Involvement of Vacuolar Processing Enzyme CgVPE1 in Vacuole Rupture in the Programmed Cell Death during the Development of the Secretory Cavity in Citrus grandis ‘Tomentosa’ Fruits
Abstract
:1. Introduction
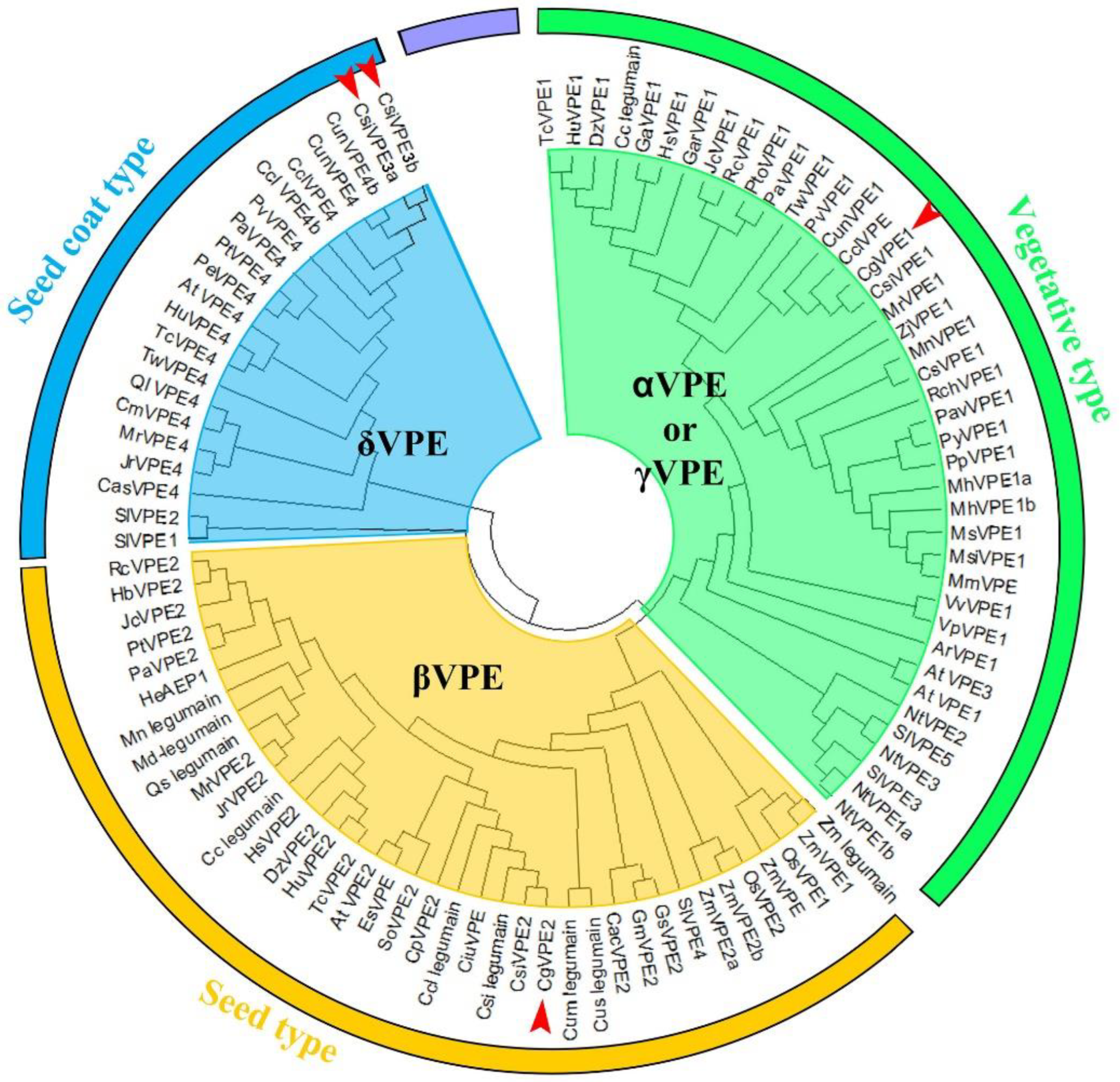
2. Results
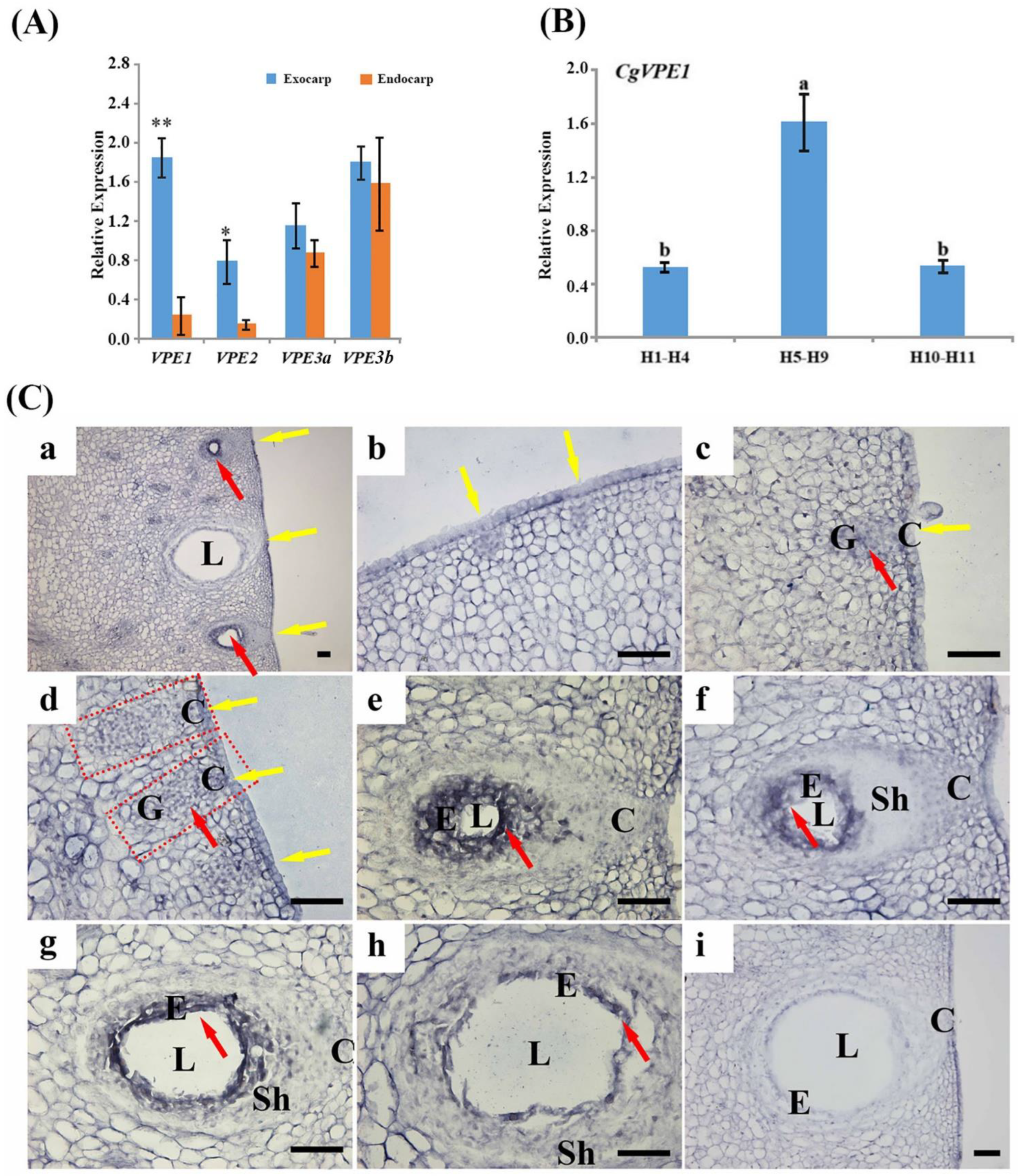
2.1. Expression and Localization of CgVPE1 during the Development of the Secretory Cavity in C. grandis ‘Tomentosa’ Fruit
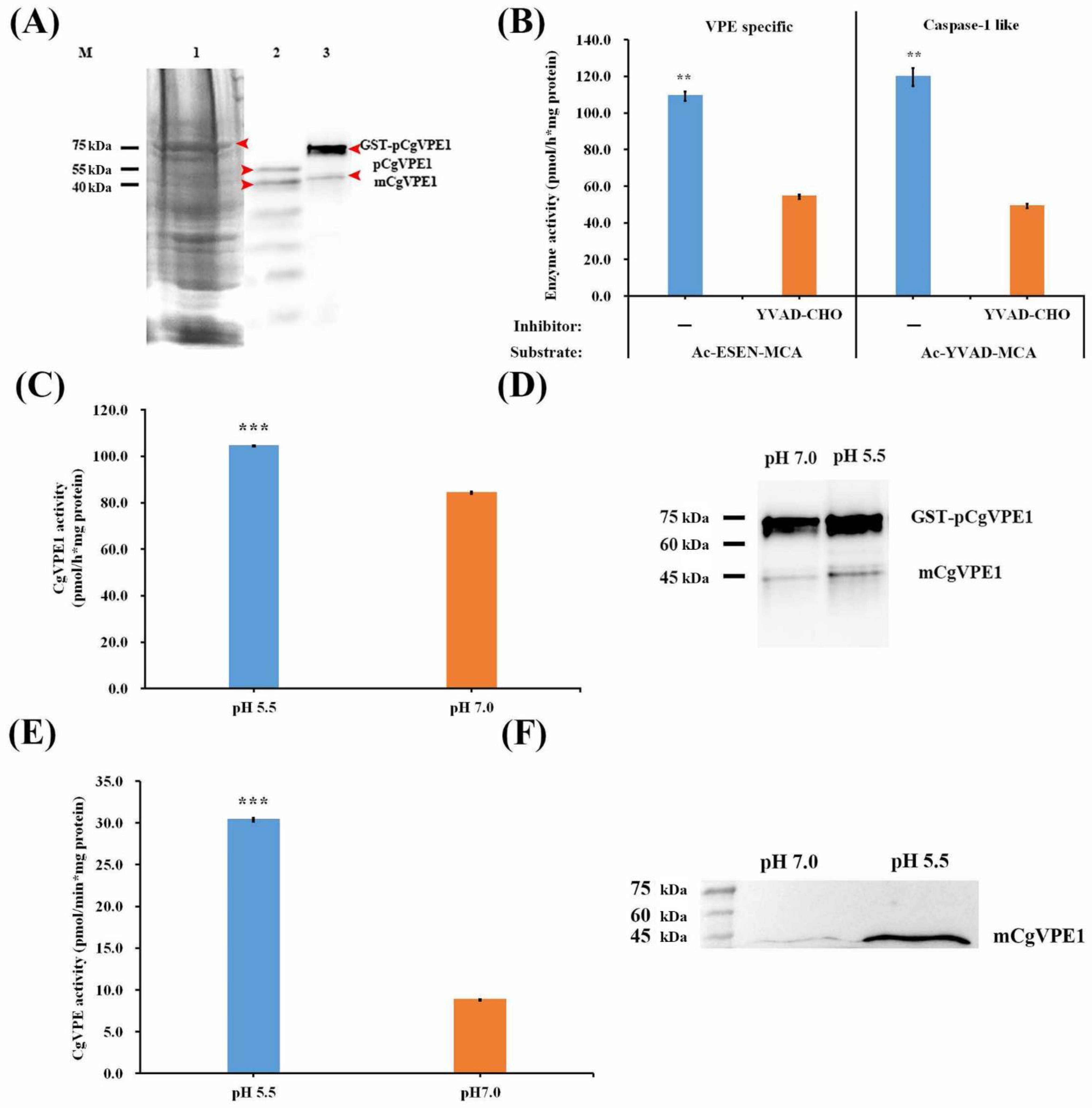
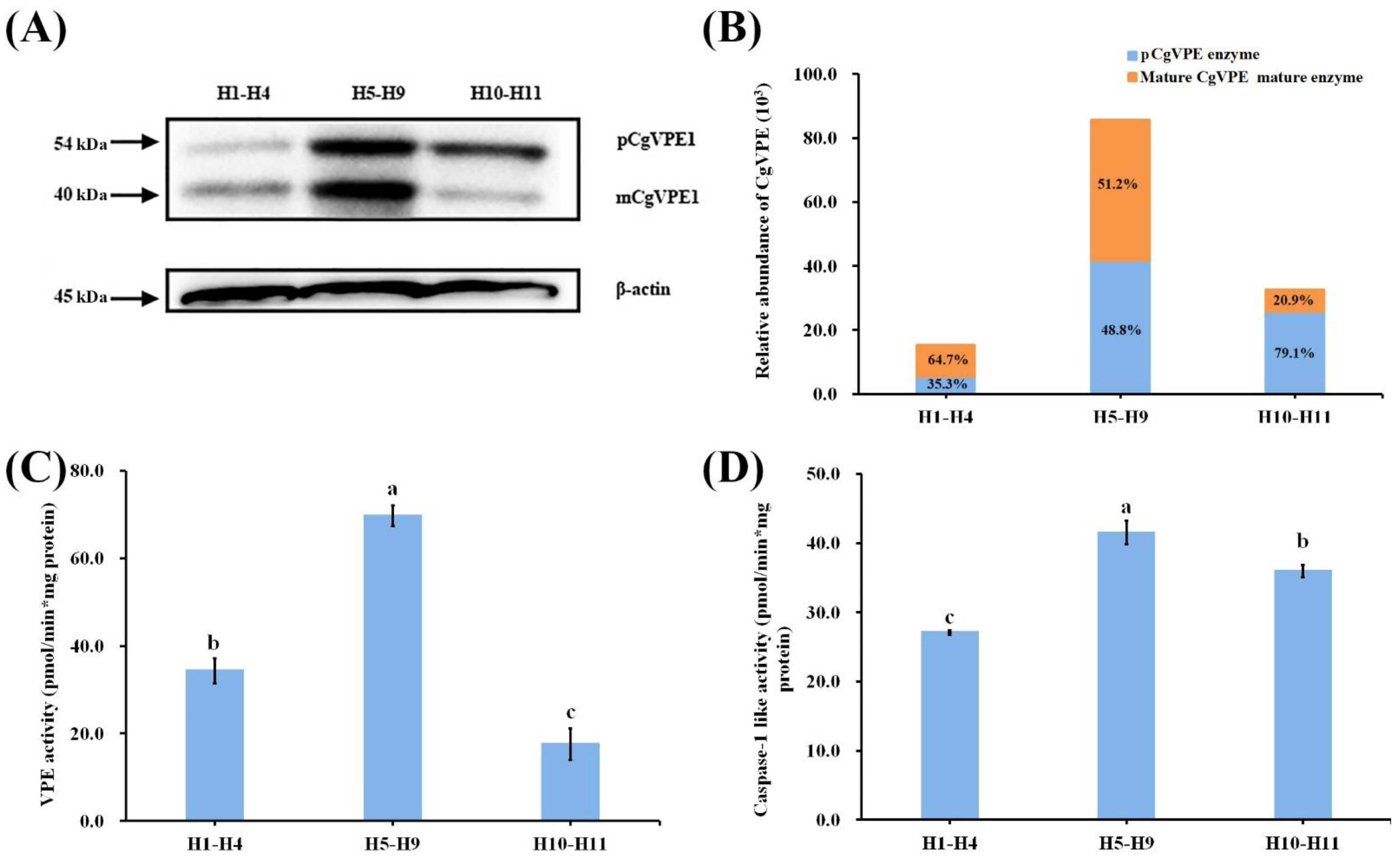
2.2. CgVPE1 Was a Typical Vacuolar Processing Enzyme and Had Caspase-1-like Activity
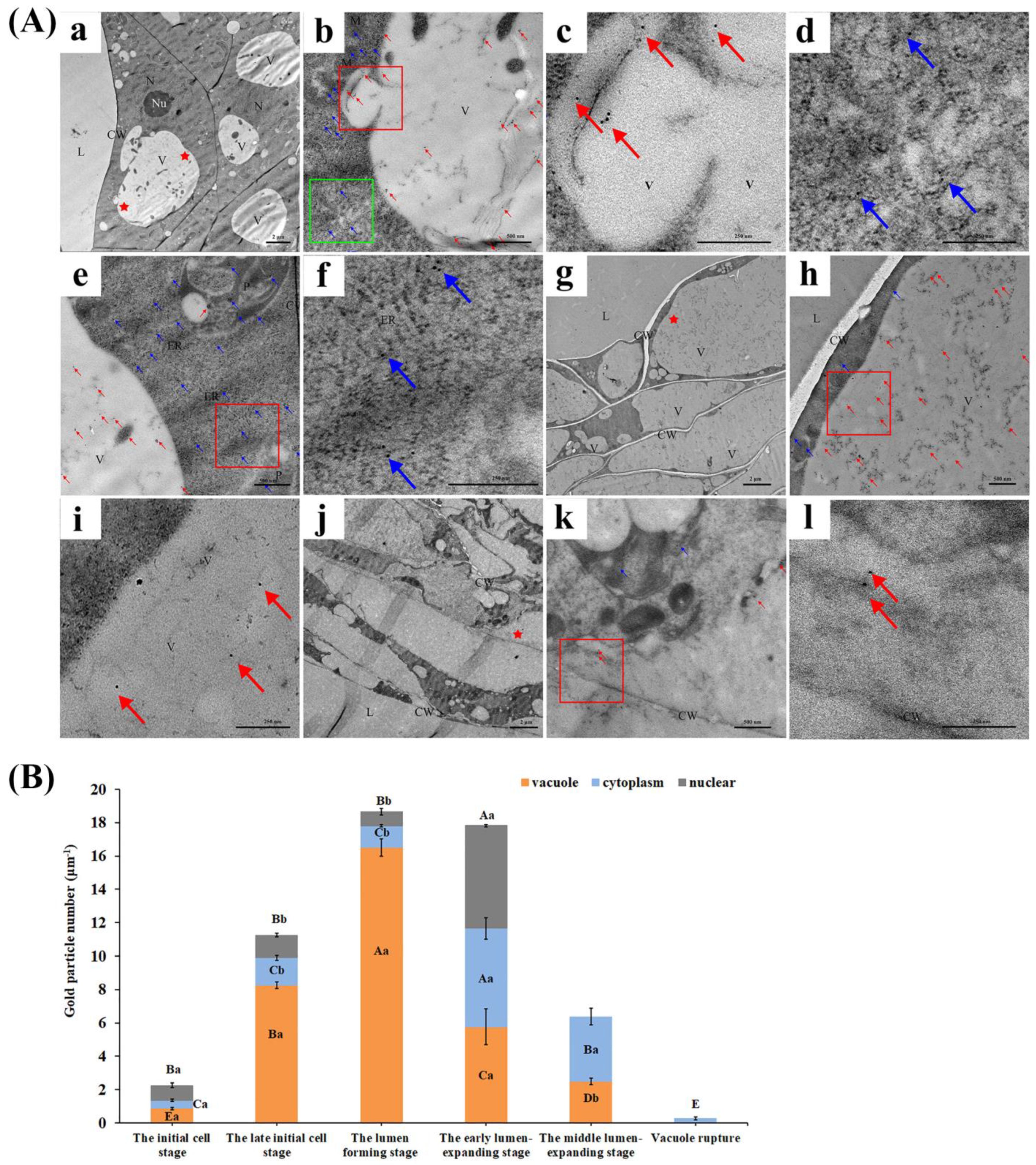
2.3. Expression and Localization of CgVPE1 during the Development of the Secretory Cavity in C. grandis ‘Tomentosa’ Fruits
3. Discussion
3.1. CgVPE1 Was a Vegetative Type with VPE and Caspase-1-like Activities
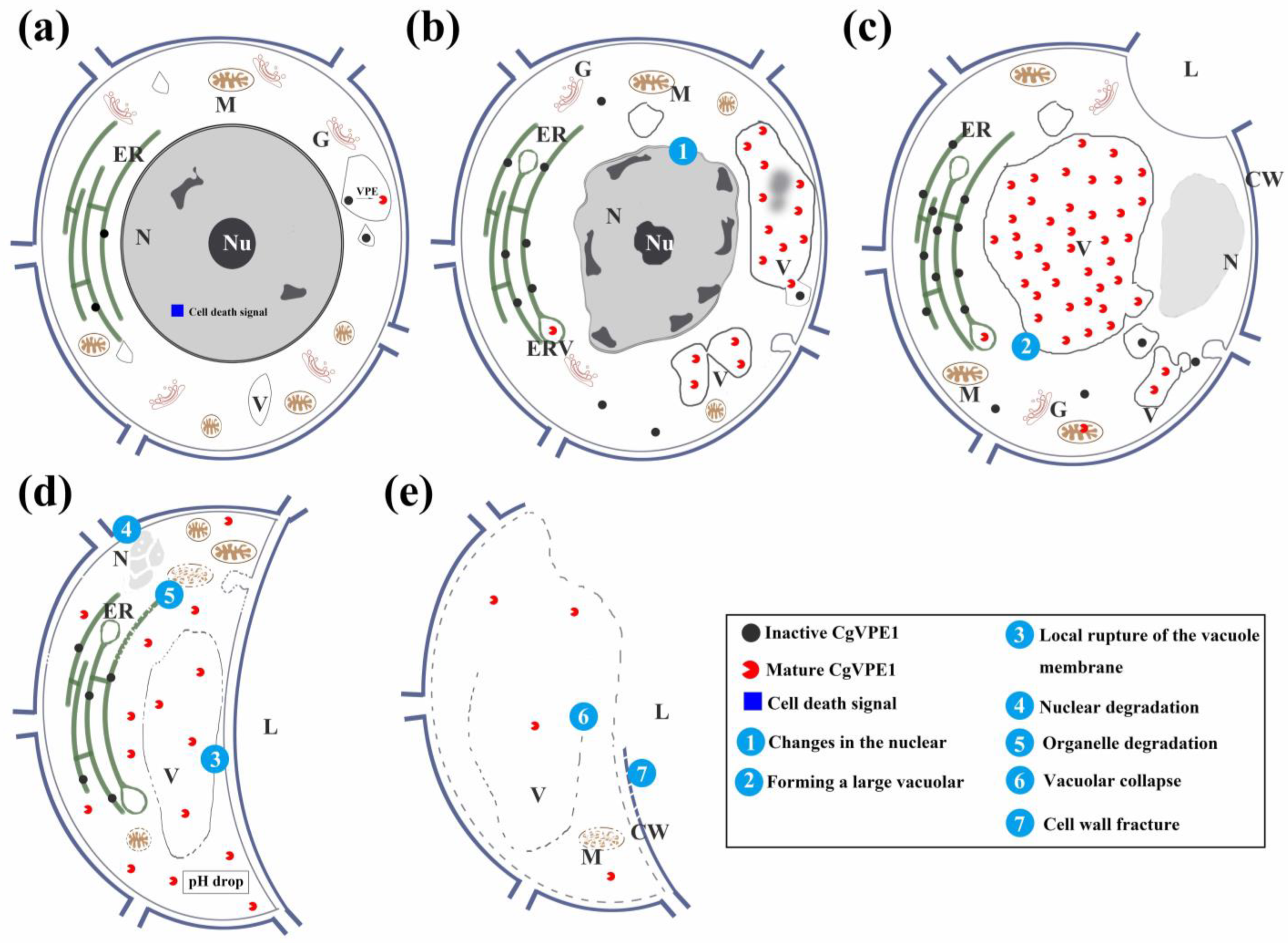
3.2. CgVPE1 Plays a Crucial Role in Tonoplast Collapse during the Developmental PCD of Secretory Cavity Cells in C. grandis ‘Tomentosa’ Fruits
4. Materials and Methods
4.1. Experimental Materials
4.2. Experimental Method
4.2.1. Transmission Electron Microscopy (TEM)
4.2.2. qRT-PCR Analyses
4.2.3. Cloning and Sequence Analysis of CgVPE1 cDNAs
4.2.4. In Situ Hybridization (ISH)
4.2.5. Western Blot
4.2.6. Immunofluorescence Localization of CgVPE1
4.2.7. Immunocytochemical Localization
4.2.8. Expression and Purification of Recombinant Protein
4.2.9. Enzyme Assays
4.2.10. Self-Activation Assay
4.2.11. Statistic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Hautegem, T.; Waters, A.J.; Goodrich, J.; Nowack, M.K. Only in dying, life: Programmed cell death during plant development. Trends Plant Sci. 2015, 20, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Reape, T.J.; McCabe, P.F. Apoptotic-like regulation of programmed cell death in plants. Apoptosis 2010, 15, 249–256. [Google Scholar] [CrossRef]
- Watanabe, N.; Lam, E. Programmed cell death in plants: Apoptotic but not quite. In Essentials of Apoptosis; Humana Press: Totowa, NJ, USA, 2009; pp. 301–321. [Google Scholar]
- Hara-Nishimura, I.; Hatsugai, N. The role of vacuole in plant cell death. Cell Death Differ. 2011, 18, 1298–1304. [Google Scholar] [CrossRef] [Green Version]
- Kuroyanagi, M.; Yamada, K.; Hatsugai, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 2005, 280, 32914–32920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakaune, S.; Yamada, K.; Kondo, M.; Kato, T.; Tabata, S.; Nishimura, M.; Hara-Nishimura, I. A vacuolar processing enzyme, ΔVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell 2005, 17, 876–887. [Google Scholar] [CrossRef] [Green Version]
- Kuriyama, H.; Fukuda, H. Developmental programmed cell death in plants. Curr. Opin. Plant Biol. 2002, 5, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Kuroyanagi, M.; Yamada, K.; Meshi, T.; Tsuda, S.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 2004, 305, 855–858. [Google Scholar] [CrossRef]
- Abdullah, W.M.A.N.W.; Saidi, N.B.; Yusof, M.T.; Wee, C.-Y.; Loh, H.-S.; Ong-Abdullah, J.; Lai, K.-S. Vacuolar processing enzymes modulating susceptibility response to Fusarium oxysporum f. sp. Cubense Tropical Race 4 infections in Banana. Front. Plant Sci. 2022, 12, 769855. [Google Scholar] [CrossRef]
- Hara-Nishimura, I.; Inoue, K.; Nishimura, M. A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett. 1991, 294, 89–93. [Google Scholar] [CrossRef] [Green Version]
- Hatsugai, N.; Kuroyanagi, M.; Nishimura, M.; Hara-Nishimura, I. A cellular suicide strategy of plants: Vacuole-mediated cell death. Apoptosis 2006, 11, 905–911. [Google Scholar] [CrossRef]
- Hiraiwa, N.; Nishimura, M.; Hara-Nishimura, I. Vacuolar processing enzyme is self-catalytically activated by sequential removal of the C-terminal and N-terminal propeptides. FEBS Lett. 1999, 447, 213–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara-Nishimura, I.; Shimada, T.; Hiraiwa, N.; Nishimura, M. Vacuolar processing enzyme responsible for maturation of seed proteins. J. Plant Physiol. 1995, 145, 632–640. [Google Scholar] [CrossRef]
- Yamada, K.; Shimada, T.; Nishimura, M.; Hara-Nishimura, I. A VPE family supporting various vacuolar functions in plants. Physiol. Plant. 2005, 123, 369–375. [Google Scholar] [CrossRef]
- Vorster, B.J.; Cullis, C.A.; Kunert, K.J. Plant vacuolar processing enzymes. Front. Plant Sci. 2019, 10, 479. [Google Scholar] [CrossRef] [Green Version]
- Julián, I.; Gandullo, J.; Santos-Silva, L.K.; Diaz, I.; Martinez, M. Phylogenetically distant barley legumains have a role in both seed and vegetative tissues. J. Exp. Bot. 2013, 64, 2929–2941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatsugai, N.; Yamada, K.; Goto-Yamada, S.; Hara-Nishimura, I. Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 2015, 6, 234. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Cai, J.; Wang, P.; Tian, S.; Qin, G. Post-transcriptional regulation of fruit ripening and disease resistance in tomato by the vacuolar protease SlVPE3. Genome Biol. 2017, 18, 47. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Wang, X.; Tian, J.; Zhang, X.; Yu, T.; Li, Y.; Li, D. Genome-wide analysis of VPE family in four Gossypium species and transcriptional expression of VPEs in the upland cotton seedlings under abiotic stresses. Funct. Integr. Genom. 2022, 22, 179–192. [Google Scholar] [CrossRef]
- Mino, M.; Murata, N.; Date, S.; Inoue, M. Cell death in seedlings of the interspecific hybrid of Nicotiana gossei and N. tabacum; possible role of knob-like bodies formed on tonoplast in vacuolar-collapse-mediated cell death. Plant Cell Rep. 2007, 26, 407–419. [Google Scholar] [CrossRef]
- Teper-Bamnolker, P.; Buskila, Y.; Lopesco, Y.; Ben-Dor, S.; Saad, I.; Holdengreber, V.; Belausov, E.; Zemach, H.; Ori, N.; Lers, A.; et al. Release of apical dominance in potato tuber is accompanied by programmed cell death in the apical bud meristem. Plant Physiol. 2012, 158, 2053–2067. [Google Scholar] [CrossRef] [Green Version]
- Misas-Villamil, J.C.; Toenges, G.; Kolodziejek, I.; Sadaghiani, A.M.; Kaschani, F.; Colby, T.; Bogyo, M.; van der Hoorn, R.A. Activity profiling of vacuolar processing enzymes reveals a role for VPE during oomycete infection. Plant J. 2013, 73, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Qiang, X.; Zechmann, B.; Reitz, M.U.; Kogel, K.-H.; Schäfer, P. The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress–triggered caspase-dependent cell death. Plant Cell 2012, 24, 794–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, G.C.; Reis, P.A.B.; Calil, I.P.; Carvalho, H.H.; Aragão, F.J.L.; Fontes, E.P.B. GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. USA 2013, 110, 19627–19632. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yue, H.; Xing, D. MAP Kinase 6-mediated activation of vacuolar processing enzyme modulates heat shock-induced programmed cell death in Arabidopsis. New Phytol. 2012, 195, 85–96. [Google Scholar] [CrossRef]
- Deng, M.; Bian, H.; Xie, Y.; Kim, Y.; Wang, W.; Lin, E.; Zeng, Z.; Guo, F.; Pan, J.; Han, N.; et al. Bcl-2 suppresses hydrogen peroxide-induced programmed cell death via OsVPE2 and OsVPE3, but not via OsVPE1 and OsVPE4, in rice. FEBS J. 2011, 278, 4797–4810. [Google Scholar] [CrossRef]
- Liang, S.; Wang, H.; Yang, M.; Wu, H. Sequential actions of pectinases and cellulases during secretory cavity formation in Citrus fruits. Trees-Struct. Funct. 2009, 23, 19–27. [Google Scholar] [CrossRef]
- Tong, P.; Huai, B.; Chen, Y.; Bai, M.; Wu, H. CisPG21 and CisCEL16 are involved in the regulation of the degradation of cell walls during secretory cavity cell programmed cell death in the fruits of Citrus sinensis (L.) Osbeck. Plant Sci. 2020, 297, 110540. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, H. Programmed cell death involved in the schizolysigenous formation of the secretory cavity in Citrus sinensis L. (Osbeck). Chin. Sci. Bull. 2010, 55, 2160–2168. [Google Scholar] [CrossRef]
- Liu, P.; Liang, S.; Yao, N.; Wu, H. Programmed cell death of secretory cavity cells in fruits of Citrus grandis cv. Tomentosa is associated with activation of caspase 3-like protease. Trees-Struct. Funct. 2012, 26, 1821–1835. [Google Scholar] [CrossRef]
- Zheng, P.; Bai, M.; Chen, Y.; Liu, P.W.; Gao, L.; Liang, S.J.; Wu, H. Programmed cell death of secretory cavity cells of Citrus fruits is associated with Ca2+ accumulation in the nucleus. Trees-Struct. Funct. 2014, 28, 1137–1144. [Google Scholar] [CrossRef]
- Bai, M.; Liang, M.; Huai, B.; Gao, H.; Tong, P.; Shen, R.; He, H.; Wu, H. Ca2+-dependent nuclease is involved in DNA degradation during the formation of the secretory cavity by programmed cell death in fruit of Citrus grandis ‘Tomentosa’. J. Exp. Bot. 2020, 71, 4812–4827. [Google Scholar] [CrossRef] [PubMed]
- Huai, B.; Bai, M.; Tong, P.; He, H.; Liang, M.; Chen, C.; Wu, H. CgPBA1 may be involved in nuclear degradation during secretory cavity formation by programmed cell death in Citrus grandis ‘Tomentosa’ fruits. Plant Physiol. Biochem. 2021, 160, 306–314. [Google Scholar] [CrossRef]
- Liang, M.J.; Bai, M.; Wu, H. Zn2+ dependent nuclease is involved in nuclear degradation during the programmed cell death of secretory cavity formation in Citrus grandis ‘Tomentosa’ fruits. Cells 2021, 10, 3222. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Granell, A. A Putative vacuolar processing protease is regulated by ethylene and also during fruit ripening in Citrus fruit. Plant Physiol. 1995, 109, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Huai, B.; Liang, M.J.; Bai, M.; He, H.J.; Chen, J.Z.; Wu, H. Localization of CgVPE1 in secondary cell wall formation during tracheary element differentiation in the pericarp of Citrus grandis ‘Tomentosa’ fruits. Planta 2022, 256, 89. [Google Scholar] [CrossRef]
- Kuroyanagi, M.; Nishimura, M.; Hara-Nishimura, I. Activation of Arabidopsis vacuolar processing enzyme by self-catalytic removal of an auto-inhibitory domain of the C-terminal propeptide. Plant Cell Physiol. 2002, 43, 143–151. [Google Scholar] [CrossRef] [Green Version]
- del Pozo, O.; Lam, E. Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr. Biol. 1998, 8, 1129–1132. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Wang, M.; Bai, Y.; Zeng, Z.; Guo, F.; Han, N.; Bian, H.; Wang, J.; Pan, J.; Zhu, M. Bcl-2 suppresses activation of VPEs by inhibiting cytosolic Ca2+ level with elevated K+ effluxin NaCl-induced PCD in rice. Plant Physiol. Biochem. 2014, 80, 168–175. [Google Scholar] [CrossRef]
- Kariya, K.; Demiral, T.; Sasaki, T.; Tsuchiya, Y.; Turkan, I.; Sano, T.; Hasezawa, S.; Yamamoto, Y. A novel mechanism of aluminium-induced cell death involving vacuolar processing enzyme and vacuolar collapse in tobacco cell line BY-2. J. Inorg. Biochem. 2013, 128, 196–201. [Google Scholar] [CrossRef]
- Kinoshita, T.; Nishimura, M.; Hara-Nishimura, I. Homologues of a vacuolar processing enzyme that are expressed in different organs in Arabidopsis thaliana. Plant Mol. Biol. 1995, 29, 81–89. [Google Scholar] [CrossRef]
- Kinoshita, T.; Nishimura, M.; Hara-Nishimura, I. The sequence and expression of the γ-VPE gene, one member of a family of three genes for vacuolar processing enzymes in Arabidopsis thaliana. Plant Cell Physiol. 1995, 36, 1555–1562. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Yamada, K.; Hiraiwa, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J. 1999, 19, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Hara-Nishimura, I.; Kinoshita, T.; Hiraiwa, N.; Nishimura, M. Vacuolar processing enzymes in protein-storage vacuoles and lytic vacuoles. J. Plant Physiol. 1998, 152, 668–674. [Google Scholar] [CrossRef]
- Vaux, D.L.; Korsmeyer, S.J. Cell death in development. Cell 1999, 96, 245–254. [Google Scholar] [CrossRef] [Green Version]
- Danon, A.; Delorme, V.; Mailhac, N.; Gallois, P. Plant programmed cell death: A common way to die. Plant Physiol. Biochem. 2000, 38, 647–655. [Google Scholar] [CrossRef]
- Hara-Nishimura, I.; Hatsugai, N.; Nakaune, S.; Kuroyanagi, M.; Nishimura, M. Vacuolar processing enzyme: An executor of plant cell death. Curr. Opin. Plant Biol. 2005, 8, 404–408. [Google Scholar] [CrossRef]
- Jones, A.M. Programmed cell death in development and defense. Plant Physiol. 2001, 125, 95–97. [Google Scholar] [CrossRef] [Green Version]
- Hiraiwa, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. An aspartic proteinase is involved in the maturation of storage proteins in concert with the vacuolar processing enzyme. Eur. J. Biochem. 1997, 246, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Wada, Y. Vacuoles in mammals: A subcellular structure indispensable for early embryogenesis. Bioarchitecture 2013, 3, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, D. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999, 6, 1028–1042. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Basak, A.K.; Goto-Yamada, S.; Tarnawska-Glatt, K.; Hara-Nishimura, I. Vacuolar processing enzymes in the plant life cycle. New Phytol. 2020, 226, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Guo, X.; Zhang, J.; Liu, Y.; Wang, B.; Li, H.; Lu, H. βVPE is involved in tapetal degradation and pollen development by activating proprotease maturation in Arabidopsis thaliana. J. Exp. Bot. 2019, 71, 1943–1955. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, J.; Yin, B.; Liu, Y.; Wang, B.; Li, H.; Lu, H. γVPE plays an important role in programmed cell death for xylem fiber cells by activating protease CEP1 maturation in Arabidopsis thaliana. Int. J. Biol. Macro-Mol. 2019, 137, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Van Durme, M.; Nowack, M.K. Mechanisms of developmentally controlled cell death in plants. Curr. Opin. Plant Biol. 2016, 29, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Zauner, F.B.; Dall, E.; Regl, C.; Grassi, L.; Huber, C.G.; Cabrele, C.; Brandstetter, H. Crystal structure of plant legumain reveals a unique two-chain state with pH-dependent activity regulation. Plant Cell 2018, 30, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Bosch, M.; Poulter, N.S.; Perry, R.M.; Wilkins, K.A.; Franklin-Tong, V.E. Characterization of a legumain/vacuolar processing enzyme and YVADase activity in Papaver pollen. Plant Mol. Biol. 2010, 74, 381–393. [Google Scholar] [CrossRef]
- Teper-Bamnolker, P.; Buskila, Y.; Belausov, E.; Wolf, D.; Doron-Faigenboim, A.; Ben-Dor, S.; Van der Hoorn, R.A.; Lers, A.; Eshel, D. Vacuolar processing enzyme activates programmed cell death in the apical meristem inducing loss of apical dominance. Plant Cell Environ. 2017, 40, 2381–2392. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huai, B.; Liang, M.; Lin, J.; Tong, P.; Bai, M.; He, H.; Liang, X.; Chen, J.; Wu, H. Involvement of Vacuolar Processing Enzyme CgVPE1 in Vacuole Rupture in the Programmed Cell Death during the Development of the Secretory Cavity in Citrus grandis ‘Tomentosa’ Fruits. Int. J. Mol. Sci. 2023, 24, 11681. https://doi.org/10.3390/ijms241411681
Huai B, Liang M, Lin J, Tong P, Bai M, He H, Liang X, Chen J, Wu H. Involvement of Vacuolar Processing Enzyme CgVPE1 in Vacuole Rupture in the Programmed Cell Death during the Development of the Secretory Cavity in Citrus grandis ‘Tomentosa’ Fruits. International Journal of Molecular Sciences. 2023; 24(14):11681. https://doi.org/10.3390/ijms241411681
Chicago/Turabian StyleHuai, Bin, Minjian Liang, Junjun Lin, Panpan Tong, Mei Bai, Hanjun He, Xiangxiu Liang, Jiezhong Chen, and Hong Wu. 2023. "Involvement of Vacuolar Processing Enzyme CgVPE1 in Vacuole Rupture in the Programmed Cell Death during the Development of the Secretory Cavity in Citrus grandis ‘Tomentosa’ Fruits" International Journal of Molecular Sciences 24, no. 14: 11681. https://doi.org/10.3390/ijms241411681
APA StyleHuai, B., Liang, M., Lin, J., Tong, P., Bai, M., He, H., Liang, X., Chen, J., & Wu, H. (2023). Involvement of Vacuolar Processing Enzyme CgVPE1 in Vacuole Rupture in the Programmed Cell Death during the Development of the Secretory Cavity in Citrus grandis ‘Tomentosa’ Fruits. International Journal of Molecular Sciences, 24(14), 11681. https://doi.org/10.3390/ijms241411681







