A Transcriptome-Wide Analysis of Psoriasis: Identifying the Potential Causal Genes and Drug Candidates
Abstract
1. Introduction
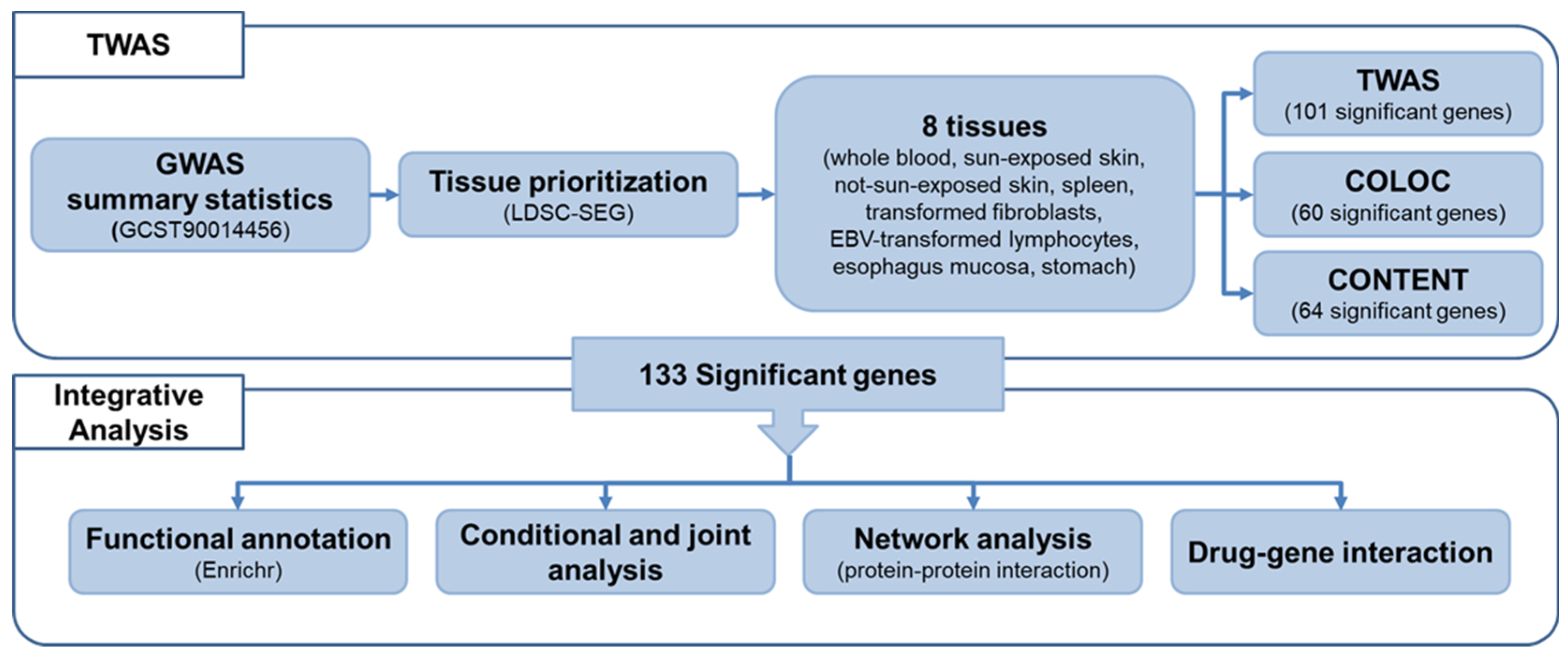
2. Results
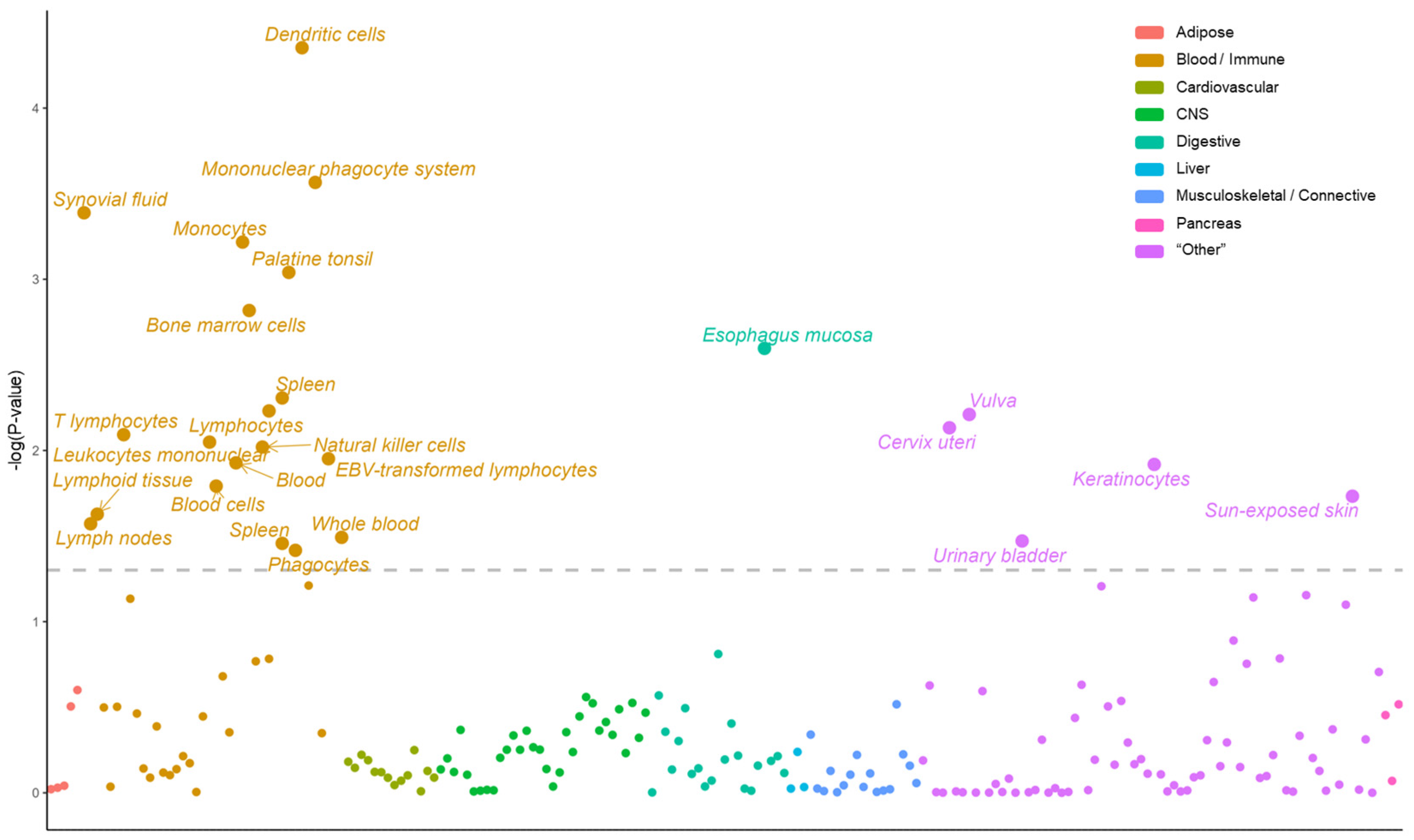
2.1. Prioritizing Genetically Relevant Tissue for Psoriasis
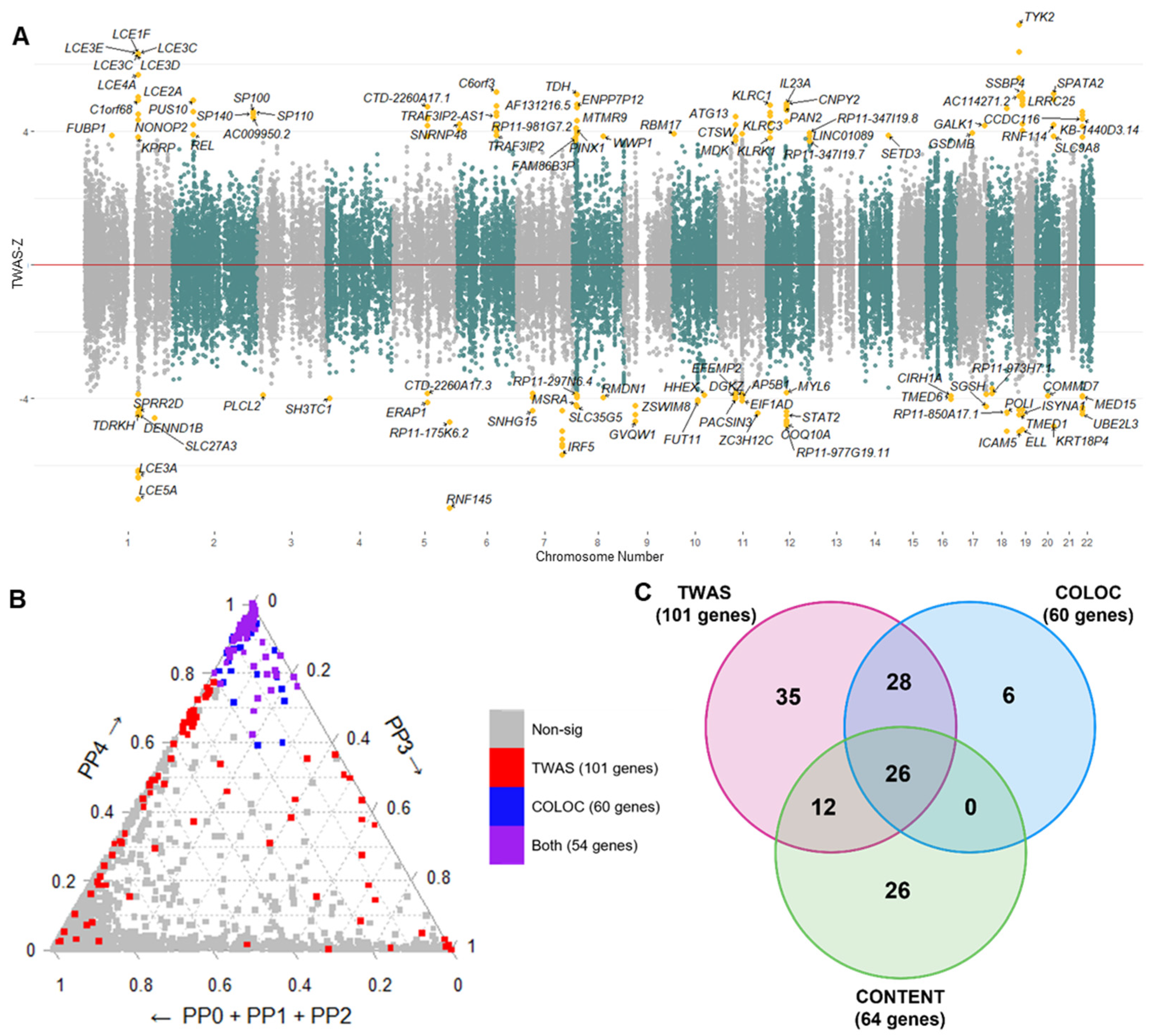
2.2. Transcriptome-Wide Associations for Psoriasis
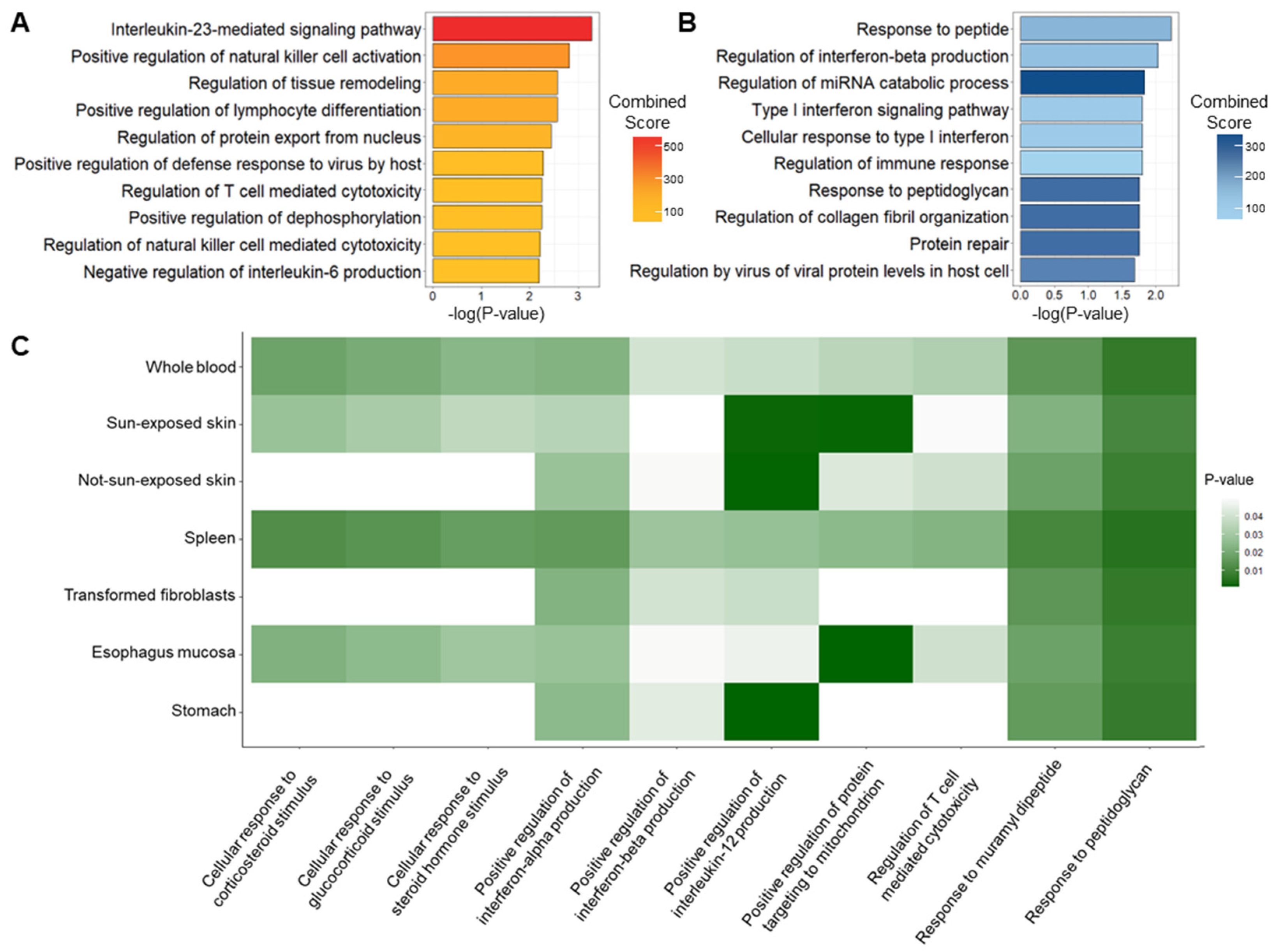
2.3. Biological Enrichment of Genetic Signatures of Psoriasis
2.4. Conditional and Joint Analysis of TWAS Signals
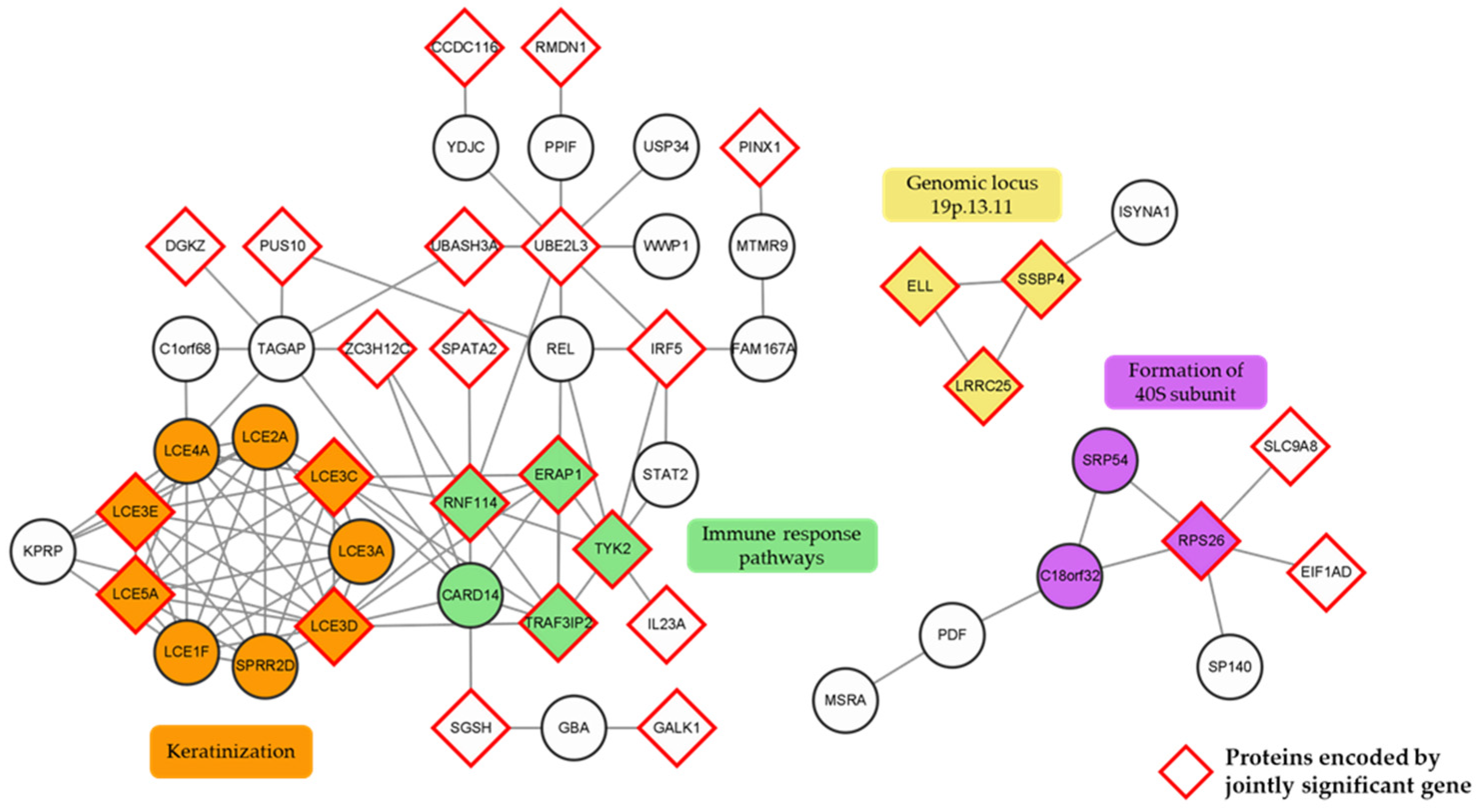
2.5. Protein–Protein Interaction Network Analysis and Cluster Identification
2.6. Potential Drug/Chemical Compound Candidates for Psoriasis
3. Discussion
4. Materials and Methods
4.1. Data Collection and Pre-Processing
4.2. LDSC-SEG
4.3. Transcriptome-Wide Association Analysis
4.4. Colocalization
4.5. Multi-Tissue Signals Using CONTENT
4.6. Functional Annotation of Significant Psoriasis Genes
4.7. Conditional and Joint Analysis
4.8. Network Analysis with Clustering
4.9. Gene–Drug Interaction Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boehncke, W.-H.; Schön, M.P. Psoriasis. Lancet 2015, 386, 983–994. [Google Scholar] [CrossRef]
- Raychaudhuri, S.K.; Maverakis, E.; Raychaudhuri, S.P. Diagnosis and classification of psoriasis. Autoimmun. Rev. 2014, 13, 490–495. [Google Scholar] [CrossRef]
- Parisi, R.; Symmons, D.P.; Griffiths, C.E.; Ashcroft, D.M. Global Epidemiology of Psoriasis: A Systematic Review of Incidence and Prevalence. J. Investig. Dermatol. 2013, 133, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Parisi, R.; Iskandar, I.Y.; Kontopantelis, E.; Augustin, M.; Griffiths, C.E.; Ashcroft, D.M. National, regional, and worldwide epidemiology of psoriasis: Systematic analysis and modelling study. BMJ 2020, 369, m1590. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, K.; Olsen, A.O.; Wilsgaard, T.; Furberg, A.S. Is the prevalence of psoriasis increasing? A 30-year follow-up of a population-based cohort. Br. J. Dermatol. 2013, 168, 1303–1310. [Google Scholar] [CrossRef]
- Rendon, A.; Schäkel, K. Psoriasis Pathogenesis and Treatment. Int. J. Mol. Sci. 2019, 20, 1475. [Google Scholar] [CrossRef]
- Szepietowski, J.C.; Reich, A. Pruritus in psoriasis: An update. Eur. J. Pain 2016, 20, 41–46. [Google Scholar] [CrossRef]
- Teng, Y.; Xie, W.; Tao, X.; Liu, N.; Yu, Y.; Huang, Y.; Xu, D.; Fan, Y. Infection-provoked psoriasis: Induced or aggravated (Review). Exp. Ther. Med. 2021, 21, 567. [Google Scholar] [CrossRef]
- Barrea, L.; Nappi, F.; Di Somma, C.; Savanelli, M.C.; Falco, A.; Balato, A.; Balato, N.; Savastano, S. Environmental Risk Factors in Psoriasis: The Point of View of the Nutritionist. Int. J. Environ. Res. Public Health 2016, 13, 743. [Google Scholar] [CrossRef] [PubMed]
- Farber, E.M.; Nall, M.L.; Watson, W. Natural History of Psoriasis in 61 Twin Pairs. Arch. Dermatol. 1974, 109, 207–211. [Google Scholar] [CrossRef]
- Ran, D.; Cai, M.; Zhang, X. Genetics of psoriasis: A basis for precision medicine. Precis. Clin. Med. 2019, 2, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Huang, W.; Yang, S.; Sun, L.D.; Zhang, F.Y.; Zhu, Q.X.; Zhang, F.R.; Zhang, C.; Du, W.H.; Pu, X.M.; et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 2009, 41, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Stawczyk-Macieja, M.; Rębała, K.; Szczerkowska-Dobosz, A.; Wysocka, J.; Cybulska, L.; Kapińska, E.; Haraś, A.; Miniszewska, P.; Nowicki, R. Evaluation of Psoriasis Genetic Risk Based on Five Susceptibility Markers in a Population from Northern Poland. PLoS ONE 2016, 11, e0163185. [Google Scholar] [CrossRef]
- Bergboer, J.G.; Tjabringa, G.S.; Kamsteeg, M.; van Vlijmen-Willems, I.M.; Rodijk-Olthuis, D.; Jansen, P.A.; Thuret, J.Y.; Narita, M.; Ishida-Yamamoto, A.; Zeeuwen, P.L.; et al. Psoriasis Risk Genes of the Late Cornified Envelope-3 Group Are Distinctly Expressed Compared with Genes of Other LCE Groups. Am. J. Pathol. 2011, 178, 1470–1477. [Google Scholar] [CrossRef] [PubMed]
- Niehues, H.; Tsoi, L.C.; van der Krieken, D.A.; Jansen, P.A.; Oortveld, M.A.; Rodijk-Olthuis, D.; van Vlijmen, I.M.; Hendriks, W.J.; Helder, R.W.; Bouwstra, J.A.; et al. Psoriasis-Associated Late Cornified Envelope (LCE) Proteins Have Antibacterial Activity. J. Investig. Dermatol. 2017, 137, 2380–2388. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Chandran, V. The Genetics of Psoriasis and Psoriatic Arthritis. Clin. Rev. Allergy Immunol. 2013, 44, 149–156. [Google Scholar] [CrossRef]
- Elder, J.T. Genome-wide association scan yields new insights into the immunopathogenesis of psoriasis. Genes Immun. 2009, 10, 201–209. [Google Scholar] [CrossRef]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.; Jansen, R.; De Geus, E.J.; Boomsma, D.I.; Wright, F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef]
- Thompson, M.; Gordon, M.G.; Lu, A.; Tandon, A.; Halperin, E.; Gusev, A.; Ye, C.J.; Balliu, B.; Zaitlen, N. Multi-context genetic modeling of transcriptional regulation resolves novel disease loci. Nat. Commun. 2022, 13, 5704. [Google Scholar] [CrossRef]
- Hu, Y.; Li, M.; Lu, Q.; Weng, H.; Wang, J.; Zekavat, S.M.; Yu, Z.; Li, B.; Gu, J.; Muchnik, S.; et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 2019, 51, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Gusev, A.; Mancuso, N.; Won, H.; Kousi, M.; Finucane, H.K.; Reshef, Y.; Song, L.; Safi, A.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; McCarroll, S.; et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018, 50, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Valette, K.; Li, Z.; Bon-Baret, V.; Chignon, A.; Bérubé, J.C.; Eslami, A.; Lamothe, J.; Gaudreault, N.; Joubert, P.; Obeidat, M.E.; et al. Prioritization of candidate causal genes for asthma in susceptibility loci derived from UK Biobank. Commun. Biol. 2021, 4, 700. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Tan, S.; Liu, L.; Cheng, S.; Li, P.; Li, W.; Liu, H.; Zhang, F.E.; Wang, S.; Ning, Y.; et al. Transcriptome-wide association study identifies susceptibility genes for rheumatoid arthritis. Arthritis Res. Ther. 2021, 23, 38. [Google Scholar] [CrossRef]
- Díez-Obrero, V.; Moratalla-Navarro, F.; Ibáñez-Sanz, G.; Guardiola, J.; Rodríguez-Moranta, F.; Obón-Santacana, M.; Díez-Villanueva, A.; Dampier, C.H.; Devall, M.; Carreras-Torres, R.; et al. Transcriptome-Wide Association Study for Inflammatory Bowel Disease Reveals Novel Candidate Susceptibility Genes in Specific Colon Subsites and Tissue Categories. J. Crohn’s Colitis 2022, 16, 275–285. [Google Scholar] [CrossRef]
- Glanville, K.P.; Coleman, J.R.; O’Reilly, P.F.; Galloway, J.; Lewis, C.M. Investigating Pleiotropy Between Depression and Autoimmune Diseases Using the UK Biobank. Biol. Psychiatry Glob. Open Sci. 2021, 1, 48–58. [Google Scholar] [CrossRef]
- Finucane, H.K.; Reshef, Y.A.; Anttila, V.; Slowikowski, K.; Gusev, A.; Byrnes, A.; Gazal, S.; Loh, P.R.; Lareau, C.; Shoresh, N.; et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018, 50, 621–629. [Google Scholar] [CrossRef]
- Wainberg, M.; Sinnott-Armstrong, N.; Mancuso, N.; Barbeira, A.N.; Knowles, D.A.; Golan, D.; Ermel, R.; Ruusalepp, A.; Quertermous, T.; Hao, K.; et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019, 51, 592–599. [Google Scholar] [CrossRef]
- GTEx Consortium; Ardlie, K.G.; Deluca, D.S.; Segrè, A.V.; Sullivan, T.J.; Young, T.R.; Gelfand, E.T.; Trowbridge, C.A.; Maller, J.B.; Tukiainen, T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar]
- Fehrmann, R.S.; Karjalainen, J.M.; Krajewska, M.; Westra, H.J.; Maloney, D.; Simeonov, A.; Pers, T.H.; Hirschhorn, J.N.; Jansen, R.C.; Schultes, E.A.; et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 2015, 47, 115–125. [Google Scholar] [CrossRef]
- Pers, T.H.; Karjalainen, J.M.; Chan, Y.; Westra, H.J.; Wood, A.R.; Yang, J.; Lui, J.C.; Vedantam, S.; Gustafsson, S.; Esko, T.; et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015, 6, 5890. [Google Scholar] [CrossRef]
- Hägg, D.; Sundström, A.; Eriksson, M.; Schmitt-Egenolf, M. Severity of Psoriasis Differs Between Men and Women: A Study of the Clinical Outcome Measure Psoriasis Area and Severity Index (PASI) in 5438 Swedish Register Patients. Am. J. Clin. Dermatol. 2017, 18, 583–590. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Bordas; García, P.; Puig, S.; Pujol, R.; Smandía. Prevalence of psoriasis in Spain (Epiderma Project: Phase I). J. Eur. Acad. Dermatol. Venereol. 2001, 15, 20–23. [Google Scholar] [CrossRef]
- Gelfand, J.M.; Weinstein, R.; Porter, S.B.; Neimann, A.L.; Berlin, J.A.; Margolis, D.J. Prevalence and Treatment of Psoriasis in the United Kingdom: A Population-Based Study. Arch. Dermatol. 2005, 141, 1537–1541. [Google Scholar] [CrossRef]
- Casciano, F.; Pigatto, P.D.; Secchiero, P.; Gambari, R.; Reali, E. T Cell Hierarchy in the Pathogenesis of Psoriasis and Associated Cardiovascular Comorbidities. Front. Immunol. 2018, 9, 1390. [Google Scholar] [CrossRef]
- Diani, M.; Casciano, F.; Marongiu, L.; Longhi, M.; Altomare, A.; Pigatto, P.D.; Secchiero, P.; Gambari, R.; Banfi, G.; Manfredi, A.A.; et al. Increased frequency of activated CD8+ T cell effectors in patients with psoriatic arthritis. Sci. Rep. 2019, 9, 10870. [Google Scholar] [CrossRef]
- Chan, T.C.; Hawkes, J.E.; Krueger, J.G. Interleukin 23 in the skin: Role in psoriasis pathogenesis and selective interleukin 23 blockade as treatment. Ther. Adv. Chronic Dis. 2018, 9, 111–119. [Google Scholar] [CrossRef]
- Puig, L. The role of IL 23 in the treatment of psoriasis. Expert Rev. Clin. Immunol. 2017, 13, 525–534. [Google Scholar] [CrossRef]
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and Gut Microbiome in Psoriasis: Gaining Insight into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front. Microbiol. 2020, 11, 589726. [Google Scholar] [CrossRef]
- Benhadou, F.; Mintoff, D.; Schnebert, B.; Thio, H.B. Psoriasis and Microbiota: A Systematic Review. Diseases 2018, 6, 47. [Google Scholar] [CrossRef]
- Ergen, E.N.; Yusuf, N. Inhibition of interleukin-12 and/or interleukin-23 for the treatment of psoriasis: What is the evidence for an effect on malignancy? Exp. Dermatol. 2018, 27, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Stringer, S.; Frei, O.; Umićević Mirkov, M.; de Leeuw, C.; Polderman, T.J.C.; van der Sluis, S.; Andreassen, O.A.; Neale, B.M.; Posthuma, D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019, 51, 1339–1348. [Google Scholar] [CrossRef]
- Schön, M.P. Adaptive and Innate Immunity in Psoriasis and Other Inflammatory Disorders. Front. Immunol. 2019, 10, 1764. [Google Scholar] [CrossRef] [PubMed]
- Lowes, M.A.; Suárez-Fariñas, M.; Krueger, J.G. Immunology of psoriasis. Annu. Rev. Immunol. 2014, 32, 227–255. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Zhu, Y.-j.; Zou, S.; Zhou, P.; Hu, Y.-w.; Zhao, Q.-x.; Gu, L.-n.; Zhang, H.-z.; Wang, Z.; Li, J. Metabolic Syndrome and Psoriasis: Mechanisms and Future Directions. Front. Immunol. 2021, 12, 711060. [Google Scholar] [CrossRef]
- Gisondi, P.; Fostini, A.C.; Fossà, I.; Girolomoni, G.; Targher, G. Psoriasis and the metabolic syndrome. Clin. Dermatol. 2018, 36, 21–28. [Google Scholar] [CrossRef]
- Saalbach, A.; Kunz, M. Impact of Chronic Inflammation in Psoriasis on Bone Metabolism. Front. Immunol. 2022, 13, 925503. [Google Scholar] [CrossRef]
- Ferreira, B.I.; Abreu, J.L.; Reis, J.P.; Figueiredo, A.M. Psoriasis and Associated Psychiatric Disorders: A Systematic Review on Etiopathogenesis and Clinical Correlation. J. Clin. Aesthet. Dermatol. 2016, 9, 36–43. [Google Scholar]
- Pietrzak, D.; Pietrzak, A.; Krasowska, D.; Borzęcki, A.; Franciszkiewicz-Pietrzak, K.; Polkowska-Pruszyńska, B.; Baranowska, M.; Reich, K. Digestive system in psoriasis: An update. Arch. Dermatol. Res. 2017, 309, 679–693. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef]
- Ishida-Yamamoto, A.; Iizuka, H. Structural organization of cornified cell envelopes and alterations in inherited skin disorders. Exp. Dermatol. 1998, 7, 1–10. [Google Scholar] [CrossRef]
- Tian, S.; Chen, S.; Feng, Y.; Li, Y. The Interactions of Small Proline-Rich Proteins with Late Cornified Envelope Proteins are Involved in the Pathogenesis of Psoriasis. Clin. Cosmet. Investig. Dermatol. 2021, 14, 1355–1365. [Google Scholar] [CrossRef]
- Kvist-Hansen, A.; Hansen, P.R.; Skov, L. Systemic Treatment of Psoriasis with JAK Inhibitors: A Review. Dermatol. Ther. 2020, 10, 29–42. [Google Scholar] [CrossRef]
- Goldminz, A.M.; Au, S.C.; Kim, N.; Gottlieb, A.B.; Lizzul, P.F. NF-κB: An essential transcription factor in psoriasis. J. Dermatol. Sci. 2013, 69, 89–94. [Google Scholar] [CrossRef]
- Liao, Y.H.; Jee, S.H.; Sheu, B.C.; Huang, Y.L.; Tseng, M.P.; Hsu, S.M.; Tsai, T.F. Increased expression of the natural killer cell inhibitory receptor CD94/NKG2A and CD158b on circulating and lesional T cells in patients with chronic plaque psoriasis. Br. J. Dermatol. 2006, 155, 318–324. [Google Scholar] [CrossRef]
- Batista, M.D.; Ho, E.L.; Kuebler, P.J.; Milush, J.M.; Lanier, L.L.; Kallas, E.G.; York, V.A.; Chang, D.; Liao, W.; Unemori, P.; et al. Skewed distribution of natural killer cells in psoriasis skin lesions. Exp. Dermatol. 2013, 22, 64–66. [Google Scholar] [CrossRef]
- van Hall, T.; André, P.; Horowitz, A.; Ruan, D.F.; Borst, L.; Zerbib, R.; Narni-Mancinelli, E.; van der Burg, S.H.; Vivier, E. Monalizumab: Inhibiting the novel immune checkpoint NKG2A. J. ImmunoTherapy Cancer 2019, 7, 263. [Google Scholar] [CrossRef]
- Song, J.; Kim, D.; Lee, S.; Jung, J.; Joo, J.W.J.; Jang, W. Integrative transcriptome-wide analysis of atopic dermatitis for drug repositioning. Commun. Biol. 2022, 5, 615. [Google Scholar] [CrossRef]
- Kim, G.; Jang, G.; Song, J.; Kim, D.; Lee, S.; Joo, J.W.J.; Jang, W. A transcriptome-wide association study of uterine fibroids to identify potential genetic markers and toxic chemicals. PLoS ONE 2022, 17, e0274879. [Google Scholar] [CrossRef]
- Liao, C.; Laporte, A.D.; Spiegelman, D.; Akçimen, F.; Joober, R.; Dion, P.A.; Rouleau, G.A. Transcriptome-wide association study of attention deficit hyperactivity disorder identifies associated genes and phenotypes. Nat. Commun. 2019, 10, 4450. [Google Scholar] [CrossRef]
- Gusev, A.; Lawrenson, K.; Lin, X.; Lyra, P.C.; Kar, S.; Vavra, K.C.; Segato, F.; Fonseca, M.A.S.; Lee, J.M.; Pejovic, T.; et al. A transcriptome-wide association study of high-grade serous epithelial ovarian cancer identifies new susceptibility genes and splice variants. Nat. Genet. 2019, 51, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Barbeira, A.N.; Dickinson, S.P.; Bonazzola, R.; Zheng, J.; Wheeler, H.E.; Torres, J.M.; Torstenson, E.S.; Shah, K.P.; Garcia, T.; Edwards, T.L.; et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018, 9, 1825. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Yao, S.; Wu, H.; Ke, X.; Zhou, X.; Geng, S.; Dong, S.; Chen, H.; Yang, T.; Cheng, Y.; et al. A transcriptome-wide association study identifies novel susceptibility genes for psoriasis. Hum. Mol. Genet. 2021, 31, 300–308. [Google Scholar] [CrossRef]
- Balato, N.; Napolitano, M.; Ayala, F.; Patruno, C.; Megna, M.; Tarantino, G. Nonalcoholic fatty liver disease, spleen and psoriasis: New aspects of low-grade chronic inflammation. World J. Gastroenterol. 2015, 21, 6892–6897. [Google Scholar] [CrossRef] [PubMed]
- Gisondi, P.; Del Giglio, M.; Cozzi, A.; Girolomoni, G. Psoriasis, the liver, and the gastrointestinal tract. Dermatol. Ther. 2010, 23, 155–159. [Google Scholar] [CrossRef]
- Cai, Y.; Fleming, C.; Yan, J. New insights of T cells in the pathogenesis of psoriasis. Cell. Mol. Immunol. 2012, 9, 302–309. [Google Scholar] [CrossRef]
- Polese, B.; Zhang, H.; Thurairajah, B.; King, I.L. Innate Lymphocytes in Psoriasis. Front. Immunol. 2020, 11, 242. [Google Scholar] [CrossRef]
- Krueger, J.G.; Bowcock, A. Psoriasis pathophysiology: Current concepts of pathogenesis. Ann. Rheum. Dis. 2005, 64 (Suppl. S2), ii30. [Google Scholar] [CrossRef]
- He, Y.; Chhetri, S.B.; Arvanitis, M.; Srinivasan, K.; Aguet, F.; Ardlie, K.G.; Barbeira, A.N.; Bonazzola, R.; Im, H.K.; Brown, C.D.; et al. sn-spMF: Matrix factorization informs tissue-specific genetic regulation of gene expression. Genome Biol. 2020, 21, 235. [Google Scholar] [CrossRef]
- Baliwag, J.; Barnes, D.H.; Johnston, A. Cytokines in psoriasis. Cytokine 2015, 73, 342–350. [Google Scholar] [CrossRef]
- Jitlada, M.; Urairack, S.; Mayumi, K.; Mamitaro, O. Pathogenic Role of Cytokines and Effect of Their Inhibition in Psoriasis, in Psoriasis; Anca, C., Ed.; IntechOpen: Rijeka, Croatia, 2017; Chapter 3. [Google Scholar]
- Brembilla, N.C.; Senra, L.; Boehncke, W.-H. The IL-17 Family of Cytokines in Psoriasis: IL-17A and Beyond. Front. Immunol. 2018, 9, 1682. [Google Scholar] [CrossRef]
- Zhang, L.J. Type1 Interferons Potential Initiating Factors Linking Skin Wounds with Psoriasis Pathogenesis. Front. Immunol. 2019, 10, 1440. [Google Scholar] [CrossRef]
- Dhuriya, Y.K.; Sharma, D. Necroptosis: A regulated inflammatory mode of cell death. J. Neuroinflamm. 2018, 15, 199. [Google Scholar] [CrossRef]
- Seo, J.; Nam, Y.W.; Kim, S.; Oh, D.-B.; Song, J. Necroptosis molecular mechanisms: Recent findings regarding novel necroptosis regulators. Exp. Mol. Med. 2021, 53, 1007–1017. [Google Scholar] [CrossRef]
- Wei, R.; Xu, L.W.; Liu, J.; Li, Y.; Zhang, P.; Shan, B.; Lu, X.; Qian, L.; Wu, Z.; Dong, K.; et al. SPATA2 regulates the activation of RIPK1 by modulating linear ubiquitination. Genes Dev. 2017, 31, 1162–1176. [Google Scholar] [CrossRef]
- Yuan, J.; Amin, P.; Ofengeim, D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 2019, 20, 19–33. [Google Scholar] [CrossRef]
- Liu, L.; Tang, Z.; Zeng, Y.; Liu, Y.; Zhou, L.; Yang, S.; Wang, D. Role of necroptosis in infection-related, immune-mediated, and autoimmune skin diseases. J. Dermatol. 2021, 48, 1129–1138. [Google Scholar] [CrossRef]
- Duan, X.; Liu, X.; Liu, N.; Huang, Y.; Jin, Z.; Zhang, S.; Ming, Z.; Chen, H. Inhibition of keratinocyte necroptosis mediated by RIPK1/RIPK3/MLKL provides a protective effect against psoriatic inflammation. Cell Death Dis. 2020, 11, 134. [Google Scholar] [CrossRef]
- Khoury, M.K.; Gupta, K.; Franco, S.R.; Liu, B. Necroptosis in the Pathophysiology of Disease. Am. J. Pathol. 2020, 190, 272–285. [Google Scholar] [CrossRef]
- Feng, Y.; Duan, T.; Du, Y.; Jin, S.; Wang, M.; Cui, J.; Wang, R.F. LRRC25 Functions as an Inhibitor of NF-κB Signaling Pathway by Promoting p65/RelA for Autophagic Degradation. Sci. Rep. 2017, 7, 13448. [Google Scholar] [CrossRef]
- Du, Y.; Duan, T.; Feng, Y.; Liu, Q.; Lin, M.; Cui, J.; Wang, R.F. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 2018, 37, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, O.; Hanel, A.; Carlberg, C. Key Vitamin D Target Genes with Functions in the Immune System. Nutrients 2020, 12, 1140. [Google Scholar] [CrossRef]
- Cantorna, M.T.; Snyder, L.; Lin, Y.-D.; Yang, L. Vitamin D and 1,25(OH)2D Regulation of T cells. Nutrients 2015, 7, 3011–3021. [Google Scholar] [CrossRef] [PubMed]
- Dubertret, L.; Wallach, D.; Souteyrand, P.; Perussel, M.; Kalis, B.; Meynadier, J.; Chevrant-Breton, J.; Beylot, C.; Bazex, J.A.; Jurgensen, H.J. Efficacy and safety of calcipotriol (MC 903) ointment in psoriasis vulgaris: A randomized, double-blind, right/left comparative, vehicle-controlled study. J. Am. Acad. Dermatol. 1992, 27 Pt 1, 983–988. [Google Scholar] [CrossRef]
- Sîrbe, C.; Rednic, S.; Grama, A.; Pop, T.L. An Update on the Effects of Vitamin D on the Immune System and Autoimmune Diseases. Int. J. Mol. Sci. 2022, 23, 9784. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Brown, K.R.; Jurisica, I. Online predicted human interaction database. Bioinformatics 2005, 21, 2076–2082. [Google Scholar] [CrossRef]
- Rual, J.F.; Venkatesan, K.; Hao, T.; Hirozane-Kishikawa, T.; Dricot, A.; Li, N.; Berriz, G.F.; Gibbons, F.D.; Dreze, M.; Ayivi-Guedehoussou, N.; et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005, 437, 1173–1178. [Google Scholar] [CrossRef]
- Hussain, S.; Berki, D.M.; Choon, S.-E.; Burden, A.D.; Allen, M.H.; Arostegui, J.I.; Chaves, A.; Duckworth, M.; Irvine, A.D.; Mockenhaupt, M.; et al. IL36RN mutations define a severe autoinflammatory phenotype of generalized pustular psoriasis. J. Allergy Clin. Immunol. 2015, 135, 1067–1070.e9. [Google Scholar] [CrossRef]
- Dietrich, D.; Gabay, C. IL-36 has proinflammatory effects in skin but not in joints. Nat. Rev. Rheumatol. 2014, 10, 639–640. [Google Scholar] [CrossRef]
- Li, J.; Bansal, V.; Tiwari, M.; Chen, Y.; Sen, G.L. ELL Facilitates RNA Polymerase II-Mediated Transcription of Human Epidermal Proliferation Genes. J. Investig. Dermatol. 2021, 141, 1352–1356.e3. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, Y.; Cui, L.; Shi, Y.; Guo, C. Advances in the pathogenesis of psoriasis: From keratinocyte perspective. Cell Death Dis. 2022, 13, 81. [Google Scholar] [CrossRef]
- Tian, F.; Chen, Z.; Xu, T. Efficacy and safety of tofacitinib for the treatment of chronic plaque psoriasis: A systematic review and meta-analysis. J. Int. Med. Res. 2019, 47, 2342–2350. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, L.; Wang, L.; Jiang, X. The efficacy and safety of tofacitinib, peficitinib, solcitinib, baricitinib, abrocitinib and deucravacitinib in plaque psoriasis—A network meta-analysis. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1937–1946. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Damsky, W.; King, B.A. JAK inhibitors in dermatology: The promise of a new drug class. J. Am. Acad. Dermatol. 2017, 76, 736–744. [Google Scholar] [CrossRef]
- Wang, H.; Dou, Y.; Tian, J.; Li, F.; Wang, S.; Wang, Z. Research on medical speciality of traditional Chinese medicines using dot-immunoblotting method based on polyclonal antibody prepared from traditional Chinese medicines with hot/cold nature. Zhongguo Zhong Yao Za Zhi Zhongguo Zhongyao Zazhi China J. Chin. Mater. Med. 2009, 34, 438–442. [Google Scholar]
- Kim, Y.R.; Park, B.K.; Kim, Y.H.; Shim, I.; Kang, I.C.; Lee, M.Y. Antidepressant Effect of Fraxinus rhynchophylla Hance Extract in a Mouse Model of Chronic Stress-Induced Depression. Biomed. Res. Int. 2018, 2018, 8249563. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Y.; Ekunwe, S.I.N.; Yi, X.; Liu, X.; Wang, H.; Pan, Y. Antioxidant activity and inhibition effect on the growth of human colon carcinoma (HT-29) cells of esculetin from Cortex Fraxini. Med. Chem. Res. 2011, 20, 968–974. [Google Scholar] [CrossRef]
- Kirsch, G.; Abdelwahab, A.B.; Chaimbault, P. Natural and Synthetic Coumarins with Effects on Inflammation. Molecules 2016, 21, 1322. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Li, X.; Liu, L.; Xiao, X.; Zhang, L.; Zhang, S.; Lin, P.; Wang, X.; Wang, Y.; Li, Q. Attenuation of doxorubicin-induced cardiotoxicity by esculetin through modulation of Bmi-1 expression. Exp. Ther. Med. 2017, 14, 2216–2220. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gu, J.; Qian, H. Esculetin Attenuates the Growth of Lung Cancer by Downregulating Wnt Targeted Genes and Suppressing NF-κB. Arch. Bronconeumol. 2018, 54, 128–133. [Google Scholar] [CrossRef]
- Hong, S.H.; Jeong, H.K.; Han, M.H.; Park, C.; Choi, Y.H. Esculetin suppresses lipopolysaccharide-induced inflammatory mediators and cytokines by inhibiting nuclear factor-κB translocation in RAW 264.7 macrophages. Mol. Med. Rep. 2014, 10, 3241–3246. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Q.; Liu, H.; Lu, C.; Liang, C.L.; Qiu, F.; Han, L.; Dai, Z. Esculetin Ameliorates Psoriasis-Like Skin Disease in Mice by Inducing CD4(+)Foxp3(+) Regulatory T Cells. Front. Immunol. 2018, 9, 2092. [Google Scholar] [CrossRef]
- Tian, Q.; Wang, L.; Sun, X.; Zeng, F.; Pan, Q.; Xue, M. Scopoletin exerts anticancer effects on human cervical cancer cell lines by triggering apoptosis, cell cycle arrest, inhibition of cell invasion and PI3K/AKT signalling pathway. J. BUON 2019, 24, 997–1002. [Google Scholar]
- Bulik-Sullivan, B.K.; Loh, P.-R.; Finucane, H.K.; Ripke, S.; Yang, J.; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M.; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Wallace, C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 2021, 17, e1009440. [Google Scholar] [CrossRef]
- Hormozdiari, F.; van de Bunt, M.; Segrè, A.V.; Li, X.; Joo, J.W.J.; Bilow, M.; Sul, J.H.; Sankararaman, S.; Pasaniuc, B.; Eskin, E. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am. J. Hum. Genet. 2016, 99, 1245–1260. [Google Scholar] [CrossRef]
- Li, Y.I.; Wong, G.; Humphrey, J.; Raj, T. Prioritizing Parkinson’s disease genes using population-scale transcriptomic data. Nat. Commun. 2019, 10, 994. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. an automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef]
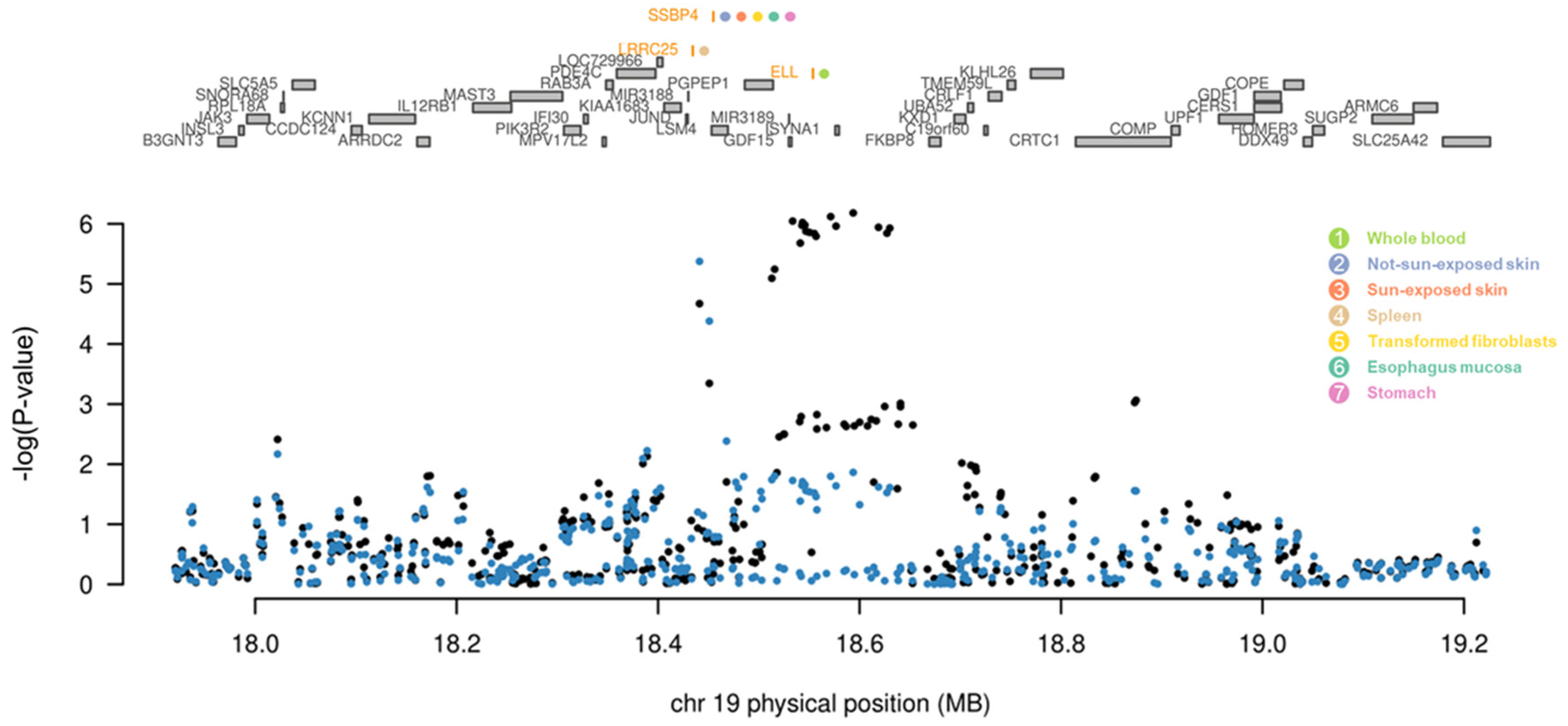
| Gene | Z (TWAS) | P (TWAS) | Z (Joint) | P (Joint) | Tissue | Direction of Regulation |
|---|---|---|---|---|---|---|
| ELL | 4.3 | 1.6 × 10−5 | 4.3 | 1.6 × 10−5 | Whole blood | Up-regulated |
| LRRC25 | −5 | 5.8 × 10−7 | −5 | 5.8 × 10−7 | Spleen | Down-regulated |
| SSBP4 | −4.9 | 9.5 × 10−7 | −4.9 | 9.5 × 10−7 | Sun-exposed skin | Down-regulated |
| −4.8 | 1.6 × 10−6 | −4.8 | 1.6 × 10−6 | Not-sun-exposed skin | Down-regulated | |
| −5.0 | 6.6 × 10−7 | −5.0 | 6.6 × 10−7 | Transformed fibroblasts | Down-regulated | |
| −5.0 | 6.4 × 10−7 | −5.0 | 6.4 × 10−7 | Esophagus mucosa | Down-regulated | |
| −5.2 | 2.6 × 10−7 | −5.2 | 2.6 × 10−7 | Stomach | Down-regulated |
| Gene | Drug Candidate | Interaction Score | Gene | Drug Candidate | Interaction Score |
|---|---|---|---|---|---|
| KLRC1 | Monalizumab | 127.30 | TRAF3IP2 | Nevirapine | 2.77 |
| SGSH | N-sulfoglucosamine sulfohydrolase recombinant | 63.65 | GALK1 | Pyrantel pamoate | 1.33 |
| Tricetin | 0.66 | ||||
| Suramin hexasodium | 0.48 | ||||
| IL23A | Guselkumab | 31.83 | TYK2 | Brepocitinib | 1.03 |
| Tildrakizumab | 10.61 | Tofacitinib | 0.91 | ||
| Brazikumab | 10.61 | Peficitinib | 0.82 | ||
| Risankizumab | 10.61 | Delgocitinib | 0.51 | ||
| Briakinumab | 7.07 | Tofacitinib citrate | 0.51 | ||
| Ustekinumab | 6.37 | BMS-911543 | 0.51 | ||
| ERAP1 | Umbelliferone | 5.30 | Oclacitinib | 0.51 | |
| Scopoletin | 1.77 | Solcitinib | 0.51 | ||
| Esculetin | 1.77 | Cerdulatinib | 0.46 | ||
| Tosedostat | 0.56 | Upadacitinib | 0.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, Y.; Song, J.; Lee, Y.; Choi, E.; Won, Y.; Kim, B.; Jang, W. A Transcriptome-Wide Analysis of Psoriasis: Identifying the Potential Causal Genes and Drug Candidates. Int. J. Mol. Sci. 2023, 24, 11717. https://doi.org/10.3390/ijms241411717
Jeong Y, Song J, Lee Y, Choi E, Won Y, Kim B, Jang W. A Transcriptome-Wide Analysis of Psoriasis: Identifying the Potential Causal Genes and Drug Candidates. International Journal of Molecular Sciences. 2023; 24(14):11717. https://doi.org/10.3390/ijms241411717
Chicago/Turabian StyleJeong, Yeonbin, Jaeseung Song, Yubin Lee, Eunyoung Choi, Youngtae Won, Byunghyuk Kim, and Wonhee Jang. 2023. "A Transcriptome-Wide Analysis of Psoriasis: Identifying the Potential Causal Genes and Drug Candidates" International Journal of Molecular Sciences 24, no. 14: 11717. https://doi.org/10.3390/ijms241411717
APA StyleJeong, Y., Song, J., Lee, Y., Choi, E., Won, Y., Kim, B., & Jang, W. (2023). A Transcriptome-Wide Analysis of Psoriasis: Identifying the Potential Causal Genes and Drug Candidates. International Journal of Molecular Sciences, 24(14), 11717. https://doi.org/10.3390/ijms241411717






