interaction, for the treatment of patients suffering from myeloid malignancies [197].
4.1.1. Checkpoint Inhibitor Blockade in dMMR-MSI CRCs
4.1.2. Checkpoint Inhibitor Blockade in pMMR-MSS CRCs
4.2. Cellular Immunotherapies
4.2.1. Immunotherapies with CAR T Cells in CRC
- (i)
- NCT03018405 (THINK) using CAR T cells.
- (ii)
- NCT03692429 (alloSHRINK) using CAR T cells after chemotherapy.
- (iii)
- NCT03370198 (LINK) using CAR T cells by hepatic transarterial infusion.
- (iv)
- NCT03310008 (SHRINK) using CAR T cells plus FOLFOX (folinic acid, fluorouracil and oxaliplatin).
- (v)
- NCT04107142 (CTM-N2D-101) using CAR γδ T cells.
4.2.2. Immunotherapies with γδ T Cells in CRC
5. Therapeutic Options: Turning “Cold” CRCs into “Hot” CRCs
5.1. Possibilities to Increase the Infiltration of T Cells
5.1.1. Oncogenic Pathway Inhibitors
5.1.2. Anti-Angiogenic Therapy
5.1.3. TGF-β Inhibitors
5.1.4. CXCR4 Inhibitors
5.2. Enhancement of Neo-Antigens
5.3. Better Priming and Activation of T Cells
5.3.1. Oncolytic Viruses (OVs)
5.3.2. Chemotherapy and Radiotherapy
5.3.3. Cancer Vaccines
5.4. Enhancement of T Cell Trafficking to the Tumor
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACT | Adoptive Cell Transfer |
| ALL | Acute Lymphoblastic Leukemia |
| APC | Antigen-Presenting Cell |
| ATP | Adenosine Triphosphate |
| CAF | Cancer-Associated Fibroblast |
| CAR | Chimeric Antigen Receptor |
| CDK | Cyclin-Dependent Kinase |
| CD4 | Cluster of Differentiation Antigen 4 |
| CD8 | Cluster of Differentiation Antigen 8 |
| CEA | Carcino-Embryonic Antigen |
| CIN | Chromosomal Instability |
| CMS | Consensus Molecular Subtype |
| CRC | Colorectal Cancer |
| CRT | Calreticulin |
| CTL | Cytotoxic T Lymphocyte |
| CTLA-4 | Cytotoxic T-Lymphocyte-Associated Protein 4 |
| DC | Dendritic Cell |
| dMMR | Mismatch Repair deficient |
| DAMP | Damage Associated Molecular Pattern |
| DLN | Draining Lymphnode |
| DNMTi | DNA Methyltransferase Inhibitor |
| EGF | Epidermal Growth Factor |
| ERV | Endogenous Retrovirus |
| FDA | Food and Drug Administration |
| FOXP3 | Forkhead Box P3 |
| GZMB | Granzyme B |
| HMGB | High Mobility Group Box |
| ICB | Checkpoint Inhibitor Blockade |
| ICD | Immunogenic Cell Death |
| iCMS | intrinsic Consensus Molecular Subtype |
| ICR | Immunologic Constant of Rejection |
| IDO-1 | Indoleamine 2-3 Dioxygenase-1 |
| IEL | Intraepithelial Lymphocyte |
| IFN-γ | Interferon-Gamma |
| IL-17 | Interleukin-17 |
| KRAS | Kirsten Rat Sarcoma Virus Oncogene |
| LTR | Long-Terminal Repeats |
| mCRC | Metastatic Colorectal Cancer |
| mOS | Median Overall Survival |
| MDSC | Myeloid-Derived Suppressor Cell |
| MHC | Major Histocompatibility Complex |
| MHC I | Major Histocompatibility Complex 1 |
| miRNA | Micro-RNA |
| MMR | Mismatch Repair |
| MSI | Microsatellite Instability |
| MSS | Microsatellite stable |
| ncRNA | Non-Coding RNA |
| NF | Nonfibrosis |
| NK | Natural Killer Cell |
| OV | Oncolytic Virus |
| PDAC | Pancreatic Ductal Adenocarcinoma |
| pMMR | Mismatch Repair proficient |
| PBMC | Peripheral Blood Mononuclear Cell |
| PDAC | Pancreatic Ductal Adenocarcinoma |
| PD-1 | Programmed Death 1 |
| RB | Retinoblastoma |
| SBRT | Stereotactic Body Radiotherapy |
| SITC | Society for Immunotherapy of Cancer |
| STAT | Signal transducer and activator of transcription |
| TAM | Tumor-Associated Macrophage |
| Tfh | Follicular helper cell |
| TGF-β | Transforming Growth Factor Beta |
| TGFBR | TGF-β Receptor |
| TIM-3 | Mucin-containing protein 3 |
| TIL | Tumor-Infiltrating Lymphocyte |
| TME | Tumor Microenvironment |
| TNF-α | Tumor Necrosis Factor-alpha |
| Treg | T Regulatory Cell |
| VEGF | Vascular Endothelial Growth Factor |
| WNT | Wingless Pathway |
| IMGT Database | |
| Atezolizumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=526 |
| Avelumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=512 |
| Bevacizumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=24 |
| Ipilimumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=180 |
| Lirilumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=423 |
| Nivolumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=424 |
| Pembrolizumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=472 |
| Ramucirumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=295 |
| Relatlimab | https://www.imgt.org/mAb-DB/mAbcard?AbId=781 |
| Tocilizumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=96 |
| Urelumab | https://www.imgt.org/mAb-DB/mAbcard?AbId=373 |
References
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.M.; Weaver, C. Janeway’s Immunobiology; Ninth International Student Edition; Garland Science, Taylor & Francis: New York, NY, USA, 2017. [Google Scholar]
- Finn, O.J. Cancer immunology. N. Engl. J. Med. 2008, 358, 2704–2715. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A., Jr. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef]
- Roth, A.D.; Tejpar, S.; Delorenzi, M.; Yan, P.; Fiocca, R.; Klingbiel, D.; Dietrich, D.; Biesmans, B.; Bodoky, G.; Barone, C.; et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J. Clin. Oncol. 2010, 28, 466–474. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Fridman, W.H.; Miller, I.; Sautès-Fridman, C.; Byrne, A.T. Therapeutic Targeting of the Colorectal Tumor Stroma. Gastroenterology 2020, 158, 303–321. [Google Scholar] [CrossRef]
- Luke, J.J.; Bao, R.; Sweis, R.F.; Spranger, S.; Gajewski, T.F. WNT/β-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin. Cancer Res. 2019, 25, 3074–3083. [Google Scholar] [CrossRef] [Green Version]
- Angelova, M.; Charoentong, P.; Hackl, H.; Fischer, M.L.; Snajder, R.; Krogsdam, A.M.; Waldner, M.J.; Bindea, G.; Mlecnik, B.; Galon, J.; et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015, 16, 64. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Wang, K.; Mucida, D.; Stewart, C.A.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.Y.; Osterreicher, C.H.; Hung, K.E.; et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012, 491, 254–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joanito, I.; Wirapati, P.; Zhao, N.; Nawaz, Z.; Yeo, G.; Lee, F.; Eng, C.L.P.; Macalinao, D.C.; Kahraman, M.; Srinivasan, H.; et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 2022, 54, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Roelands, J.; Kuppen, P.J.K.; Ahmed, E.I.; Mall, R.; Masoodi, T.; Singh, P.; Monaco, G.; Raynaud, C.; de Miranda, N.F.; Ferraro, L.; et al. An integrated tumor, immune and microbiome atlas of colon cancer. Nat. Med. 2023, 29, 1273–1286. [Google Scholar] [CrossRef]
- Wang, E.; Worschech, A.; Marincola, F.M. The immunologic constant of rejection. Trends Immunol. 2008, 29, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crino, L.; Eberhardt, W.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, C.; Thomas, L.; Bondarenko, I.; O’Day, S.; Weber, J.; Garbe, C.; Lebbe, C.; Baurain, J.-F.; Testori, A.; Grob, J.-J.; et al. Ipilimumab plus Dacarbazine for Previously Untreated Metastatic Melanoma. N. Engl. J. Med. 2011, 364, 2517–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, J.; Li, S.; Zhu, Z.; Liu, Q.; Zhang, R.; Yang, Y.; Li, X. Exploring immunotherapy in colorectal cancer. J. Hematol. Oncol. 2022, 15, 95. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [Green Version]
- Galon, J.; Bruni, D. The Role of the Immune Infiltrate in Distinct Cancer Types and Its Clinical Implications: Lymphocytic Infiltration in Colorectal Cancer. Cancer Treat. Res. 2020, 180, 197–211. [Google Scholar]
- Galon, J.; Bruni, D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity 2020, 52, 55–81. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheon, H.; Borden, E.C.; Stark, G.R. Interferons and Their Stimulated Genes in the Tumor Microenvironment. Semin. Oncol. 2014, 41, 156–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braumüller, H.; Mauerer, B.; Andris, J.; Berlin, C.; Wieder, T.; Kesselring, R. The Cytokine Network in Colorectal Cancer: Implications for New Treatment Strategies. Cells 2022, 12, 138. [Google Scholar] [CrossRef]
- Braoudaki, M.; Ahmad, M.S.; Mustafov, D.; Seriah, S.; Siddiqui, M.N.; Siddiqui, S.S. Chemokines and chemokine receptors in colorectal cancer; multifarious roles and clinical impact. Semin. Cancer Biol. 2022, 86, 436–449. [Google Scholar] [CrossRef]
- Bule, P.; Aguiar, S.I.; Aires-Da-Silva, F.; Dias, J.N.R. Chemokine-Directed Tumor Microenvironment Modulation in Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 9804. [Google Scholar] [CrossRef] [PubMed]
- Zumwalt, T.J.; Arnold, M.; Goel, A.; Boland, C.R. Active secretion of CXCL10 and CCL5 from colorectal cancer microenvironments associates with GranzymeB+ CD8+ T-cell infiltration. Oncotarget 2015, 6, 2981–2991. [Google Scholar] [CrossRef] [Green Version]
- Becht, E.; de Reynies, A.; Giraldo, N.A.; Pilati, C.; Buttard, B.; Lacroix, L.; Selves, J.; Sautes-Fridman, C.; Laurent-Puig, P.; Fridman, W.H. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin. Cancer Res. 2016, 22, 4057–4066. [Google Scholar] [CrossRef] [Green Version]
- Saito, T.; Nishikawa, H.; Wada, H.; Nagano, Y.; Sugiyama, D.; Atarashi, K.; Maeda, Y.; Hamaguchi, M.; Ohkura, N.; Sato, E.; et al. Two FOXP3+CD4+ T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 2016, 22, 679–684. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Dieu-Nosjean, M.-C.; Sautès-Fridman, C.; Fridman, W.H. Cancer immune contexture and immunotherapy. Curr. Opin. Immunol. 2016, 39, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Atreya, I.; Neurath, M.F. How the Tumor Micromilieu Modulates the Recruitment and Activation of Colorectal Cancer-Infiltrating Lymphocytes. Biomedicines 2022, 10, 2940. [Google Scholar] [CrossRef]
- Hurtado, C.G.; Wan, F.; Housseau, F.; Sears, C.L. Roles for Interleukin 17 and Adaptive Immunity in Pathogenesis of Colorectal Cancer. Gastroenterology 2018, 155, 1706–1715. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Vadgama, J.V.; Wang, P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun. Signal. 2020, 18, 82. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt signaling in colorectal cancer: Pathogenic role and therapeutic target. Mol. Cancer 2022, 21, 144. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Fearon, D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015, 348, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Mlecnik, B.; Bifulco, C.; Bindea, G.; Marliot, F.; Lugli, A.; Lee, J.J.; Zlobec, I.; Rau, T.T.; Berger, M.D.; Nagtegaal, I.D.; et al. Multicenter International Society for Immunotherapy of Cancer Study of the Consensus Immunoscore for the Prediction of Survival and Response to Chemotherapy in Stage III Colon Cancer. J. Clin. Oncol. 2020, 38, 3638–3651. [Google Scholar] [CrossRef]
- Mlecnik, B.; Lugli, A.; Bindea, G.; Marliot, F.; Bifulco, C.; Lee, J.-K.J.; Zlobec, I.; Rau, T.T.; Berger, M.D.; Nagtegaal, I.D.; et al. Multicenter International Study of the Consensus Immunoscore for the Prediction of Relapse and Survival in Early-Stage Colon Cancer. Cancers 2023, 15, 418. [Google Scholar] [CrossRef]
- Oliveira, G.; Wu, C.J. Dynamics and specificities of T cells in cancer immunotherapy. Nat. Rev. Cancer 2023, 23, 295–316. [Google Scholar] [CrossRef]
- Geginat, J.; Paroni, M.; Maglie, S.; Alfen, J.S.; Kastirr, I.; Gruarin, P.; De Simone, M.; Pagani, M.; Abrignani, S. Plasticity of Human CD4 T Cell Subsets. Front. Immunol. 2014, 5, 630. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Chong, M.M.W.; Littman, D.R. Plasticity of CD4+ T Cell Lineage Differentiation. Immunity 2009, 30, 646–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaumik, S.; Basu, R. Cellular and Molecular Dynamics of Th17 Differentiation and its Developmental Plasticity in the Intestinal Immune Response. Front. Immunol. 2017, 8, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speiser, D.E.; Chijioke, O.; Schaeuble, K.; Munz, C. CD4+ T cells in cancer. Nat. Cancer 2023, 4, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, A.; Saito, T. CD4 CTL, a Cytotoxic Subset of CD4+ T Cells, Their Differentiation and Function. Front. Immunol. 2017, 8, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, N.; Ueno, H. Regulation of human helper T cell subset differentiation by cytokines. Curr. Opin. Immunol. 2015, 34, 130–136. [Google Scholar] [CrossRef] [Green Version]
- Braumuller, H.; Wieder, T.; Brenner, E.; Assmann, S.; Hahn, M.; Alkhaled, M.; Schilbach, K.; Essmann, F.; Kneilling, M.; Griessinger, C.; et al. T-helper-1-cell cytokines drive cancer into senescence. Nature 2013, 494, 361–365. [Google Scholar] [CrossRef] [Green Version]
- Brenner, E.; Schörg, B.F.; Ahmetlić, F.; Wieder, T.; Hilke, F.J.; Simon, N.; Schroeder, C.; Demidov, G.; Riedel, T.; Fehrenbacher, B.; et al. Cancer immune control needs senescence induction by interferon-dependent cell cycle regulator pathways in tumours. Nat. Commun. 2020, 11, 1335. [Google Scholar] [CrossRef] [Green Version]
- Geginat, J.; Paroni, M.; Facciotti, F.; Gruarin, P.; Kastirr, I.; Caprioli, F.; Pagani, M.; Abrignani, S. The CD4-centered universe of human T cell subsets. Semin. Immunol. 2013, 25, 252–262. [Google Scholar] [CrossRef]
- DiToro, D.; Basu, R. Emerging Complexity in CD4+ T Lineage Programming and Its Implications in Colorectal Cancer. Front. Immunol. 2021, 12, 694833. [Google Scholar] [CrossRef]
- Richardson, J.R.; Schöllhorn, A.; Gouttefangeas, C.; Schuhmacher, J. CD4+ T Cells: Multitasking Cells in the Duty of Cancer Immunotherapy. Cancers 2021, 13, 596. [Google Scholar] [CrossRef]
- Tanaka, A.; Sakaguchi, S. Targeting Treg cells in cancer immunotherapy. Eur. J. Immunol. 2019, 49, 1140–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Craft, J. T follicular helper cell heterogeneity: Time, space, and function. Immunol. Rev. 2019, 288, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Harbour, S.N.; Maynard, C.L.; Zindl, C.L.; Schoeb, T.R.; Weaver, C.T. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc. Natl. Acad. Sci. USA 2015, 112, 7061–7066. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Duarte, J.H.; Veldhoen, M.; Hornsby, E.; Li, Y.; Cua, D.J.; Ahlfors, H.; Wilhelm, C.; Tolaini, M.; Menzel, U.; et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 2011, 12, 255–263. [Google Scholar] [CrossRef]
- Wu, T.; Cui, L.; Liang, Z.; Liu, C.; Liu, Y.; Li, J. Elevated serum IL-22 levels correlate with chemoresistant condition of colorectal cancer. Clin. Immunol. 2013, 147, 38–39. [Google Scholar] [CrossRef]
- Huber, S.; Gagliani, N.; Zenewicz, L.A.; Huber, F.J.; Bosurgi, L.; Hu, B.; Hedl, M.; Zhang, W.; O’connor, W.; Murphy, A.J.; et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 2012, 491, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Kirchberger, S.; Royston, D.J.; Boulard, O.; Thornton, E.; Franchini, F.; Szabady, R.L.; Harrison, O.; Powrie, F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J. Exp. Med. 2013, 210, 917–931. [Google Scholar] [CrossRef]
- Perez, L.G.; Kempski, J.; McGee, H.M.; Pelzcar, P.; Agalioti, T.; Giannou, A.; Konczalla, L.; Brockmann, L.; Wahib, R.; Xu, H.; et al. TGF-β signaling in Th17 cells promotes IL-22 production and colitis-associated colon cancer. Nat. Commun. 2020, 11, 2608. [Google Scholar] [CrossRef]
- Sawant, D.V.; Vignali, D.A. Once a Treg, always a Treg? Immunol. Rev. 2014, 259, 173–191. [Google Scholar] [CrossRef]
- Rentschler, M.; Braumüller, H.; Briquez, P.S.; Wieder, T. Cytokine-Induced Senescence in the Tumor Microenvironment and Its Effects on Anti-Tumor Immune Responses. Cancers 2022, 14, 1364. [Google Scholar] [CrossRef]
- Bruni, D.; Angell, H.K.; Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef] [PubMed]
- Muller-Hermelink, N.; Braumuller, H.; Pichler, B.; Wieder, T.; Mailhammer, R.; Schaak, K.; Ghoreschi, K.; Yazdi, A.; Haubner, R.; Sander, C.A.; et al. TNFR1 signaling and IFN-γ signaling determine whether T cells induce tumor dormancy or promote multistage carcinogenesis. Cancer Cell 2008, 13, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wozniakova, M.; Skarda, J.; Raska, M. The Role of Tumor Microenvironment and Immune Response in Colorectal Cancer Development and Prognosis. Pathol. Oncol. Res. 2022, 28, 1610502. [Google Scholar] [CrossRef] [PubMed]
- Carretero, R.; Sektioglu, I.M.; Garbi, N.; Salgado, O.C.; Beckhove, P.; Hammerling, G.J. Eosinophils orchestrate cancer rejection by normalizing tumor vessels and enhancing infiltration of CD8+ T cells. Nat. Immunol. 2015, 16, 609–617. [Google Scholar] [CrossRef]
- Liu, M.; Kuo, F.; Capistrano, K.J.; Kang, D.; Nixon, B.G.; Shi, W.; Chou, C.; Do, M.H.; Stamatiades, E.G.; Gao, S.; et al. TGF-β suppresses type 2 immunity to cancer. Nature 2020, 587, 115–120. [Google Scholar] [CrossRef]
- Chang, S.H. T helper 17 (Th17) cells and interleukin-17 (IL-17) in cancer. Arch. Pharmacal Res. 2019, 42, 549–559. [Google Scholar] [CrossRef]
- Li, J.; Zeng, M.; Yan, K.; Yang, Y.; Li, H.; Xu, X. IL-17 promotes hepatocellular carcinoma through inhibiting apoptosis induced by IFN-γ. Biochem. Biophys. Res. Commun. 2020, 522, 525–531. [Google Scholar] [CrossRef]
- Amicarella, F.; Muraro, M.G.; Hirt, C.; Cremonesi, E.; Padovan, E.; Mele, V.; Governa, V.; Han, J.; Huber, X.; Droeser, R.A.; et al. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut 2017, 66, 692–704. [Google Scholar] [CrossRef] [Green Version]
- De Simone, V.; Pallone, F.; Monteleone, G.; Stolfi, C. Role of TH17 cytokines in the control of colorectal cancer. Oncoimmunology 2013, 2, e26617. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wang, X.; Gupta, A.; Fang, X.; Wang, L.; Zhang, C. Expression of IL-17 with tumor budding as a prognostic marker in oral squamous cell carcinoma. Am. J. Transl. Res. 2019, 11, 1876–1883. [Google Scholar]
- Chen, J.-G.; Xia, J.-C.; Liang, X.-T.; Pan, K.; Wang, W.; Lv, L.; Zhao, J.-J.; Wang, Q.-J.; Li, Y.-Q.; Chen, S.-P.; et al. Intratumoral Expression of IL-17 and Its Prognostic Role in Gastric Adenocarcinoma Patients. Int. J. Biol. Sci. 2011, 7, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Punt, S.; Fleuren, G.J.; Kritikou, E.; Lubberts, E.; Trimbos, J.B.; Jordanova, E.S.; Gorter, A. Angels and demons: Th17 cells represent a beneficial response, while neutrophil IL-17 is associated with poor prognosis in squamous cervical cancer. Oncoimmunology 2015, 4, e984539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duhen, T.; Geiger, R.; Jarrossay, D.; Lanzavecchia, A.; Sallusto, F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 2009, 10, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Pickert, G.; Neufert, C.; Leppkes, M.; Zheng, Y.; Wittkopf, N.; Warntjen, M.; Lehr, H.A.; Hirth, S.; Weigmann, B.; Wirtz, S.; et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 2009, 206, 1465–1472. [Google Scholar] [CrossRef] [PubMed]
- Tosti, N.; Cremonesi, E.; Governa, V.; Basso, C.; Kancherla, V.; Coto-Llerena, M.; Amicarella, F.; Weixler, B.; Daster, S.; Sconocchia, G.; et al. Infiltration by IL22-Producing T Cells Promotes Neutrophil Recruitment and Predicts Favorable Clinical Outcome in Human Colorectal Cancer. Cancer Immunol. Res. 2020, 8, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Revilla, S.A.; Kranenburg, O.; Coffer, P.J. Colorectal Cancer-Infiltrating Regulatory T Cells: Functional Heterogeneity, Metabolic Adaptation, and Therapeutic Targeting. Front. Immunol. 2022, 13, 903564. [Google Scholar] [CrossRef]
- Brunkow, M.E.; Jeffery, E.W.; Hjerrild, K.A.; Paeper, B.; Clark, L.B.; Yasayko, S.-A.; Wilkinson, J.E.; Galas, D.; Ziegler, S.F.; Ramsdell, F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001, 27, 68–73. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Pillars Article: Control of Regulatory T Cell Development by the Transcription Factor Foxp3. J. Immunol. 2017, 198, 981–985. [Google Scholar]
- Sakaguchi, S.; Sakaguchi, N.; Asano, M.; Itoh, M.; Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995, 155, 1151–1164. [Google Scholar] [CrossRef]
- Lenardo, M.J. Interleukin-2 programs mouse αβ T lymphocytes for apoptosis. Nature 1991, 353, 858–861. [Google Scholar] [CrossRef]
- Wang, H.Y.; Wang, R.F. Regulatory T cells and cancer. Curr. Opin. Immunol. 2007, 19, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Shang, B.; Liu, Y.; Jiang, S.J.; Liu, Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: A systematic review and meta-analysis. Sci. Rep. 2015, 5, 15179. [Google Scholar] [CrossRef] [Green Version]
- Nishikawa, H.; Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Curr. Opin. Immunol. 2014, 27, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinicrope, F.A.; Rego, R.L.; Ansell, S.M.; Knutson, K.L.; Foster, N.R.; Sargent, D.J. Intraepithelial effector (CD3+)/regulatory (FoxP3+) T-cell ratio predicts a clinical outcome of human colon carcinoma. Gastroenterology 2009, 137, 1270–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraiva, M.; O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, S.; Schramm, C.; Lehr, H.A.; Mann, A.; Schmitt, S.; Becker, C.; Protschka, M.; Galle, P.R.; Neurath, M.F.; Blessing, M. Cutting edge: TGF-β signaling is required for the in vivo expansion and immunosuppressive capacity of regulatory CD4+CD25+ T cells. J. Immunol. 2004, 173, 6526–6531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collison, L.W.; Pillai, M.R.; Chaturvedi, V.; Vignali, D.A.A. Regulatory T Cell Suppression Is Potentiated by Target T Cells in a Cell Contact, IL-35- and IL-10-Dependent Manner. J. Immunol. 2009, 182, 6121–6128. [Google Scholar] [CrossRef] [Green Version]
- Olguín, J.E.; Medina-Andrade, I.; Rodríguez, T.; Rodríguez-Sosa, M.; Terrazas, L.I. Relevance of Regulatory T Cells during Colorectal Cancer Development. Cancers 2020, 12, 1888. [Google Scholar] [CrossRef]
- Schneider, H.; Valk, E.; Dias, S.d.R.; Wei, B.; Rudd, C.E. CTLA-4 up-regulation of lymphocyte function-associated antigen 1 adhesion and clustering as an alternate basis for coreceptor function. Proc. Natl. Acad. Sci. USA 2005, 102, 12861–12866. [Google Scholar] [CrossRef]
- Gutiérrez-Melo, N.; Baumjohann, D. T follicular helper cells in cancer. Trends Cancer 2023, 9, 309–325. [Google Scholar] [CrossRef]
- Eisenbarth, S.C.; Baumjohann, D.; Craft, J.; Fazilleau, N.; Ma, C.S.; Tangye, S.G.; Vinuesa, C.G.; Linterman, M.A. CD4+ T cells that help B cells—A proposal for uniform nomenclature. Trends Immunol. 2021, 42, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Cao, Y.; Dong, L.; Huang, Y.; Zhang, Z.; Bi, Y.; Liu, G. Regulation of follicular T helper cell differentiation in antitumor immunity. Int. J. Cancer 2022, 153, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Schurch, C.M.; Bhate, S.S.; Barlow, G.L.; Phillips, D.J.; Noti, L.; Zlobec, I.; Chu, P.; Black, S.; Demeter, J.; McIlwain, D.R.; et al. Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front. Cell 2020, 182, 1341–1359.e19. [Google Scholar] [CrossRef] [PubMed]
- Niogret, J.; Berger, H.; Rebe, C.; Mary, R.; Ballot, E.; Truntzer, C.; Thibaudin, M.; Derangere, V.; Hibos, C.; Hampe, L.; et al. Follicular helper-T cells restore CD8+-dependent antitumor immunity and anti-PD-L1/PD-1 efficacy. J. Immunother. Cancer 2021, 9, e002157. [Google Scholar] [CrossRef] [PubMed]
- Hiam-Galvez, K.J.; Allen, B.M.; Spitzer, M.H. Systemic immunity in cancer. Nat. Rev. Cancer 2021, 21, 345–359. [Google Scholar] [CrossRef]
- Bai, Z.; Zhou, Y.; Ye, Z.; Xiong, J.; Lan, H.; Wang, F. Tumor-Infiltrating Lymphocytes in Colorectal Cancer: The Fundamental Indication and Application on Immunotherapy. Front. Immunol. 2021, 12, 808964. [Google Scholar] [CrossRef]
- Duval, A.; Hamelin, R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: Toward a new concept of target genes for instability. Cancer Res. 2002, 62, 2447–2454. [Google Scholar]
- Hamelin, R.; Chalastanis, A.; Colas, C.; El Bchiri, J.; Mercier, D.; Schreurs, A.-S.; Simon, V.; Svrcek, M.; Zaanan, A.; Borie, C.; et al. Clinical and molecular consequences of microsatellite instability in human cancers. Bull. Cancer 2008, 95, 121–132. [Google Scholar]
- Xiao, Y.; Freeman, G.J. The Microsatellite Instable Subset of Colorectal Cancer Is a Particularly Good Candidate for Checkpoint Blockade Immunotherapy. Cancer Discov. 2015, 5, 16–18. [Google Scholar] [CrossRef] [Green Version]
- Talaat, I.M.; Elemam, N.M.; Zaher, S.; Saber-Ayad, M. Checkpoint molecules on infiltrating immune cells in colorectal tumor microenvironment. Front. Med. 2022, 9, 955599. [Google Scholar] [CrossRef]
- Whiteside, T.L.; Demaria, S.; Rodriguez-Ruiz, M.E.; Zarour, H.M.; Melero, I. Emerging Opportunities and Challenges in Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 1845–1855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarour, H.M. Reversing T-cell Dysfunction and Exhaustion in Cancer. Clin. Cancer Res. 2016, 22, 1856–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Vries, N.L.; van de Haar, J.; Veninga, V.; Chalabi, M.; Ijsselsteijn, M.E.; van der Ploeg, M.; van den Bulk, J.; Ruano, D.; van den Berg, J.G.; Haanen, J.B.; et al. γδ T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature 2023, 613, 743–750. [Google Scholar] [CrossRef]
- Suzuki, T.; Hayman, L.; Kilbey, A.; Edwards, J.; Coffelt, S.B. Gut γδ T cells as guardians, disruptors, and instigators of cancer. Immunol. Rev. 2020, 298, 198–217. [Google Scholar] [CrossRef] [PubMed]
- Rock, E.P.; Sibbald, P.R.; Davis, M.M.; Chien, Y.H. CDR3 length in antigen-specific immune receptors. J. Exp. Med. 1994, 179, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Schild, H.; Mavaddat, N.; Litzenberger, C.; Ehrich, E.W.; Davis, M.M.; Bluestone, J.A.; Matis, L.; Draper, R.K.; Chien, Y.H. The nature of major histocompatibility complex recognition by γδ T cells. Cell 1994, 76, 29–37. [Google Scholar] [CrossRef]
- Sebestyen, Z.; Prinz, I.; Déchanet-Merville, J.; Silva-Santos, B.; Kuball, J. Translating gammadelta (γδ) T cells and their receptors into cancer cell therapies. Nat. Rev. Drug Discov. 2020, 19, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Scheper, W.; van Dorp, S.; Kersting, S.; Pietersma, F.; Lindemans, C.; Hol, S.; Heijhuurs, S.; Sebestyen, Z.; Grunder, C.; Marcu-Malina, V.; et al. γδ T cells elicited by CMV reactivation after allo-SCT cross-recognize CMV and leukemia. Leukemia 2013, 27, 1328–1338. [Google Scholar] [CrossRef] [Green Version]
- Ravens, S.; Schultze-Florey, C.; Raha, S.; Sandrock, I.; Drenker, M.; Oberdorfer, L.; Reinhardt, A.; Ravens, I.; Beck, M.; Geffers, R.; et al. Human γδ T cells are quickly reconstituted after stem-cell transplantation and show adaptive clonal expansion in response to viral infection. Nat. Immunol. 2017, 18, 393–401. [Google Scholar] [CrossRef]
- Davey, M.S.; Willcox, C.R.; Joyce, S.P.; Ladell, K.; Kasatskaya, S.A.; McLaren, J.E.; Hunter, S.; Salim, M.; Mohammed, F.; Price, D.A.; et al. Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat. Commun. 2017, 8, 14760. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, N.E.; Eberl, M. Human γδ T-Cell Control of Mucosal Immunity and Inflammation. Front. Immunol. 2018, 9, 985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todaro, M.; D’Asaro, M.; Caccamo, N.; Iovino, F.; Francipane, M.G.; Meraviglia, S.; Orlando, V.; La Mendola, C.; Gulotta, G.; Salerno, A.; et al. Efficient killing of human colon cancer stem cells by γδ T lymphocytes. J. Immunol. 2009, 182, 7287–7296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikulak, J.; Oriolo, F.; Bruni, E.; Roberto, A.; Colombo, F.S.; Villa, A.; Bosticardo, M.; Bortolomai, I.; Lo Presti, E.; Meraviglia, S.; et al. NKp46-expressing human gut-resident intraepithelial Vδ1 T cell subpopulation exhibits high antitumor activity against colorectal cancer. JCI Insight 2019, 4, e125884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vantourout, P.; Hayday, A. Six-of-the-best: Unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 2013, 13, 88–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilshaw, P.; Baker, K. A unique surface antigen on intraepithelial lymphocytes in the mouse. Immunol. Lett. 1988, 18, 149–154. [Google Scholar] [CrossRef]
- Russell, G.J.; Parker, C.M.; Cepek, K.L.; Mandelbrot, D.A.; Sood, A.; Mizoguchi, E.; Ebert, E.C.; Brenner, M.B.; Bhan, A.K. Distinct structural and functional epitopes of the αEβ7 integrin. Eur. J. Immunol. 1994, 24, 2832–2841. [Google Scholar] [CrossRef]
- Silva-Santos, B.; Mensurado, S.; Coffelt, S.B. γδ T cells: Pleiotropic immune effectors with therapeutic potential in cancer. Nat. Rev. Cancer 2019, 19, 392–404. [Google Scholar] [CrossRef] [Green Version]
- Tie, G.; Yan, J.; Khair, L.; Messina, J.A.; Deng, A.; Kang, J.; Fazzio, T.; Messina, L.M. Hypercholesterolemia Increases Colorectal Cancer Incidence by Reducing Production of NKT and γδ T Cells from Hematopoietic Stem Cells. Cancer Res. 2017, 77, 2351–2362. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, S.; Kudoh, S.; Katayama, S. Enhanced formation of azoxymethane-induced colorectal adenocarcinoma in γδ T lymphocyte-deficient mice. Jpn. J. Cancer Res. 2001, 92, 880–885. [Google Scholar] [CrossRef]
- Wu, P.; Wu, D.; Ni, C.; Ye, J.; Chen, W.; Hu, G.; Wang, Z.; Wang, C.; Zhang, Z.; Xia, W.; et al. γδ T17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity 2014, 40, 785–800. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.S.; Shah, S.U.; Shrikhande, S.V.; Goel, M.; Dikshit, R.P.; Chiplunkar, S.V. IL17 producing γδ T cells induce angiogenesis and are associated with poor survival in gallbladder cancer patients. Int. J. Cancer 2016, 139, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Corvaisier, M.; Moreau-Aubry, A.; Diez, E.; Bennouna, J.; Mosnier, J.F.; Scotet, E.; Bonneville, M.; Jotereau, F. Vγ9Vδ2 T cell response to colon carcinoma cells. J. Immunol. 2005, 175, 5481–5488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosolini, M.; Pont, F.; Poupot, M.; Vergez, F.; Nicolau-Travers, M.L.; Vermijlen, D.; Sarry, J.E.; Dieli, F.; Fournie, J.J. Assessment of tumor-infiltrating TCRVγ9Vδ2 γδ lymphocyte abundance by deconvolution of human cancers microarrays. Oncoimmunology 2017, 6, e1284723. [Google Scholar] [CrossRef]
- Pont, F.; Familiades, J.; Dejean, S.; Fruchon, S.; Cendron, D.; Poupot, M.; Poupot, R.; L’Faqihi-Olive, F.; Prade, N.; Ycart, B.; et al. The gene expression profile of phosphoantigen-specific human γδ T lymphocytes is a blend of αβ T-cell and NK-cell signatures. Eur. J. Immunol. 2012, 42, 228–240. [Google Scholar] [CrossRef]
- Wu, D.; Wu, P.; Wu, X.; Ye, J.; Wang, Z.; Zhao, S.; Ni, C.; Hu, G.; Xu, J.; Han, Y.; et al. Ex vivo expanded human circulating Vδ1 γδ T cells exhibit favorable therapeutic potential for colon cancer. Oncoimmunology 2015, 4, e992749. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, C.H.; Kramm, K.; Gálvez-Jirón, F.; Pola, V.; Bustamante, M.; Contreras, H.R.; Sabag, A.; Garrido-Tapia, M.; Hernández, C.J.; Zúñiga, R.; et al. Clinical significance of tumor expression of major histocompatibility complex class I-related chains A and B (MICA/B) in gastric cancer patients. Oncol. Rep. 2015, 35, 1309–1317. [Google Scholar] [CrossRef] [Green Version]
- Chabab, G.; Boissiere-Michot, F.; Mollevi, C.; Ramos, J.; Lopez-Crapez, E.; Colombo, P.E.; Jacot, W.; Bonnefoy, N.; Lafont, V. Diversity of Tumor-Infiltrating, γδ T-Cell Abundance in Solid Cancers. Cells 2020, 9, 1537. [Google Scholar] [CrossRef]
- Reis, B.S.; Darcy, P.W.; Khan, I.Z.; Moon, C.S.; Kornberg, A.E.; Schneider, V.S.; Alvarez, Y.; Eleso, O.; Zhu, C.; Schernthanner, M.; et al. TCR-Vγδ usage distinguishes protumor from antitumor intestinal γδ T cell subsets. Science 2022, 377, 276–284. [Google Scholar] [CrossRef]
- Tan, L.; Sandrock, I.; Odak, I.; Aizenbud, Y.; Wilharm, A.; Barros-Martins, J.; Tabib, Y.; Borchers, A.; Amado, T.; Gangoda, L.; et al. Single-Cell Transcriptomics Identifies the Adaptation of Scart1+ Vγ6+ T Cells to Skin Residency as Activated Effector Cells. Cell Rep. 2019, 27, 3657–3671.e4. [Google Scholar] [CrossRef] [Green Version]
- Bayatipoor, H.; Mehdizadeh, S.; Jafarpour, R.; Shojaei, Z.; Pashangzadeh, S.; Motallebnezhad, M. Role of NKT cells in cancer immunotherapy—From bench to bed. Med. Oncol. 2022, 40, 29. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Kumar, V. Type II NKT cells: A distinct CD1d-restricted immune regulatory NKT cell subset. Immunogenetics 2016, 68, 665–676. [Google Scholar] [CrossRef]
- Godfrey, D.I.; MacDonald, H.R.; Kronenberg, M.; Smyth, M.J.; Van Kaer, L. NKT cells: What’s in a name? Nat. Rev. Immunol. 2004, 4, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Godfrey, D.I. NKT cells and tumor immunity—A double-edged sword. Nat. Immunol. 2000, 1, 459–460. [Google Scholar] [CrossRef]
- Burks, J.; Olkhanud, P.B.; Berzofsky, J.A. The role of NKT cells in gastrointestinal cancers. Oncoimmunology 2022, 11, 2009666. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Crowe, N.Y.; Pellicci, D.G.; Kyparissoudis, K.; Kelly, J.M.; Takeda, K.; Yagita, H.; Godfrey, D.I. Sequential production of interferon-γ by NK1.1+ T cells and natural killer cells is essential for the antimetastatic effect of α-galactosylceramide. Blood 2002, 99, 1259–1266. [Google Scholar] [CrossRef]
- Tachibana, T.; Onodera, H.; Tsuruyama, T.; Mori, A.; Nagayama, S.; Hiai, H.; Imamura, M. Increased intratumor Vα24-positive natural killer T cells: A prognostic factor for primary colorectal carcinomas. Clin. Cancer Res. 2005, 11, 7322–7327. [Google Scholar] [CrossRef] [Green Version]
- Ambrosino, E.; Terabe, M.; Halder, R.C.; Peng, J.; Takaku, S.; Miyake, S.; Yamamura, T.; Kumar, V.; Berzofsky, J.A. Cross-Regulation between Type I and Type II NKT Cells in Regulating Tumor Immunity: A New Immunoregulatory Axis. J. Immunol. 2007, 179, 5126–5136. [Google Scholar] [CrossRef] [Green Version]
- Ambrosino, E.; Berzofsky, J.A.; Terabe, M. Regulation of tumor immunity: The role of NKT cells. Expert Opin. Biol. Ther. 2008, 8, 725–734. [Google Scholar] [CrossRef]
- Al-Mterin, M.A.; Murshed, K.; Elkord, E. PD-1 expression, among other immune checkpoints, on tumor-infiltrating NK and NKT cells is associated with longer disease-free survival in treatment-naive CRC patients. Cancer Immunol. Immunother. 2023, 72, 1933–1939. [Google Scholar] [CrossRef]
- Philip, M.; Schietinger, A. CD8+ T cell differentiation and dysfunction in cancer. Nat. Rev. Immunol. 2022, 22, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Raskov, H.; Orhan, A.; Christensen, J.P.; Gögenur, I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer 2020, 124, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Corsale, A.M.; Di Simone, M.; Lo Presti, E.; Dieli, F.; Meraviglia, S. γδ T cells and their clinical application in colon cancer. Front. Immunol. 2023, 14, 1098847. [Google Scholar] [CrossRef] [PubMed]
- Housseau, F.; Wu, S.; Wick, E.C.; Fan, H.; Wu, X.; Llosa, N.J.; Smith, K.N.; Tam, A.; Ganguly, S.; Wanyiri, J.W.; et al. Redundant Innate and Adaptive Sources of IL17 Production Drive Colon Tumorigenesis. Cancer Res. 2016, 76, 2115–2124. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Hao, J.; Dong, S.; Xia, S.; He, W.; Jia, H.; Zhang, S.; Wei, J.; O’Brien, R.L.; Born, W.K.; Wu, Z.; et al. Regulatory role of Vγ1 γδ T cells in tumor immunity through IL-4 production. J. Immunol. 2011, 187, 4979–4986. [Google Scholar] [CrossRef] [Green Version]
- Burnet, M. Cancer; a biological approach. I. The processes of control. Br. Med. J. 1957, 1, 779–786. [Google Scholar] [CrossRef]
- Thomas, L.; Lawrence, H. Cellular and Humoral Aspects of the Hypersensitive States; Hoeber-Harper: New York, NY, USA, 1959; pp. 529–532. [Google Scholar]
- Ochsenbein, A.F. Principles of tumor immunosurveillance and implications for immunotherapy. Cancer Gene Ther. 2002, 9, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.; Dai, L.-J.; Wu, S.-Y.; Xiao, Y.; Ma, D.; Jiang, Y.-Z.; Shao, Z.-M. Spatial architecture of the immune microenvironment orchestrates tumor immunity and therapeutic response. J. Hematol. Oncol. 2021, 14, 98. [Google Scholar] [CrossRef]
- Pardoll, D. T cells and tumours. Nature 2001, 411, 1010–1012. [Google Scholar] [CrossRef]
- Chow, M.T.; Möller, A.; Smyth, M.J. Inflammation and immune surveillance in cancer. Semin. Cancer Biol. 2012, 22, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denk, D.; Greten, F.R. Inflammation: The incubator of the tumor microenvironment. Trends Cancer 2022, 8, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Saito, K.; Shiiba, K.; Ohuchi, A.; Saigenji, K.; Nagura, H.; Ohtani, H. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998, 58, 3491–3494. [Google Scholar] [PubMed]
- Molon, B.; Ugel, S.; Del Pozzo, F.; Soldani, C.; Zilio, S.; Avella, D.; De Palma, A.; Mauri, P.; Monegal, A.; Rescigno, M.; et al. Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J. Exp. Med. 2011, 208, 1949–1962. [Google Scholar] [CrossRef]
- Anderson, K.G.; Stromnes, I.M.; Greenberg, P.D. Obstacles Posed by the Tumor Microenvironment to T cell Activity: A Case for Synergistic Therapies. Cancer Cell 2017, 31, 311–325. [Google Scholar] [CrossRef] [Green Version]
- Lanitis, E.; Dangaj, D.; Irving, M.; Coukos, G. Mechanisms regulating T-cell infiltration and activity in solid tumors. Ann. Oncol. 2017, 28, xii18–xii32. [Google Scholar] [CrossRef]
- Zhang, Y.; Guan, X.-Y.; Jiang, P. Cytokine and Chemokine Signals of T-Cell Exclusion in Tumors. Front. Immunol. 2020, 11, 594609. [Google Scholar] [CrossRef]
- Motz, G.T.; Santoro, S.P.; Wang, L.-P.; Garrabrant, T.; Lastra, R.R.; Hagemann, I.S.; Lal, P.; Feldman, M.D.; Benencia, F.; Coukos, G. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat. Med. 2014, 20, 607–615. [Google Scholar] [CrossRef]
- Baker, A.T.; Abuwarwar, M.H.; Poly, L.; Wilkins, S.; Fletcher, A.L. Cancer-Associated Fibroblasts and T Cells: From Mechanisms to Outcomes. J. Immunol. 2021, 206, 310–320. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, S.; Zhang, B.; Qiao, L.; Zhang, Y. T Cell Dysfunction and Exhaustion in Cancer. Front. Cell Dev. Biol. 2020, 8, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germann, M.; Zangger, N.; Sauvain, M.O.; Sempoux, C.; Bowler, A.D.; Wirapati, P.; Kandalaft, L.E.; Delorenzi, M.; Tejpar, S.; Coukos, G.; et al. Neutrophils suppress tumor-infiltrating T cells in colon cancer via matrix metalloproteinase-mediated activation of TGFβ. EMBO Mol. Med. 2020, 12, e10681. [Google Scholar] [CrossRef] [PubMed]
- Xia, A.; Zhang, Y.; Xu, J.; Yin, T.; Lu, X.-J. T Cell Dysfunction in Cancer Immunity and Immunotherapy. Front. Immunol. 2019, 10, 1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, A.D. New Insights into the Molecular Basis of T Cell Anergy: Anergy Factors, Avoidance Sensors, and Epigenetic Imprinting. J. Immunol. 2009, 182, 7331–7341. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, R.H. T cell anergy. Annu. Rev. Immunol. 2003, 21, 305–334. [Google Scholar] [CrossRef]
- Kuczma, M.P.; Szurek, E.A.; Cebula, A.; Ngo, V.L.; Pietrzak, M.; Kraj, P.; Denning, T.L.; Ignatowicz, L. Self and microbiota-derived epitopes induce CD4+ T cell anergy and conversion into CD4+Foxp3+ regulatory cells. Mucosal Immunol. 2021, 14, 443–454. [Google Scholar] [CrossRef]
- Overwijk, W.W.; Theoret, M.R.; Finkelstein, S.E.; Surman, D.R.; de Jong, L.A.; Vyth-Dreese, F.A.; Dellemijn, T.A.; Antony, P.A.; Spiess, P.J.; Palmer, D.C.; et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J. Exp. Med. 2003, 198, 569–580. [Google Scholar] [CrossRef]
- Ochsenbein, A.F.; Klenerman, P.; Karrer, U.; Ludewig, B.; Pericin, M.; Hengartner, H.; Zinkernagel, R.M. Immune surveillance against a solid tumor fails because of immunological ignorance. Proc. Natl. Acad. Sci. USA 1999, 96, 2233–2238. [Google Scholar] [CrossRef]
- Hegde, P.S.; Karanikas, V.; Evers, S. The Where, the When, and the How of Immune Monitoring for Cancer Immunotherapies in the Era of Checkpoint Inhibition. Clin. Cancer Res. 2016, 22, 1865–1874. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Kim, Y.; Jung, Y.W. The Function of Memory CD8+ T Cells in Immunotherapy for Human Diseases. Immune Netw. 2023, 23, e10. [Google Scholar] [CrossRef]
- Gebhardt, T.; Mueller, S.N.; Heath, W.R.; Carbone, F.R. Peripheral tissue surveillance and residency by memory T cells. Trends Immunol. 2013, 34, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Schenkel, J.M.; Herbst, R.H.; Canner, D.; Li, A.; Hillman, M.; Shanahan, S.-L.; Gibbons, G.; Smith, O.C.; Kim, J.Y.; Westcott, P.; et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity 2021, 54, 2338–2353.e6. [Google Scholar] [CrossRef] [PubMed]
- Fowell, D.J.; Kim, M. The spatio-temporal control of effector T cell migration. Nat. Rev. Immunol. 2021, 21, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Beltra, J.-C.; Manne, S.; Abdel-Hakeem, M.S.; Kurachi, M.; Giles, J.R.; Chen, Z.; Casella, V.; Ngiow, S.F.; Khan, O.; Huang, Y.J.; et al. Developmental Relationships of Four Exhausted CD8+ T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity 2020, 52, 825–841.e8. [Google Scholar] [CrossRef] [PubMed]
- Wherry, E.J.; Blattman, J.N.; Murali-Krishna, K.; van der Most, R.; Ahmed, R. Viral Persistence Alters CD8 T-Cell Immunodominance and Tissue Distribution and Results in Distinct Stages of Functional Impairment. J. Virol. 2003, 77, 4911–4927. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; He, Y.; He, W.; Wu, G.; Zhou, X.; Sheng, Q.; Zhong, W.; Lu, Y.; Ding, Y.; Lu, Q.; et al. Exhausted CD8+ T Cells in the Tumor Immune Microenvironment: New Pathways to Therapy. Front. Immunol. 2021, 11, 622509. [Google Scholar] [CrossRef]
- Siddiqui, I.; Schaeuble, K.; Chennupati, V.; Marraco, S.A.F.; Calderon-Copete, S.; Ferreira, D.P.; Carmona, S.J.; Scarpellino, L.; Gfeller, D.; Pradervand, S.; et al. Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 2019, 50, 195–211.e10. [Google Scholar] [CrossRef] [Green Version]
- Brunell, A.E.; Lahesmaa, R.; Autio, A.; Thotakura, A.K. Exhausted T cells hijacking the cancer-immunity cycle: Assets and liabilities. Front. Immunol. 2023, 14, 1151632. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Pauken, K.E. Localization, tissue biology and T cell state—Implications for cancer immunotherapy. Nat. Rev. Immunol. 2023. [Google Scholar] [CrossRef]
- Braumüller, H.; Mauerer, B.; Berlin, C.; Plundrich, D.; Marbach, P.; Cauchy, P.; Laessle, C.; Biesel, E.; Holzner, P.A.; Kesselring, R. Senescent Tumor Cells in the Peritoneal Carcinomatosis Drive Immunosenescence in the Tumor Microenvironment. Front. Immunol. 2022, 13, 908449. [Google Scholar] [CrossRef]
- Narasimhan, V.; Tan, S.; Kong, J.; Pham, T.; Michael, M.; Ramsay, R.; Warrier, S.; Heriot, A. Prognostic factors influencing survival in patients undergoing cytoreductive surgery with hyperthermic intraperitoneal chemotherapy for isolated colorectal peritoneal metastases: A systematic review and meta-analysis. Color. Dis. 2020, 22, 1482–1495. [Google Scholar] [CrossRef] [PubMed]
- Wieder, T.; Brenner, E.; Braumüller, H.; Röcken, M. Immunotherapy of melanoma: Efficacy and mode of action. JDDG J. Dtsch. Dermatol. Ges. 2016, 14, 28–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [Green Version]
- Wieder, T.; Brenner, E.; Braumüller, H.; Bischof, O.; Röcken, M. Cytokine-induced senescence for cancer surveillance. Cancer Metastasis Rev. 2017, 36, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Wieder, T.; Eigentler, T.; Brenner, E.; Rocken, M. Immune checkpoint blockade therapy. J. Allergy Clin. Immunol. 2018, 142, 1403–1414. [Google Scholar] [CrossRef]
- Heinzerling, L.; de Toni, E.N.; Schett, G.; Hundorfean, G.; Zimmer, L. Checkpoint Inhibitors. Dtsch. Arztebl. Int. 2019, 116, 119–126. [Google Scholar] [CrossRef]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of Antitumor Immunity by CTLA-4 Blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef] [Green Version]
- Okazaki, T.; Chikuma, S.; Iwai, Y.; Fagarasan, S.; Honjo, T. A rheostat for immune responses: The unique properties of PD-1 and their advantages for clinical application. Nat. Immunol. 2013, 14, 1212–1218. [Google Scholar] [CrossRef] [Green Version]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Postow, M.A.; Chesney, J.; Pavlick, A.C.; Robert, C.; Grossmann, K.; McDermott, D.; Linette, G.P.; Meyer, N.; Giguere, J.K.; Agarwala, S.S.; et al. Nivolumab and Ipilimumab versus Ipilimumab in Untreated Melanoma. N. Engl. J. Med. 2015, 372, 2006–2017. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, H.L.; Russell, J.; Hamid, O.; Bhatia, S.; Terheyden, P.; D’Angelo, S.P.; Shih, K.C.; Lebbé, C.; Linette, G.P.; Milella, M.; et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: A multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1374–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, J.E.; Hoffman-Censits, J.; Powles, T.; van der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y.; et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, C.C.; Fleming, G.F.; Karrison, T.G.; Liao, C.Y.; Desai, A.V.; Moroney, J.W.; Ratain, M.J.; Nanda, R.; Polite, B.N.; Hahn, O.M.; et al. Phase I Study of Stereotactic Body Radiotherapy plus Nivolumab and Urelumab or Cabiralizumab in Advanced Solid Tumors. Clin. Cancer Res. 2021, 27, 5510–5518. [Google Scholar] [CrossRef] [PubMed]
- Amaria, R.N.; Postow, M.; Burton, E.M.; Tetzlaff, M.T.; Ross, M.I.; Torres-Cabala, C.; Glitza, I.C.; Duan, F.; Milton, D.R.; Busam, K.; et al. Neoadjuvant relatlimab and nivolumab in resectable melanoma. Nature 2022, 611, 155–160. [Google Scholar] [CrossRef]
- Yalniz, F.F.; Daver, N.; Rezvani, K.; Kornblau, S.; Ohanian, M.; Borthakur, G.; DiNardo, C.D.; Konopleva, M.; Burger, J.; Gasior, Y.; et al. A Pilot Trial of Lirilumab With or Without Azacitidine for Patients With Myelodysplastic Syndrome. Clin. Lymphoma Myeloma Leuk. 2018, 18, 658–663.e2. [Google Scholar] [CrossRef]
- Brahmer, J.R.; Drake, C.G.; Wollner, I.; Powderly, J.D.; Picus, J.; Sharfman, W.H.; Stankevich, E.; Pons, A.; Salay, T.M.; McMiller, T.L.; et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: Safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. 2010, 28, 3167–3175. [Google Scholar] [CrossRef]
- Weber, J. Review: Anti–CTLA-4 Antibody Ipilimumab: Case Studies of Clinical Response and Immune-Related Adverse Events. Oncologist 2007, 12, 864–872. [Google Scholar] [CrossRef]
- Andre, T.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Nivolumab plus low-dose ipilimumab in previously treated patients with microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: 4-year follow-up from CheckMate 142. Ann. Oncol. 2022, 33, 1052–1060. [Google Scholar] [CrossRef]
- Cousin, S.; Cantarel, C.; Guegan, J.-P.; Gomez-Roca, C.; Metges, J.-P.; Adenis, A.; Pernot, S.; Bellera, C.; Kind, M.; Auzanneau, C.; et al. Regorafenib-Avelumab Combination in Patients with Microsatellite Stable Colorectal Cancer (REGOMUNE): A Single-arm, Open-label, Phase II Trial. Clin. Cancer Res. 2021, 27, 2139–2147. [Google Scholar] [CrossRef]
- Sanmamed, M.F.; Rodriguez, I.; Schalper, K.A.; Onate, C.; Azpilikueta, A.; Rodriguez-Ruiz, M.E.; Morales-Kastresana, A.; Labiano, S.; Perez-Gracia, J.L.; Martin-Algarra, S.; et al. Nivolumab and Urelumab Enhance Antitumor Activity of Human T Lymphocytes Engrafted in Rag2-/-IL2Rγnull Immunodeficient Mice. Cancer Res. 2015, 75, 3466–3478. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Huang, X.; Chen, X.; Liu, J.; Wu, C.; Pu, Q.; Wang, Y.; Kang, X.; Zhou, L. Characterization of a novel anti-human lymphocyte activation gene 3 (LAG-3) antibody for cancer immunotherapy. MAbs 2019, 11, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Crisafulli, G.; Sartore-Bianchi, A.; Lazzari, L.; Pietrantonio, F.; Amatu, A.; Macagno, M.; Barault, L.; Cassingena, A.; Bartolini, A.; Luraghi, P.; et al. Temozolomide Treatment Alters Mismatch Repair and Boosts Mutational Burden in Tumor and Blood of Colorectal Cancer Patients. Cancer Discov. 2022, 12, 1656–1675. [Google Scholar] [CrossRef] [PubMed]
- Rabin, H.; Adamson, R.H.; Neubauer, R.H.; Cicmanec, J.L.; Wallen, W.C. Pilot studies with human interferon in Herpesvirus saimiri-induced lymphoma in owl monkeys. Cancer Res. 1976, 36, 715–719. [Google Scholar] [PubMed]
- Schmidt-Wolf, I.G.H.; Finke, S.; Trojaneck, B.; Denkena, A.; Lefterova, P.; Schwella, N.; Heuft, H.-G.; Prange, G.; Korte, M.; Takeya, M.; et al. Phase I clinical study applying autologous immunological effector cells transfected with the interleukin-2 gene in patients with metastatic renal cancer, colorectal cancer and lymphoma. Br. J. Cancer 1999, 81, 1009–1016. [Google Scholar] [CrossRef] [Green Version]
- Kowolik, C.M.; Topp, M.S.; Gonzalez, S.; Pfeiffer, T.; Olivares, S.; Gonzalez, N.; Smith, D.D.; Forman, S.J.; Jensen, M.C.; Cooper, L.J. CD28 Costimulation Provided through a CD19-Specific Chimeric Antigen Receptor Enhances In Vivo Persistence and Antitumor Efficacy of Adoptively Transferred T Cells. Cancer Res. 2006, 66, 10995–11004. [Google Scholar] [CrossRef] [Green Version]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Brudno, J.N.; Somerville, R.P.; Shi, V.; Rose, J.J.; Halverson, D.C.; Fowler, D.H.; Gea-Banacloche, J.C.; Pavletic, S.Z.; Hickstein, D.D.; Lu, T.L.; et al. Allogeneic T Cells That Express an Anti-CD19 Chimeric Antigen Receptor Induce Remissions of B-Cell Malignancies That Progress After Allogeneic Hematopoietic Stem-Cell Transplantation Without Causing Graft-Versus-Host Disease. J. Clin. Oncol. 2016, 34, 1112–1121. [Google Scholar] [CrossRef] [Green Version]
- Posey, A.D., Jr.; Schwab, R.D.; Boesteanu, A.C.; Steentoft, C.; Mandel, U.; Engels, B.; Stone, J.D.; Madsen, T.D.; Schreiber, K.; Haines, K.M.; et al. Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma. Immunity 2016, 44, 1444–1454. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.; Brawley, V.S.; Hegde, M.; Robertson, C.; Ghazi, A.; Gerken, C.; Liu, E.; Dakhova, O.; Ashoori, A.; Corder, A.; et al. Human Epidermal Growth Factor Receptor 2 (HER2)-Specific Chimeric Antigen Receptor-Modified T Cells for the Immunotherapy of HER2-Positive Sarcoma. J. Clin. Oncol. 2015, 33, 1688–1696. [Google Scholar] [CrossRef]
- Rafiq, S.; Yeku, O.O.; Jackson, H.J.; Purdon, T.J.; Van Leeuwen, D.G.; Drakes, D.J.; Song, M.; Miele, M.M.; Li, Z.; Wang, P.; et al. Targeted delivery of a PD-1-blocking scFv by CAR-T cells enhances anti-tumor efficacy in vivo. Nat. Biotechnol. 2018, 36, 847–856. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Z.; Yang, Z.; Wang, M.; Li, S.; Li, Y.; Zhang, R.; Xiong, Z.; Wei, Z.; Shen, J.; et al. Phase I Escalating-Dose Trial of CAR-T Therapy Targeting CEA + Metastatic Colorectal Cancers. Mol. Ther. 2017, 25, 1248–1258. [Google Scholar] [CrossRef]
- Curio, S.; Jonsson, G.; Marinović, S. A summary of current NKG2D-based CAR clinical trials. Immunother. Adv. 2021, 1, ltab018. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M.; Kruisbeek, A.M.; Fowlkes, B.J.; Coligan, J.E.; Schwartz, R.H. The unfolding story of T cell receptor gamma. FASEB J. 1987, 1, 103–109. [Google Scholar] [CrossRef]
- Pardoll, D.M.; Fowlkes, B.J.; Bluestone, J.A.; Kruisbeek, A.; Maloy, W.L.; Coligan, J.E.; Schwartz, R.H. Differential expression of two distinct T-cell receptors during thymocyte development. Nature 1987, 326, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Malkovska, V.; Cigel, F.K.; Armstrong, N.; Storer, B.E.; Hong, R. Antilymphoma activity of human γδ T-cells in mice with severe combined immune deficiency. Cancer Res. 1992, 52, 5610–5616. [Google Scholar] [PubMed]
- Chen, J.; Niu, H.; He, W.; Ba, D. Antitumor activity of expanded human tumor-infiltrating γδ T lymphocytes. Int. Arch. Allergy Immunol. 2001, 125, 256–263. [Google Scholar] [CrossRef]
- Nakajima, J.; Murakawa, T.; Fukami, T.; Goto, S.; Kaneko, T.; Yoshida, Y.; Takamoto, S.; Kakimi, K. A phase I study of adoptive immunotherapy for recurrent non-small-cell lung cancer patients with autologous γδ T cells. Eur. J. Cardiothorac. Surg. 2010, 37, 1191–1197. [Google Scholar] [CrossRef] [Green Version]
- Aoki, T.; Matsushita, H.; Hoshikawa, M.; Hasegawa, K.; Kokudo, N.; Kakimi, K. Adjuvant combination therapy with gemcitabine and autologous γδ T-cell transfer in patients with curatively resected pancreatic cancer. Cytotherapy 2017, 19, 473–485. [Google Scholar] [CrossRef]
- Tanaka, Y. Cancer immunotherapy harnessing γδ T cells and programmed death-1. Immunol. Rev. 2020, 298, 237–253. [Google Scholar] [CrossRef]
- Mensurado, S.; Blanco-Dominguez, R.; Silva-Santos, B. The emerging roles of γδ T cells in cancer immunotherapy. Nat. Rev. Clin. Oncol. 2023, 20, 178–191. [Google Scholar] [CrossRef]
- Abril-Rodriguez, G.; Torrejon, D.Y.; Liu, W.; Zaretsky, J.M.; Nowicki, T.S.; Tsoi, J.; Puig-Saus, C.; Baselga-Carretero, I.; Medina, E.; Quist, M.J.; et al. PAK4 inhibition improves PD-1 blockade immunotherapy. Nat. Cancer 2020, 1, 46–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Q.; Zheng, F.; Tang, Q.; Wu, J.-J.; Xie, J.; Huang, H.-D.; Yang, X.-B.; Hann, S.S. Repression of PDK1- and LncRNA HOTAIR-Mediated EZH2 Gene Expression Contributes to the Enhancement of Atractylenolide 1 and Erlotinib in the Inhibition of Human Lung Cancer Cells. Cell. Physiol. Biochem. 2018, 49, 1615–1632. [Google Scholar] [CrossRef] [PubMed]
- Bugter, J.M.; Fenderico, N.; Maurice, M.M. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat. Rev. Cancer 2021, 21, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Schaer, D.A.; Beckmann, R.P.; Dempsey, J.A.; Huber, L.; Forest, A.; Amaladas, N.; Li, Y.; Wang, Y.C.; Rasmussen, E.R.; Chin, D.; et al. The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep. 2018, 22, 2978–2994. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Weekes, J.; Ho, Y.-H.; Sebesan, S.; Ong, K.; Lam, A.K.-Y. Irinotecan and colorectal cancer: The role of p53, VEGF-C and α-B-crystallin expression. Int. J. Color. Dis. 2010, 25, 907. [Google Scholar] [CrossRef] [Green Version]
- Douillard, J.-Y.; Zemelka, T.; Fountzilas, G.; Barone, C.; Schlichting, M.; Heighway, J.; Eggleton, S.P.; Srimuninnimit, V. FOLFOX4 With Cetuximab vs. UFOX With Cetuximab as First-Line Therapy in Metastatic Colorectal Cancer: The Randomized Phase II FUTURE Study. Clin. Color. Cancer 2014, 13, 14–26.e1. [Google Scholar] [CrossRef] [Green Version]
- Petrelli, F.; Coinu, A.; Cabiddu, M.; Ghilardi, M.; Barni, S. KRAS as prognostic biomarker in metastatic colorectal cancer patients treated with bevacizumab: A pooled analysis of 12 published trials. Med. Oncol. 2013, 30, 650. [Google Scholar] [CrossRef]
- Cremolini, C.; Loupakis, F.; Antoniotti, C.; Lonardi, S.; Masi, G.; Salvatore, L.; Cortesi, E.; Tomasello, G.; Spadi, R.; Zaniboni, A.; et al. Early tumor shrinkage and depth of response predict long-term outcome in metastatic colorectal cancer patients treated with first-line chemotherapy plus bevacizumab: Results from phase III TRIBE trial by the Gruppo Oncologico del Nord Ovest. Ann. Oncol. 2015, 26, 1188–1194. [Google Scholar] [CrossRef]
- Nandikolla, A.G.; Rajdev, L. Targeting angiogenesis in gastrointestinal tumors: Current challenges. Transl. Gastroenterol. Hepatol. 2016, 1, 67. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Xia, G.; Li, N.; Su, R.; Chen, X.; Zhong, L. Autophagy Inhibition Promotes Bevacizumab-induced Apoptosis and Proliferation Inhibition in Colorectal Cancer Cells. J. Cancer 2018, 9, 3407–3416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Canellas, A.; Hernando-Momblona, X.; et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanpouille-Box, C.; Diamond, J.M.; Pilones, K.A.; Zavadil, J.; Babb, J.S.; Formenti, S.C.; Barcellos-Hoff, M.H.; Demaria, S. TGFβ Is a Master Regulator of Radiation Therapy-Induced Antitumor Immunity. Cancer Res. 2015, 75, 2232–2242. [Google Scholar] [CrossRef] [Green Version]
- Mortezaee, K. CXCL12/CXCR4 axis in the microenvironment of solid tumors: A critical mediator of metastasis. Life Sci. 2020, 249, 117534. [Google Scholar] [CrossRef] [PubMed]
- Feig, C.; Jones, J.O.; Kraman, M.; Wells, R.J.; Deonarine, A.; Chan, D.S.; Connell, C.M.; Roberts, E.W.; Zhao, Q.; Caballero, O.L.; et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 20212–20217. [Google Scholar] [CrossRef]
- Dunn, J.; Rao, S. Epigenetics and immunotherapy: The current state of play. Mol. Immunol. 2017, 87, 227–239. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [Green Version]
- Brocks, D.; Schmidt, C.R.; Daskalakis, M.; Jang, H.S.; Shah, N.M.; Li, D.; Li, J.; Zhang, B.; Hou, Y.; Laudato, S.; et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat. Genet. 2017, 49, 1052–1060. [Google Scholar] [CrossRef]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef] [Green Version]
- Russell, L.; Peng, K.W.; Russell, S.J.; Diaz, R.M. Oncolytic Viruses: Priming Time for Cancer Immunotherapy. Biodrugs 2019, 33, 485–501. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Twumasi-Boateng, K.; Pettigrew, J.L.; Kwok, Y.Y.E.; Bell, J.C.; Nelson, B.H. Oncolytic viruses as engineering platforms for combination immunotherapy. Nat. Rev. Cancer 2018, 18, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Demaria, S.; Golden, E.B.; Formenti, S.C. Role of Local Radiation Therapy in Cancer Immunotherapy. JAMA Oncol. 2015, 1, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhao, L.; Pol, J.; Levesque, S.; Petrazzuolo, A.; Pfirschke, C.; Engblom, C.; Rickelt, S.; Yamazaki, T.; Iribarren, K.; et al. Crizotinib-induced immunogenic cell death in non-small cell lung cancer. Nat. Commun. 2019, 10, 1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Chen, Y.; Ding, Z.-Y.; Liu, J.-Y. Safety and Efficacy of Therapeutic Cancer Vaccines Alone or in Combination With Immune Checkpoint Inhibitors in Cancer Treatment. Front. Pharmacol. 2019, 10, 1184. [Google Scholar] [CrossRef] [PubMed]
- Kucharczyk, J.; Matrana, M.R.; Santoni, M.; Massari, F.; Scarpelli, M.; Cheng, L.; López-Beltrán, A.; Cascinu, S.; Montironi, R.; Holger, M. Emerging Immunotargets in Metastatic Renal Cell Carcinoma. Curr. Drug Targets 2016, 17, 771–776. [Google Scholar] [CrossRef]
- Lulli, M.; Napoli, C.; Landini, I.; Mini, E.; Lapucci, A. Role of Non-Coding RNAs in Colorectal Cancer: Focus on Long Non-Coding RNAs. Int. J. Mol. Sci. 2022, 23, 13431. [Google Scholar] [CrossRef]
- Overman, M.J.; Morris, V.; Moinova, H.; Manyam, G.; Ensor, J.; Lee, M.S.; Eng, C.; Kee, B.; Fogelman, D.; Shroff, R.T.; et al. Phase I/II study of azacitidine and capecitabine/oxaliplatin (CAPOX) in refractory CIMP-high metastatic colorectal cancer: Evaluation of circulating methylated vimentin. Oncotarget 2016, 7, 67495–67506. [Google Scholar] [CrossRef] [Green Version]
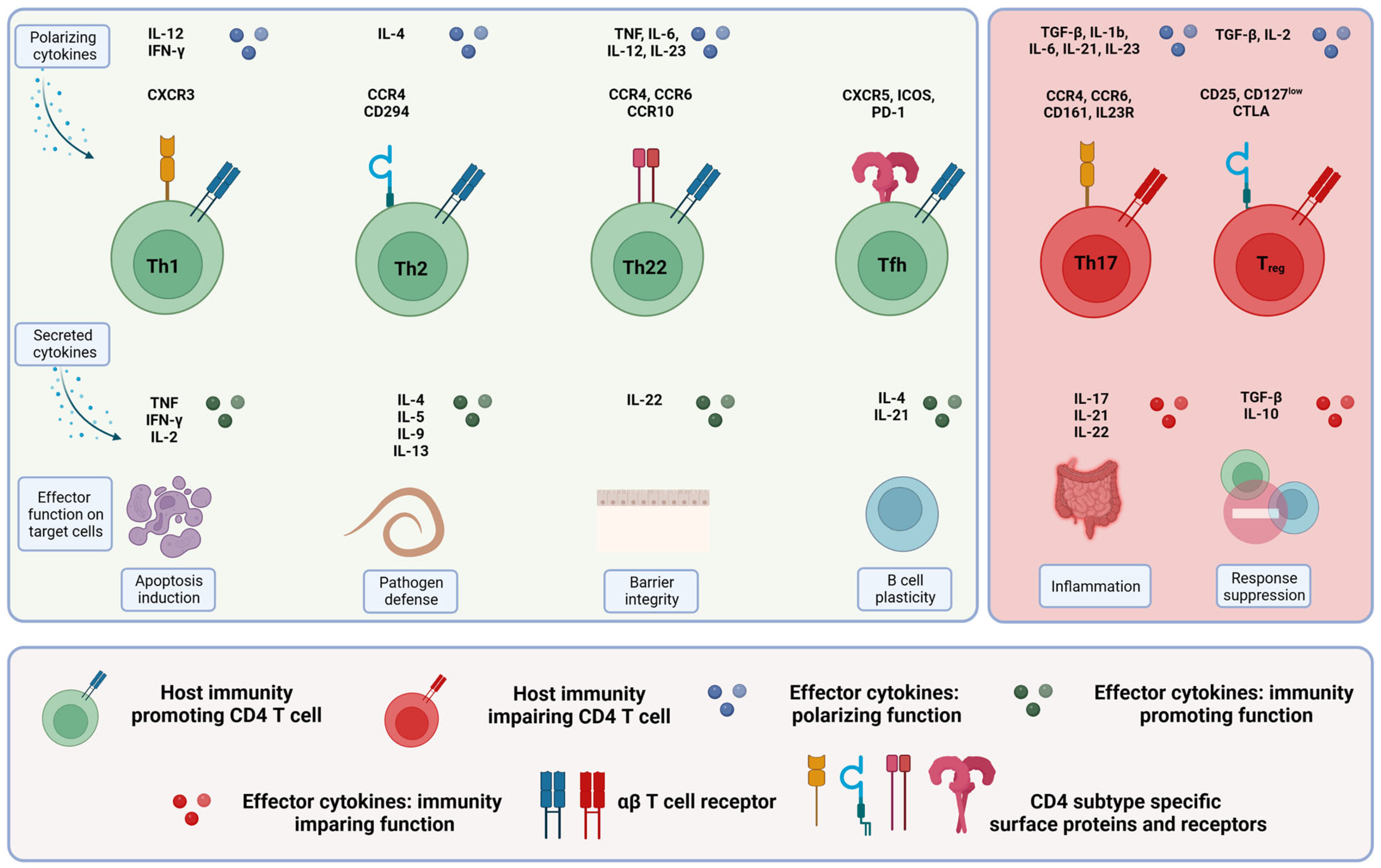

| Name | Surface Marker | Transcription Factor | Polarizing Cytokines | Function | Secreted Cytokines | References |
|---|---|---|---|---|---|---|
| Th1 | CXCR3+, CCR6− | T-BET | IL-12, IFN-γ | Destruction of infected cells by inducing apoptosis. Th1 cells promote destruction and induce senescence of cancer cells. They also display anti-angiogenic properties. | IFN-γ, TNF-α, IL-2 | [46,47,48] |
| Th2 | CXCR3−, CCR4+, CCR6−, CD294+ | GATA | IL-4 | Response to extracellular pathogens. Polarization of macrophages toward a tumor-promoting M2 phenotype. Th2 cells promote the proliferation of tumor cells. | IL-4, IL-5, IL-9, IL-13 | [49] |
| Th17 | CXCR3−, CCR4+, CCR6+, CD161+, IL23R+ | IRF4+, ROR-γt+ | TGF-β, IL-6, IL-1β, IL-21, IL-23 | Response to extracellular pathogens by recruiting neutrophils and macrophages to the site of inflammation. Promotion of cancer stemness and chemo-resistance. | IL-17, IL-21, IL-22 | [46,50] |
| Th22 | CCR10+, CCr4+, CCR6+ | AHR+, FOXO4+ | IL-6, TNF-α, IL-12, IL-23 | Regulator of epithelial barrier integrity. | IL-22 | [51] |
| Tregs | CD127low, CD25+, CTLA4+ | FOXP3+ | TGF-β, IL-2 | Maintain tolerance to self-antigens; prevent the induction of autoantibodies. Suppression of effector T cell-mediated immune responses in colorectal cancer. | IL-10, TGF-β | [46,52] |
| Tfh | CXCR5+, ICOS+, PD-1+ | BCL6+ | Orchestration of germinal center B cell responses; required for antibody class switching. | IL-21, IL-4 | [53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Z.; Wieder, T.; Mauerer, B.; Schäfer, L.; Kesselring, R.; Braumüller, H. T Cells in Colorectal Cancer: Unravelling the Function of Different T Cell Subsets in the Tumor Microenvironment. Int. J. Mol. Sci. 2023, 24, 11673. https://doi.org/10.3390/ijms241411673
Zheng Z, Wieder T, Mauerer B, Schäfer L, Kesselring R, Braumüller H. T Cells in Colorectal Cancer: Unravelling the Function of Different T Cell Subsets in the Tumor Microenvironment. International Journal of Molecular Sciences. 2023; 24(14):11673. https://doi.org/10.3390/ijms241411673
Chicago/Turabian StyleZheng, Ziwen, Thomas Wieder, Bernhard Mauerer, Luisa Schäfer, Rebecca Kesselring, and Heidi Braumüller. 2023. "T Cells in Colorectal Cancer: Unravelling the Function of Different T Cell Subsets in the Tumor Microenvironment" International Journal of Molecular Sciences 24, no. 14: 11673. https://doi.org/10.3390/ijms241411673
APA StyleZheng, Z., Wieder, T., Mauerer, B., Schäfer, L., Kesselring, R., & Braumüller, H. (2023). T Cells in Colorectal Cancer: Unravelling the Function of Different T Cell Subsets in the Tumor Microenvironment. International Journal of Molecular Sciences, 24(14), 11673. https://doi.org/10.3390/ijms241411673






