Sugar Starvation Disrupts Lipid Breakdown by Inducing Autophagy in Embryonic Axes of Lupin (Lupinus spp.) Germinating Seeds
Abstract
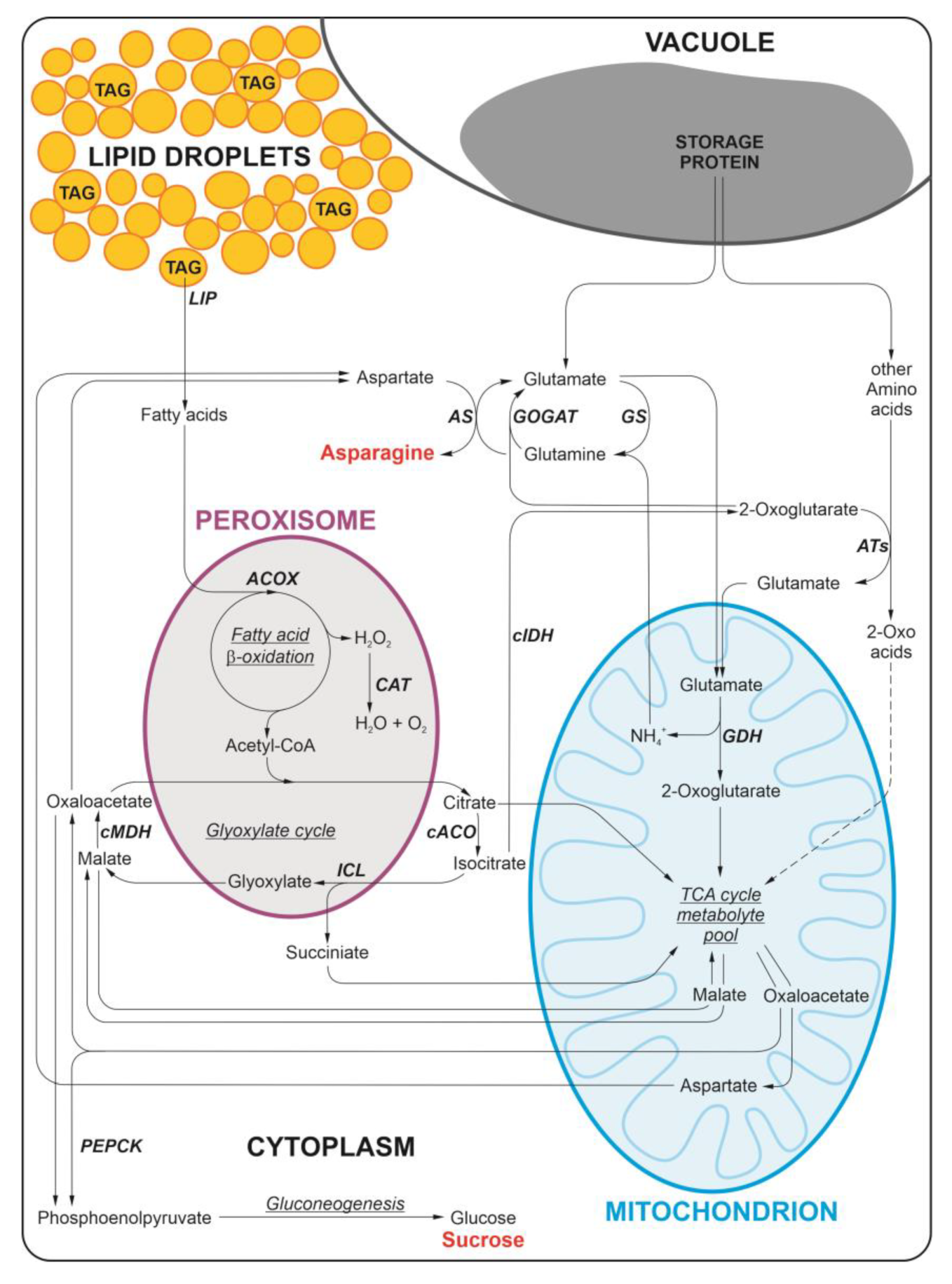
1. Introduction
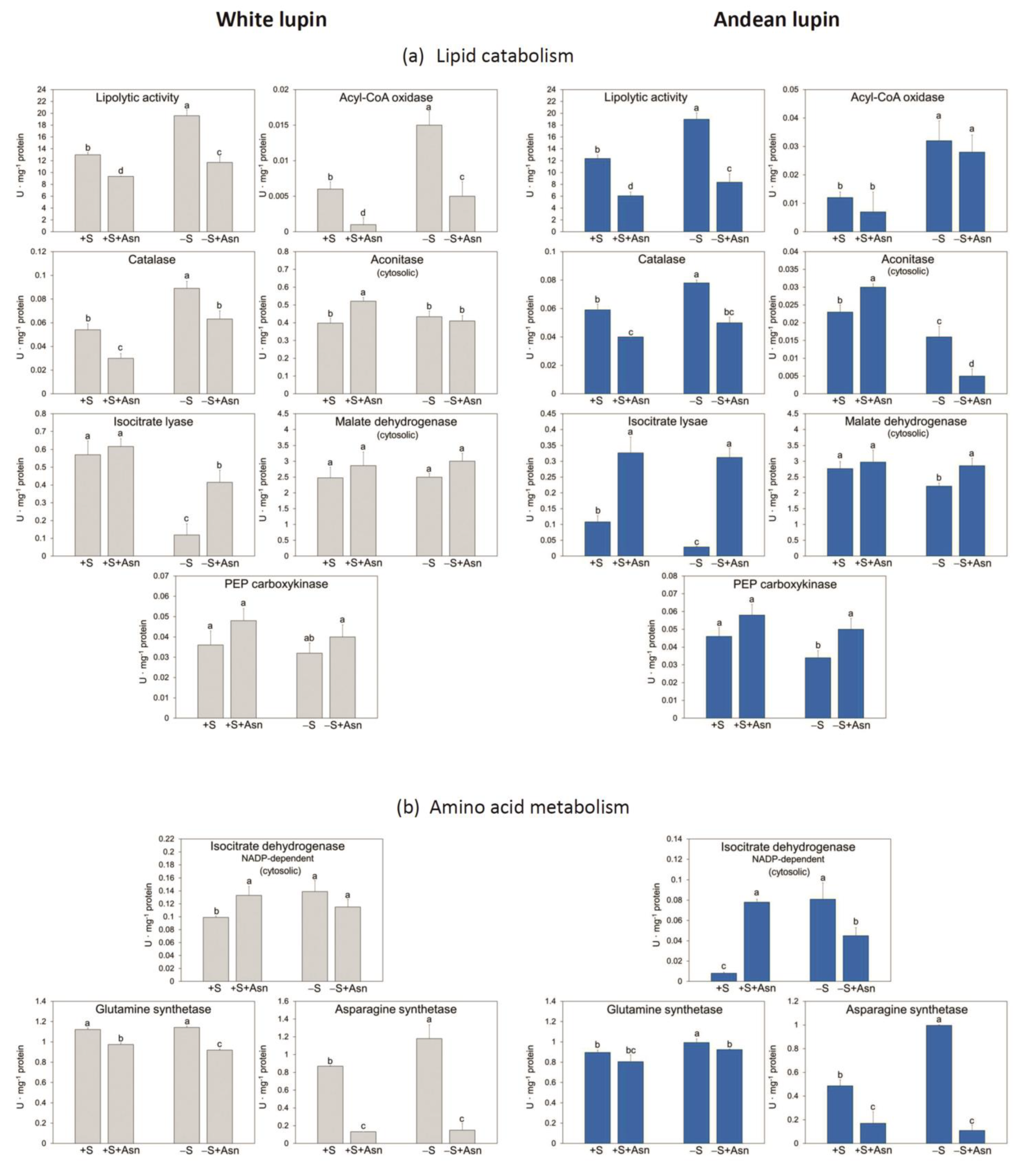
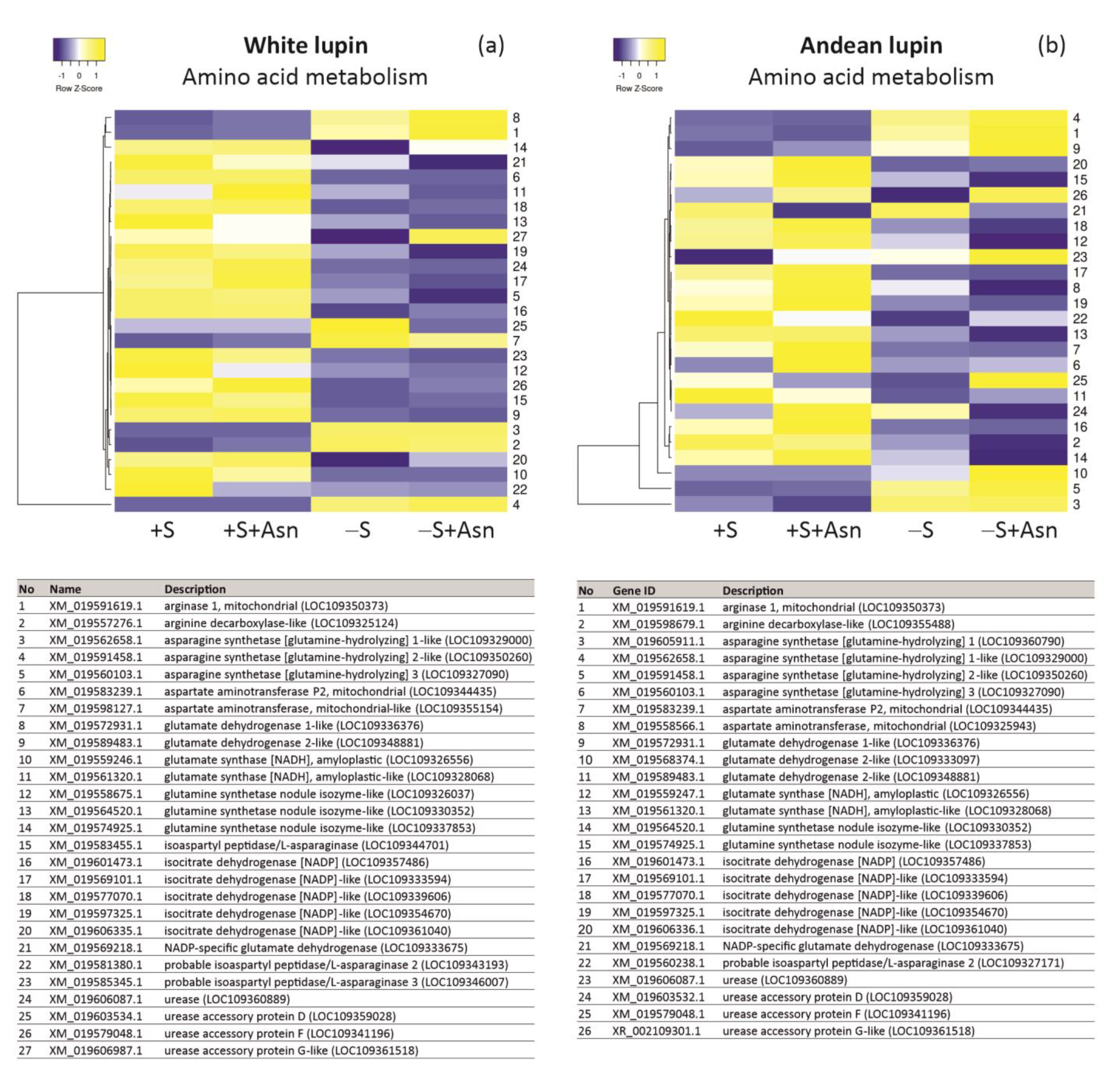
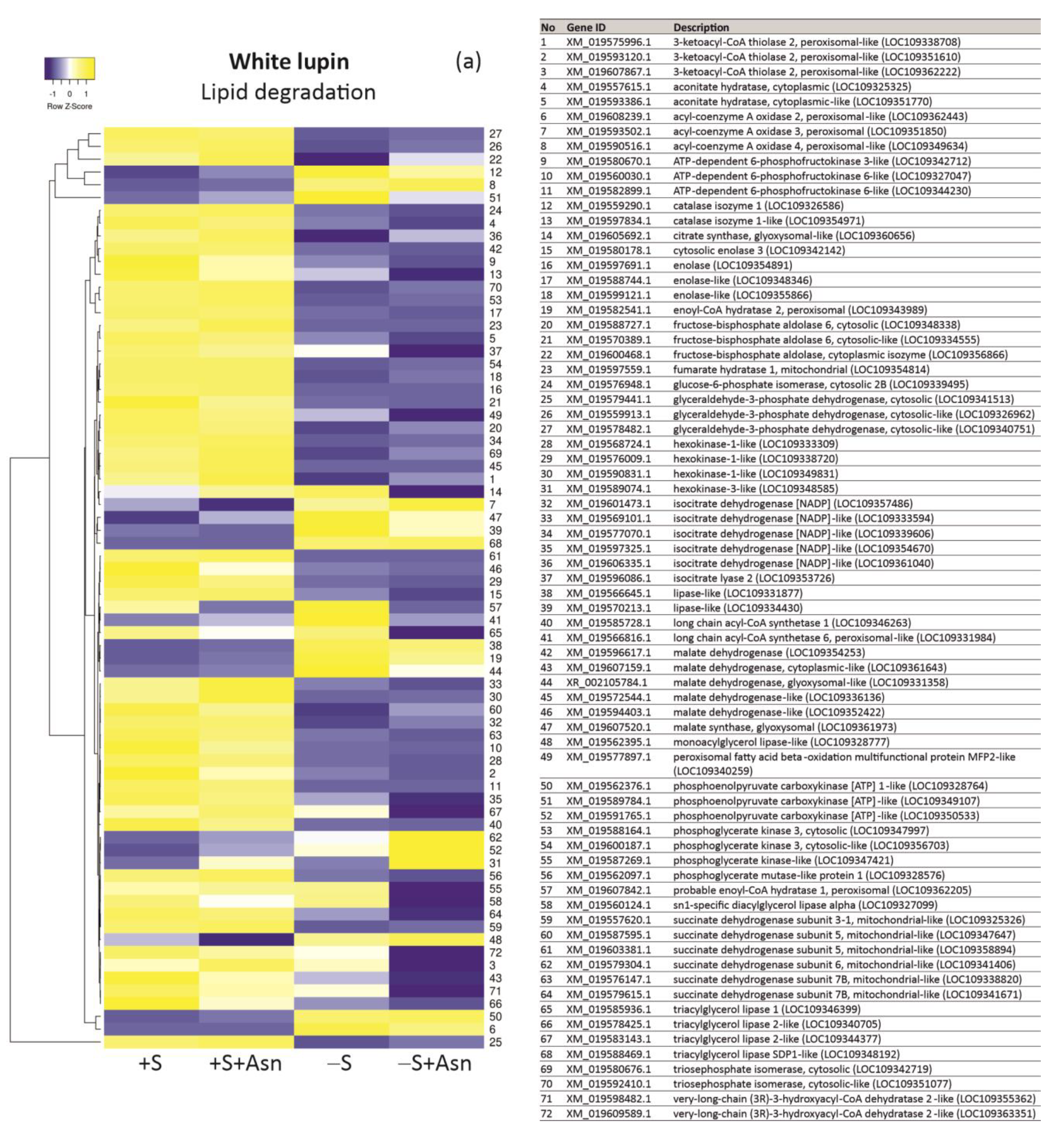
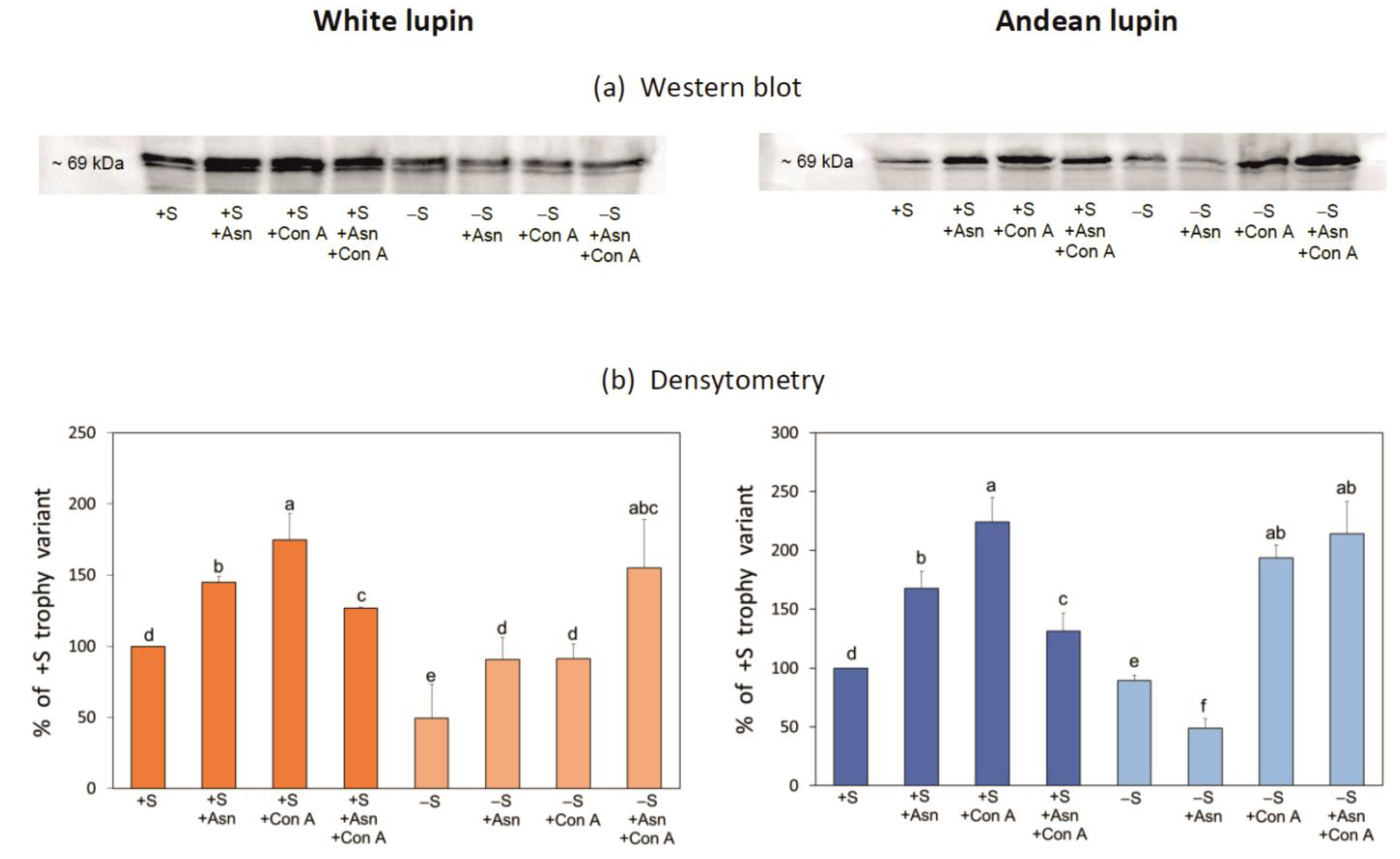
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Ultrastructure
4.3. Total Lipid and Phospholipids
4.4. Enzyme Activity Assays
4.5. Proteomics—iTRAQ
4.6. Western Blot
4.7. Transcriptomics—NGS
4.8. qRT-PCR
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ratajczak, W.; Borek, S.; Podgorski, A.; Ratajczak, L. Variability of globulin composition in cultivars and individually tested seeds of yellow lupin (Lupinus luteus L.). Acta Physiol. Plant 1999, 21, 413–417. [Google Scholar] [CrossRef]
- Borek, S.; Ratajczak, W.; Ratajczak, L. Regulation of storage lipid metabolism in developing and germinating lupin (Lupinus spp.) seeds. Acta Physiol. Plant 2015, 37, 119. [Google Scholar] [CrossRef]
- Borek, S.; Galor, A.; Paluch, E. Asparagine Enhances Starch Accumulation in Developing and Germinating Lupin Seeds. J. Plant Growth Regul. 2013, 32, 471–482. [Google Scholar] [CrossRef]
- Lehmann, T.; Ratajczak, L. The pivotal role of glutamate dehydrogenase (GDH) in the mobilization of N and C from storage material to asparagine in germinating seeds of yellow lupine. J. Plant Physiol. 2008, 165, 149–158. [Google Scholar] [CrossRef]
- Gaufichon, L.; Reisdorf-Cren, M.; Rothstein, S.J.; Chardon, F.; Suzuki, A. Biological functions of asparagine synthetase in plants. Plant Sci. 2010, 179, 141–153. [Google Scholar] [CrossRef]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine Metabolic Pathways in Arabidopsis. Plant Cell Physiol. 2016, 57, 675–689. [Google Scholar] [CrossRef]
- Morkunas, I.; Garnczarska, M.; Bednarski, W.; Ratajczak, W.; Waplak, S. Metabolic and ultrastructural responses of lupine embryo axes to sugar starvation. J. Plant Physiol. 2003, 160, 311–319. [Google Scholar] [CrossRef]
- Borek, S.; Paluch-Lubawa, E.; Pukacka, S.; Pietrowska-Borek, M.; Ratajczak, L. Asparagine slows down the breakdown of storage lipid and degradation of autophagic bodies in sugar-starved embryo axes of germinating lupin seeds. J. Plant Physiol. 2017, 209, 51–67. [Google Scholar] [CrossRef]
- Borek, S.; Ratajczak, W.; Ratajczak, L. A transfer of carbon atoms from fatty acids to sugars and amino acids in yellow lupine (Lupinus luteus L.) seedlings. J. Plant Physiol. 2003, 160, 539–545. [Google Scholar] [CrossRef]
- Borek, S.; Ratajczak, L. Storage lipids as a source of carbon skeletons for asparagine synthesis in germinating seeds of yellow lupine (Lupinus luteus L.). J. Plant Physiol. 2010, 167, 717–724. [Google Scholar] [CrossRef]
- Borek, S.; Kubala, S.; Kubala, S.; Ratajczak, L. Comparative study of storage compound breakdown in germinating seeds of three lupine species. Acta Physiol. Plant 2011, 33, 1953–1968. [Google Scholar] [CrossRef]
- Borek, S.; Kubala, S.; Kubala, S. Diverse regulation by sucrose of enzymes involved in storage lipid breakdown in germinating lupin seeds. Acta Physiol. Plant 2013, 35, 2147–2156. [Google Scholar] [CrossRef]
- Ischebeck, T.; Krawczyk, H.E.; Mullen, R.T.; Dyer, J.M.; Chapman, K.D. Lipid droplets in plants and algae: Distribution, formation, turnover and function. Semin. Cell Dev. Biol. 2020, 108, 82–93. [Google Scholar] [CrossRef]
- Zienkiewicz, K.; Zienkiewicz, A. Degradation of Lipid Droplets in Plants and Algae-Right Time, Many Paths, One Goal. Front. Plant Sci. 2020, 11, 579019. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Zaikova, K.; Yeom, S.J.; Kim, Y.S.; Lee, D.W. Biogenesis and Lipase-Mediated Mobilization of Lipid Droplets in Plants. Plants 2022, 11, 1243. [Google Scholar] [CrossRef]
- Pracharoenwattana, I.; Smith, S.M. When is a peroxisome not a peroxisome? Trends Plant Sci. 2008, 13, 522–525. [Google Scholar] [CrossRef]
- Wu, W.L.; Hsiao, Y.Y.; Lu, H.C.; Liang, C.K.; Fu, C.H.; Huang, T.H.; Chuang, M.H.; Chen, L.J.; Liu, Z.J.; Tsai, W.C. Expression regulation of MALATE SYNTHASE involved in glyoxylate cycle during protocorm development in Phalaenopsis aphrodite (Orchidaceae). Sci. Rep. 2020, 10, 10123. [Google Scholar] [CrossRef]
- Heller, R. Recherches sur la nutrition minérale des tissus végétaux ciltivés in vitro. Ann. Sci. Nat. Bot. Biol. Veg. 1954, 14, 1–223. [Google Scholar]
- Borek, S.; Kubala, S.; Kubala, S. Regulation by sucrose of storage compounds breakdown in germinating seeds of yellow lupine (Lupinus luteus L.), white lupine (Lupinus albus L.) and Andean lupine (Lupinus mutabilis Sweet): I. Mobilization of storage protein. Acta Physiol. Plant 2012, 34, 701–711. [Google Scholar] [CrossRef]
- Borek, S.; Ratajczak, W. Sugars as a metabolic regulator of storage protein mobilization in germinating seeds of yellow lupine (Lupinus luteus L.). Acta Physiol. Plant 2002, 24, 425–434. [Google Scholar] [CrossRef]
- Lehmann, T.; Skrok, A.; Dabert, M. Stress-induced changes in glutamate dehydrogenase activity imply its role in adaptation to C and N metabolism in lupine embryos. Physiol. Plant 2010, 138, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Borek, S.; Ratajczak, W.; Ratajczak, L. Ultrastructural and enzymatic research on the role of sucrose in mobilization of storage lipids in germinating yellow lupine seeds. Plant Sci. 2006, 170, 441–452. [Google Scholar] [CrossRef]
- Morkunas, I.; Borek, S.; Formela, M.; Ratajczak, L. Plant responses to sugar starvation. In Carbohydrates—Comprehensive Studies on Glycobiology and Glycotechnology; Chang, C., Ed.; IntechOpen: London, UK, 2012; Chapter 19; pp. 409–438. [Google Scholar]
- Borek, S.; Pukacka, S.; Michalski, K. Regulation by sucrose of storage compounds breakdown in germinating seeds of yellow lupine (Lupinus luteus L.), white lupine (Lupinus albus L.) and Andean lupine (Lupinus mutabilis Sweet). II. Mobilization of storage lipid. Acta Physiol. Plant 2012, 34, 1199–1206. [Google Scholar] [CrossRef]
- Aubert, S.; Gout, E.; Bligny, R.; Marty-Mazars, D.; Barrieu, F.; Alabouvette, J.; Marty, F.; Douce, R. Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: Control by the supply of mitochondria with respiratory substrates. J. Cell Biol. 1996, 133, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Moriyasu, Y. Autophagy is not a main contributor to the degradation of phospholipids in tobacco cells cultured under sucrose starvation conditions. Plant Cell Physiol. 2006, 47, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Makino, S.; Kagohashi, Y.; Sasaki, M.; Ohsumi, Y. A method for the isolation and characterization of autophagic bodies from yeast provides a key tool to investigate cargos of autophagy. J. Biol. Chem. 2022, 298, 102641. [Google Scholar] [CrossRef]
- Stefaniak, S.; Wojtyla, L.; Pietrowska-Borek, M.; Borek, S. Completing Autophagy: Formation and Degradation of the Autophagic Body and Metabolite Salvage in Plants. Int. J. Mol. Sci. 2020, 21, 2205. [Google Scholar] [CrossRef]
- Borek, S.; Stefaniak, S.; Sliwinski, J.; Garnczarska, M.; Pietrowska-Borek, M. Autophagic Machinery of Plant Peroxisomes. Int. J. Mol. Sci. 2019, 20, 4754. [Google Scholar] [CrossRef]
- Malinowska, A.; Kistowski, M.; Bakun, M.; Rubel, T.; Tkaczyk, M.; Mierzejewska, J.; Dadlez, M. Diffprot—Software for non-parametric statistical analysis of differential proteomics data. J. Proteom. 2012, 75, 4062–4073. [Google Scholar] [CrossRef]
- Borek, S.; Morkunas, I.; Ratajczak, W.; Ratajczak, L. Metabolism of amino acids in germinating yellow lupin seeds—III. Breakdown of arginine in sugar-starved organs cultivated in vitro. Acta Physiol. Plant. 2001, 23, 141–148. [Google Scholar] [CrossRef]
- Luong, A.M.; Koestel, J.; Bhati, K.K.; Batoko, H. Cargo receptors and adaptors for selective autophagy in plant cells. FEBS Lett. 2022, 596, 2104–2132. [Google Scholar] [CrossRef]
- Muhammad, D.; Smith, K.A.; Bartel, B. Plant peroxisome proteostasis-establishing, renovating, and dismantling the peroxisomal proteome. Essays Biochem. 2022, 66, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.T.; Gonzalez, K.L.; Bartel, B. Peroxisome Function, Biogenesis, and Dynamics in Plants. Plant Physiol. 2018, 176, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Farre, J.C.; Manjithaya, R.; Mathewson, R.D.; Subramani, S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 2008, 14, 365–376. [Google Scholar] [CrossRef]
- Manjithaya, R.; Nazarko, T.Y.; Farre, J.C.; Subramani, S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010, 584, 1367–1373. [Google Scholar] [CrossRef]
- Bassham, D.C. Methods for analysis of autophagy in plants. Methods 2015, 75, 181–188. [Google Scholar] [CrossRef]
- Kaur, M.; Tak, Y.; Bhatia, S.; Asthir, B.; Lorenzo, J.M.; Amarowicz, R. Crosstalk during the Carbon-Nitrogen Cycle That Interlinks the Biosynthesis, Mobilization and Accumulation of Seed Storage Reserves. Int. J. Mol. Sci. 2021, 22, 12032. [Google Scholar] [CrossRef]
- Kircher, S.; Schopfer, P. Photosynthetic sucrose acts as cotyledon-derived long-distance signal to control root growth during early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 11217–11221. [Google Scholar] [CrossRef]
- Curtis, T.Y.; Bo, V.; Tucker, A.; Halford, N.G. Construction of a network describing asparagine metabolism in plants and its application to the identification of genes affecting asparagine metabolism in wheat under drought and nutritional stress. Food Energy Secur. 2018, 7, e00126. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Pageau, K.; Lelandais, M.; Grandjean, O.; Kronenberger, J.; Valadier, M.H.; Feraud, M.; Jouglet, T.; Suzuki, A. Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 2006, 140, 444–456. [Google Scholar] [CrossRef]
- Han, M.; Zhang, C.; Suglo, P.; Sun, S.Y.; Wang, M.Y.; Su, T. l-Aspartate: An Essential Metabolite for Plant Growth and Stress Acclimation. Molecules 2021, 26, 1887. [Google Scholar] [CrossRef] [PubMed]
- Tyutereva, E.V.; Murtuzova, A.V.; Voitsekhovskaja, O.V. Autophagy and the Energy Status of Plant Cells. Russ. J. Plant Physl. 2022, 69, 19. [Google Scholar] [CrossRef]
- Baslam, M.; Mitsui, T.; Sueyoshi, K.; Ohyama, T. Recent Advances in Carbon and Nitrogen Metabolism in C3 Plants. Int. J. Mol. Sci. 2021, 22, 318. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Deedaldeechamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Theevenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Reumann, S.; Hu, J. Peroxisome biogenesis and function. Arab. Book 2009, 7, e0123. [Google Scholar] [CrossRef]
- Hu, J.P.; Baker, A.; Bartel, B.; Linka, N.; Mullen, R.T.; Reumann, S.; Zolman, B.K. Plant Peroxisomes: Biogenesis and Function. Plant Cell 2012, 24, 2279–2303. [Google Scholar] [CrossRef]
- Sandalio, L.M.; Gotor, C.; Romero, L.C.; Romero-Puertas, M.C. Multilevel Regulation of Peroxisomal Proteome by Post-Translational Modifications. Int. J. Mol. Sci. 2019, 20, 4881. [Google Scholar] [CrossRef]
- Pan, R.H.; Liu, J.; Wang, S.S.; Hu, J.P. Peroxisomes: Versatile organelles with diverse roles in plants. New Phytol. 2020, 225, 1410–1427. [Google Scholar] [CrossRef]
- Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Biogenesis and functions of lipid droplets in plants: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: From Yeast to Man. J. Lipid Res. 2012, 53, 215–226. [Google Scholar] [CrossRef]
- Huang, A.H.C. Plant Lipid Droplets and Their Associated Proteins: Potential for Rapid Advances. Plant Physiol. 2018, 176, 1894–1918. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Liu, X.F.; Su, T.; Ma, C.L.; Wang, P.P. New Insights Into the Role of Seed Oil Body Proteins in Metabolism and Plant Development. Front. Plant Sci. 2019, 10, 1568. [Google Scholar] [CrossRef] [PubMed]
- Tzen, J.T.C.; Lie, G.C.; Huang, A.H.C. Characterization of the Charged Components and Their Topology on the Surface of Plant Seed Oil Bodies. J. Biol. Chem. 1992, 267, 15626–15634. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.C.; Fan, J.L. Links between autophagy and lipid droplet dynamics. J. Exp. Bot. 2022, 73, 2848–2858. [Google Scholar] [CrossRef] [PubMed]
- Borek, S.; Pukacka, S.; Michalski, K.; Ratajczak, L. Lipid and protein accumulation in developing seeds of three lupine species: Lupinus luteus L., Lupinus albus L., and Lupinus mutabilis Sweet. J. Exp. Bot. 2009, 60, 3453–3466. [Google Scholar] [CrossRef]
- Borek, S.; Nuc, K. Sucrose controls storage lipid breakdown on gene expression level in germinating yellow lupine (Lupinus luteus L.) seeds. J. Plant Physiol. 2011, 168, 1795–1803. [Google Scholar] [CrossRef]
- Hipkin, C.R.; Syrett, P.J. Some Effects of Nitrogen-Starvation on Nitrogen and Carbohydrate-Metabolism in Ankistrodesmus braunii. Planta 1977, 133, 209–214. [Google Scholar] [CrossRef]
- Loureiro, I.; Faria, J.; Clayton, C.; Ribeiro, S.M.; Roy, N.; Santarem, N.; Tavares, J.; Cordeiro-da-Silva, A. Knockdown of Asparagine Synthetase A Renders Trypanosoma brucei Auxotrophic to Asparagine. PLoS Neglect. Trop. D 2013, 7, e2578. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Magni, C.; Scarafoni, A.; Herndl, A.; Sessa, F.; Prinsi, B.; Espen, L.; Duranti, M. Combined 2D electrophoretic approaches for the study of white lupin mature seed storage proteome. Phytochemistry 2007, 68, 997–1007. [Google Scholar] [CrossRef]
- Wojtyla, L.; Kosmala, A.; Garnczarska, M. Lupine embryo axes under salinity stress. II. Mitochondrial proteome response. Acta Physiol. Plant 2013, 35, 2383–2392. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R. Filter Aided Sample Preparation—A tutorial. Anal. Chim. Acta 2019, 1090, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.W.; Bandla, C.; Garcia-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]





| +S/+S+Asn | +S/-S | -S/-S+Asn | Protein ID/NCBI | Description White Lupin | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| q-Value | Ratio A/B | Fold Change | Peptide Number | q-Value | Ratio A/B | Fold Change | Peptide Number | q-Value | Ratio A/B | Fold Change | Peptide Number | ||
| 0.9588 | 1.04 | 1.04 | 6 | 0.0011 | 1.83 | 1.83 | 5 | - | - | - | - | XP_019412859.1 | long chain acyl-CoA synthetase 2-like |
| XP_019420557.1 | long chain acyl-CoA synthetase 2-like | ||||||||||||
| 0.9422 | 1.06 | 1.06 | 6 | 0.0000 | 0.55 | 1.82 | 11 | 0.6004 | 1.08 | 1.08 | 12 | XP_019446061.1 | acyl-coenzyme A oxidase 4, peroxisomal-like |
| 1.0000 | 1.11 | 1.11 | 3 | 0.0017 | 0.47 | 2.11 | 5 | 1.0000 | 1.07 | 1.07 | 6 | XP_019464789.1 | acetate/butyrate-CoA ligase AAE7, peroxisomal-like |
| 0.7892 | 0.99 | 1.01 | 6 | 0.0255 | 0.78 | 1.28 | 7 | 0.6844 | 1.05 | 1.05 | 4 | XP_019453051.1 | delta(3.5)-Delta(2.4)-dienoyl-CoA isomerase, peroxisomal isoform X1 |
| 1.0000 | 0.98 | 1.02 | 4 | 0.0029 | 0.67 | 1.5 | 8 | 1.0000 | 1.08 | 1.08 | 7 | XP_019438086.1 | enoyl-CoA hydratase, peroxisomal |
| 1.0000 | 0.96 | 1.04 | 6 | 0.0405 | 0.79 | 1.26 | 7 | 1.0000 | 1.02 | 1.02 | 8 | XP_019428389.1 | 3-ketoacyl-CoA thiolase 2, peroxisomal |
| 1.0000 | 0.93 | 1.08 | 8 | 0.0013 | 0.74 | 1.34 | 12 | 1.0000 | 1.02 | 1.02 | 11 | XP_019444968.1 | malate dehydrogenase, glyoxysomal isoform X1 |
| XP_019444970.1 | malate dehydrogenase, glyoxysomal isoform X2 | ||||||||||||
| 1.0000 | 1.02 | 1.02 | 6 | 0.0354 | 0.77 | 1.3 | 10 | 1.0000 | 0.85 | 1.17 | 7 | XP_019421327.1 | malate dehydrogenase, glyoxysomal-like |
| 1.0000 | 0.96 | 1.04 | 15 | 0.0004 | 0.8 | 1.25 | 19 | 0.0001 | 1.33 | 1.33 | 15 | XP_019463065.1 | malate synthase, glyoxysomal |
| 1.0000 | 0.98 | 1.02 | 7 | 0.0951 | 0.79 | 1.27 | 8 | 0.0001 | 1.46 | 1.46 | 18 | XP_019448948.1 | isocitrate lyase 2-like |
| XP_019451631.1 | isocitrate lyase 2 isoform X1 | ||||||||||||
| XP_019451632.1 | isocitrate lyase 2 isoform X2 | ||||||||||||
| 1.0000 | 1.02 | 1.02 | 11 | 0.0387 | 1.21 | 1.21 | 12 | 0.1246 | 1.15 | 1.15 | 9 | XP_019435274.1 | isocitrate dehydrogenase [NAD] catalytic subunit 5, mitochondrial |
| 0.7401 | 1.08 | 1.08 | 15 | 0.0012 | 1.25 | 1.25 | 15 | 1.0000 | 0.98 | 1.02 | 12 | XP_019436794.1 | succinate-CoA ligase [ADP-forming] subunit alpha, mitochondrial |
| XP_019457881.1 | succinate-CoA ligase [ADP-forming] subunit alpha, mitochondrial-like | ||||||||||||
| 1.0000 | 1.03 | 1.03 | 28 | 0.0444 | 1.13 | 1.13 | 30 | 1.0000 | 0.93 | 1.08 | 29 | XP_019459370.1 | succinate-CoA ligase [ADP-forming] subunit beta, mitochondrial-like |
| XP_019428639.1 | succinate-CoA ligase [ADP-forming] subunit beta, mitochondrial-like isoform X1 | ||||||||||||
| XP_019433054.1 | succinate-CoA ligase [ADP-forming] subunit beta, mitochondrial | ||||||||||||
| 0.1451 | 0.90 | 1.11 | 3 | 0.0005 | 0.6 | 1.68 | 5 | 0.3194 | 0.85 | 1.18 | 9 | XP_019417921.1 | phosphoenolpyruvate carboxykinase [ATP] 1-like |
| 0.1289 | 0.82 | 1.22 | 3 | 0.0001 | 0.51 | 1.96 | 7 | 1.0000 | 0.99 | 1.01 | 4 | XP_019445329.1 | phosphoenolpyruvate carboxykinase |
| - | - | - | - | 0.0135 | 0.47 | 2.13 | 3 | 0.6389 | 1.27 | 1.27 | 2 | XP_019420368.1 | phosphoenolpyruvate carboxykinase [ATP]-like |
| +S/+S+Asn | +S/-S | -S/-S+Asn | Protein ID/NCBI | Description Andean Lupin | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| q-Value | Ratio A/B | Fold Change | Peptide Number | q-Value | Ratio A/B | Fold Change | Peptide Number | q-Value | Ratio A/B | Fold Change | Peptide Number | ||
| 0.3273 | 1.07 | 1.07 | 12 | 0.0199 | 0.82 | 1.22 | 14 | 0.1969 | 1.21 | 1.21 | 14 | XP_019449047.1 | acyl-coenzyme A oxidase 3, peroxisomal |
| - | - | - | - | 0.9065 | 0.98 | 1.02 | 3 | 0.0123 | 0.59 | 1.69 | 6 | XP_019463387.1 | probable enoyl-CoA hydratase, peroxisomal |
| 1.0000 | 1.09 | 1.09 | 7 | 0.4980 | 0.90 | 1.11 | 4 | 0.0014 | 0.47 | 2.12 | 7 | XP_019453379.1 | catalase isozyme 1-like |
| 1.0000 | 0.96 | 1.05 | 13 | 0.1186 | 1.22 | 1.22 | 14 | 0.0004 | 0.65 | 1.53 | 14 | XP_019449188.1 | malate dehydrogenase, glyoxysomal |
| 1.0000 | 1.16 | 1.16 | 16 | 0.0001 | 0.62 | 1.62 | 17 | 0.0001 | 1.65 | 1.65 | 19 | XP_019435367.1 | citrate synthase, glyoxysomal-like |
| XP_019461237.1 | citrate synthase, glyoxysomal-like | ||||||||||||
| 0.7026 | 0.92 | 1.08 | 14 | 0.0008 | 0.68 | 1.46 | 9 | 0.4166 | 1.17 | 1.17 | 7 | XP_019451631.1 | isocitrate lyase 2 isoform X1 |
| XP_019451632.1 | isocitrate lyase 2 isoform X2 | ||||||||||||
| 1.0000 | 1.08 | 1.08 | 11 | 1.0000 | 1.04 | 1.04 | 2 | 0.0166 | 1.35 | 1.35 | 11 | XP_019423390.1 | aconitate hydratase 1 |
| XP_019444853.1 | aconitate hydratase 1-like isoform X2 | ||||||||||||
| 1.0000 | 0.97 | 1.03 | 6 | 0.0477 | 0.66 | 1.53 | 4 | 0.0807 | 0.78 | 1.28 | 9 | XP_019417921.1 | phosphoenolpyruvate carboxykinase [ATP] 1-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borek, S.; Stefaniak, S.; Nuc, K.; Wojtyla, Ł.; Ratajczak, E.; Sitkiewicz, E.; Malinowska, A.; Świderska, B.; Wleklik, K.; Pietrowska-Borek, M. Sugar Starvation Disrupts Lipid Breakdown by Inducing Autophagy in Embryonic Axes of Lupin (Lupinus spp.) Germinating Seeds. Int. J. Mol. Sci. 2023, 24, 11773. https://doi.org/10.3390/ijms241411773
Borek S, Stefaniak S, Nuc K, Wojtyla Ł, Ratajczak E, Sitkiewicz E, Malinowska A, Świderska B, Wleklik K, Pietrowska-Borek M. Sugar Starvation Disrupts Lipid Breakdown by Inducing Autophagy in Embryonic Axes of Lupin (Lupinus spp.) Germinating Seeds. International Journal of Molecular Sciences. 2023; 24(14):11773. https://doi.org/10.3390/ijms241411773
Chicago/Turabian StyleBorek, Sławomir, Szymon Stefaniak, Katarzyna Nuc, Łukasz Wojtyla, Ewelina Ratajczak, Ewa Sitkiewicz, Agata Malinowska, Bianka Świderska, Karolina Wleklik, and Małgorzata Pietrowska-Borek. 2023. "Sugar Starvation Disrupts Lipid Breakdown by Inducing Autophagy in Embryonic Axes of Lupin (Lupinus spp.) Germinating Seeds" International Journal of Molecular Sciences 24, no. 14: 11773. https://doi.org/10.3390/ijms241411773
APA StyleBorek, S., Stefaniak, S., Nuc, K., Wojtyla, Ł., Ratajczak, E., Sitkiewicz, E., Malinowska, A., Świderska, B., Wleklik, K., & Pietrowska-Borek, M. (2023). Sugar Starvation Disrupts Lipid Breakdown by Inducing Autophagy in Embryonic Axes of Lupin (Lupinus spp.) Germinating Seeds. International Journal of Molecular Sciences, 24(14), 11773. https://doi.org/10.3390/ijms241411773









