Ectopic Lipid Accumulation Correlates with Cellular Stress in Rabbit Blastocysts from Diabetic Mothers
Abstract
1. Introduction
2. Results
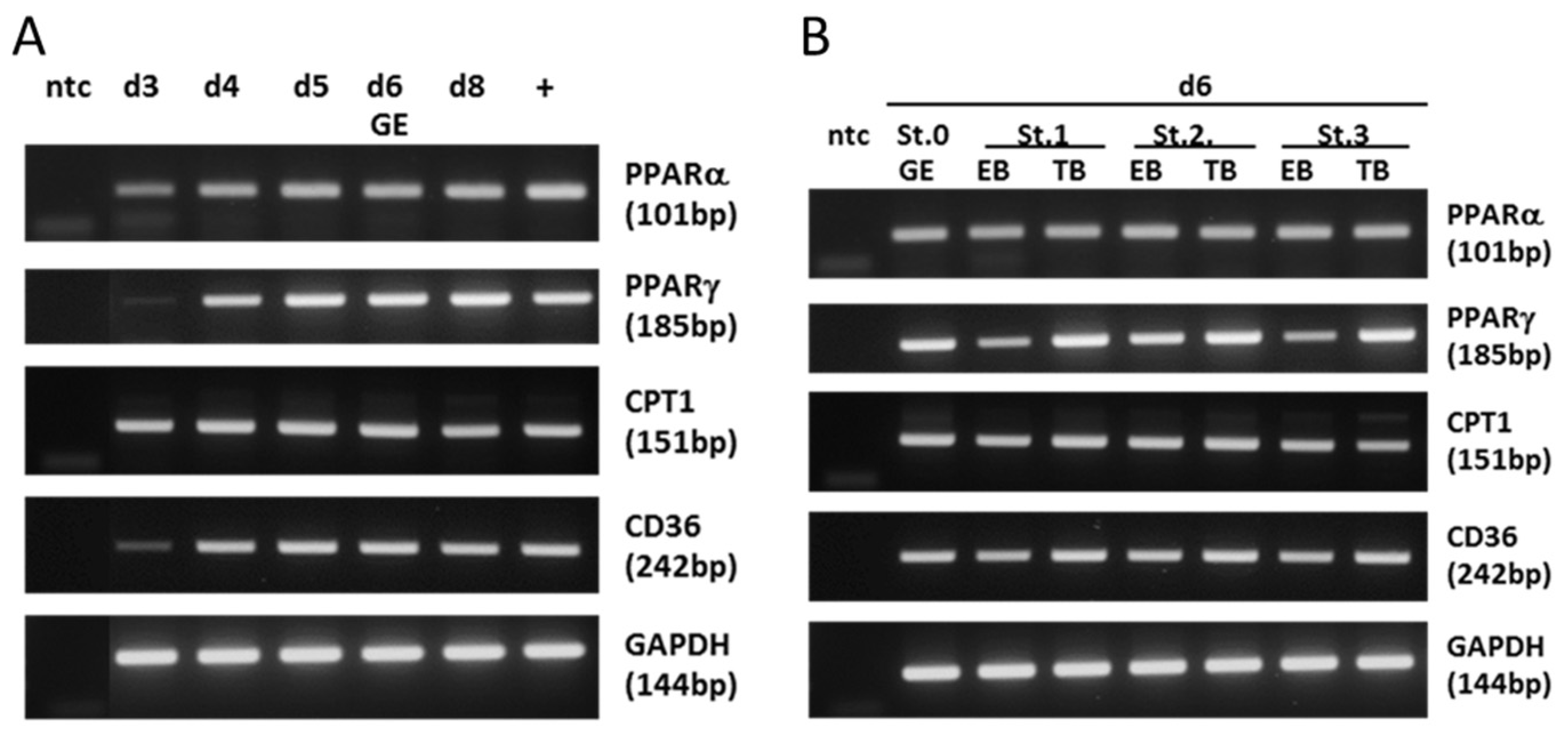
2.1. Expression of Key Lipogenic Markers in Blastocysts
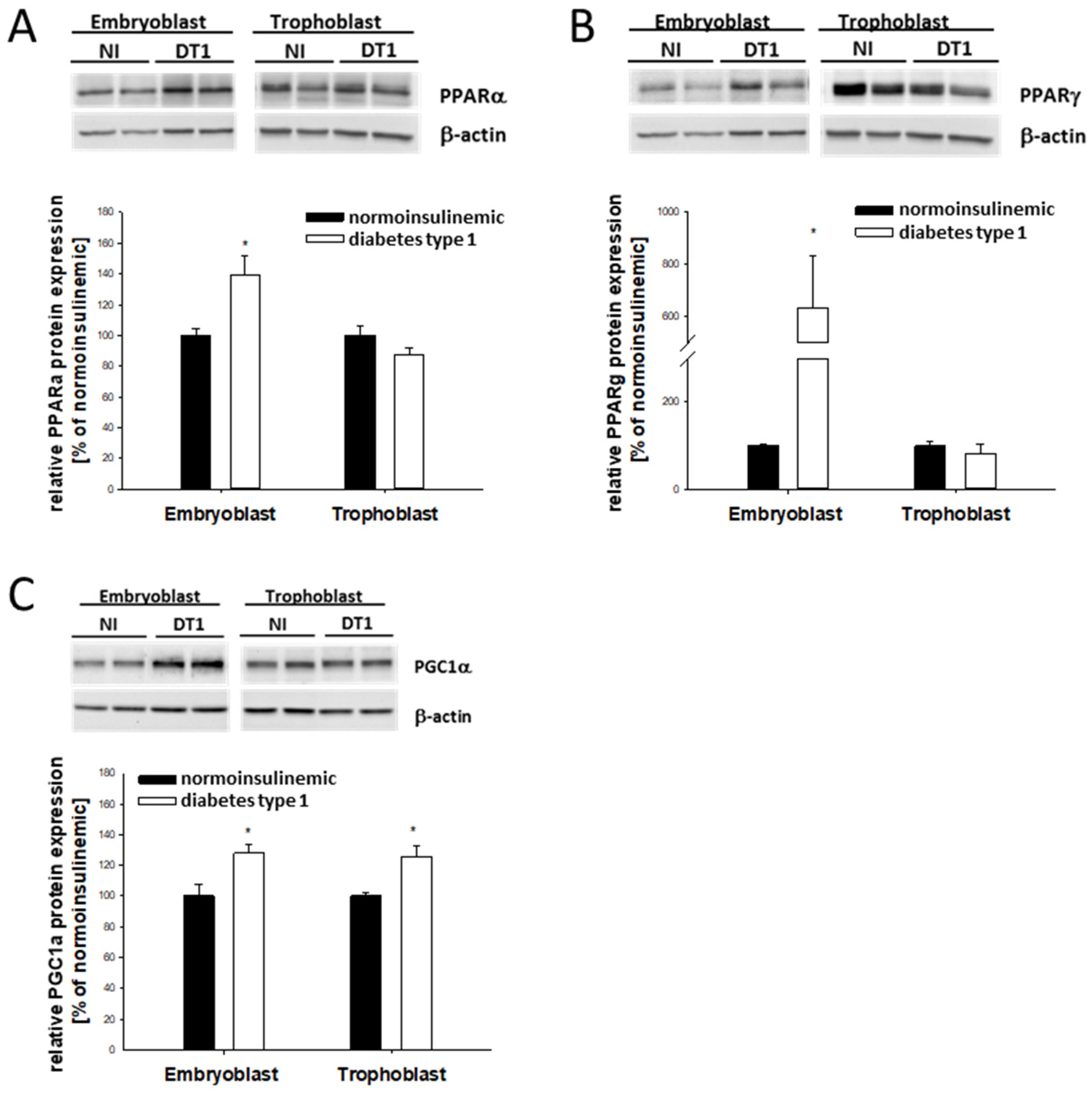
2.2. PPAR Expression in Blastocysts from Diabetic Rabbits In Vivo
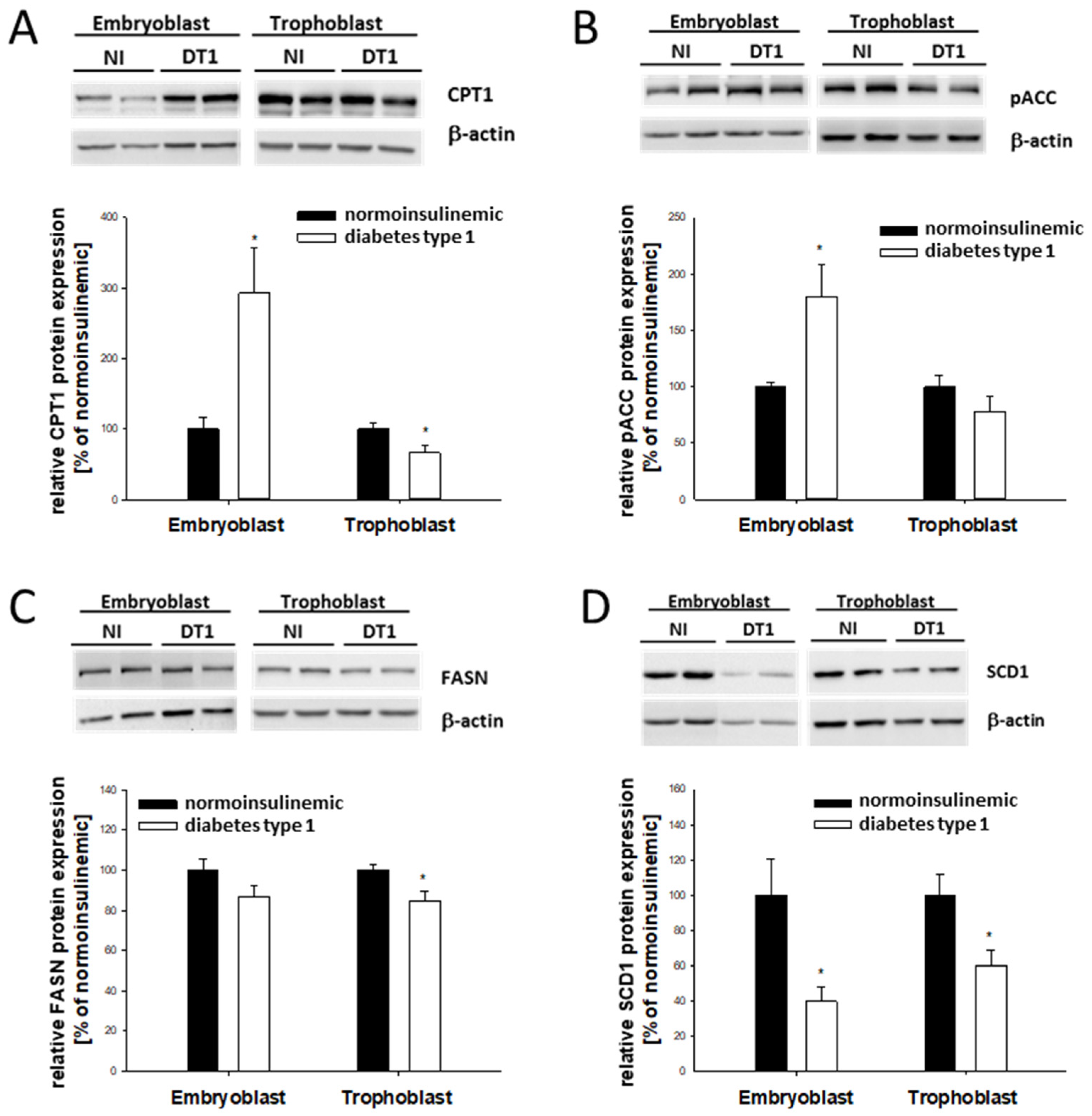
2.3. Lipogenic Marker Expression in Blastocysts from Diabetic Rabbits In Vivo
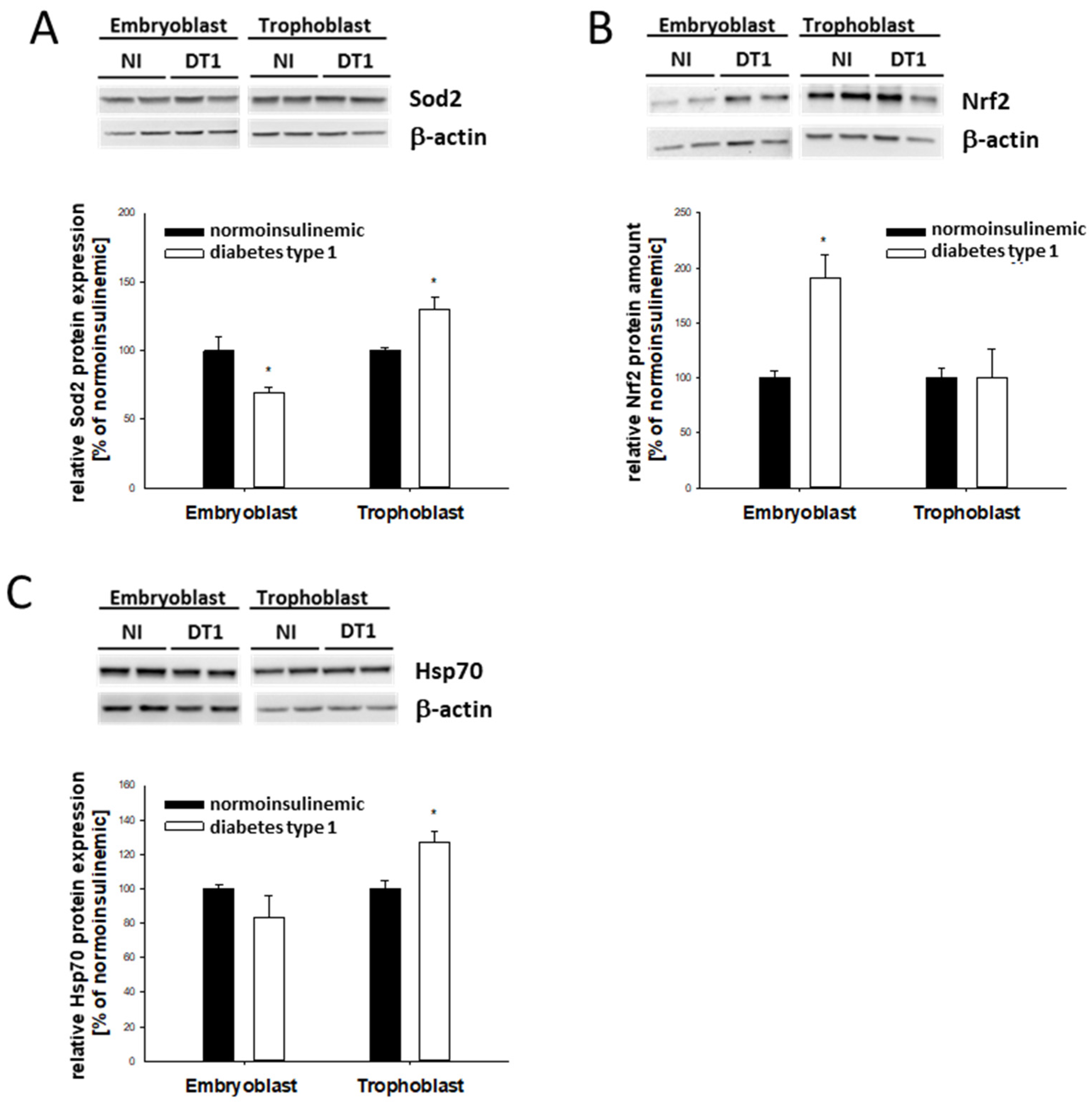
2.4. Stress Marker Expression in Blastocysts from Diabetic Rabbits In Vivo
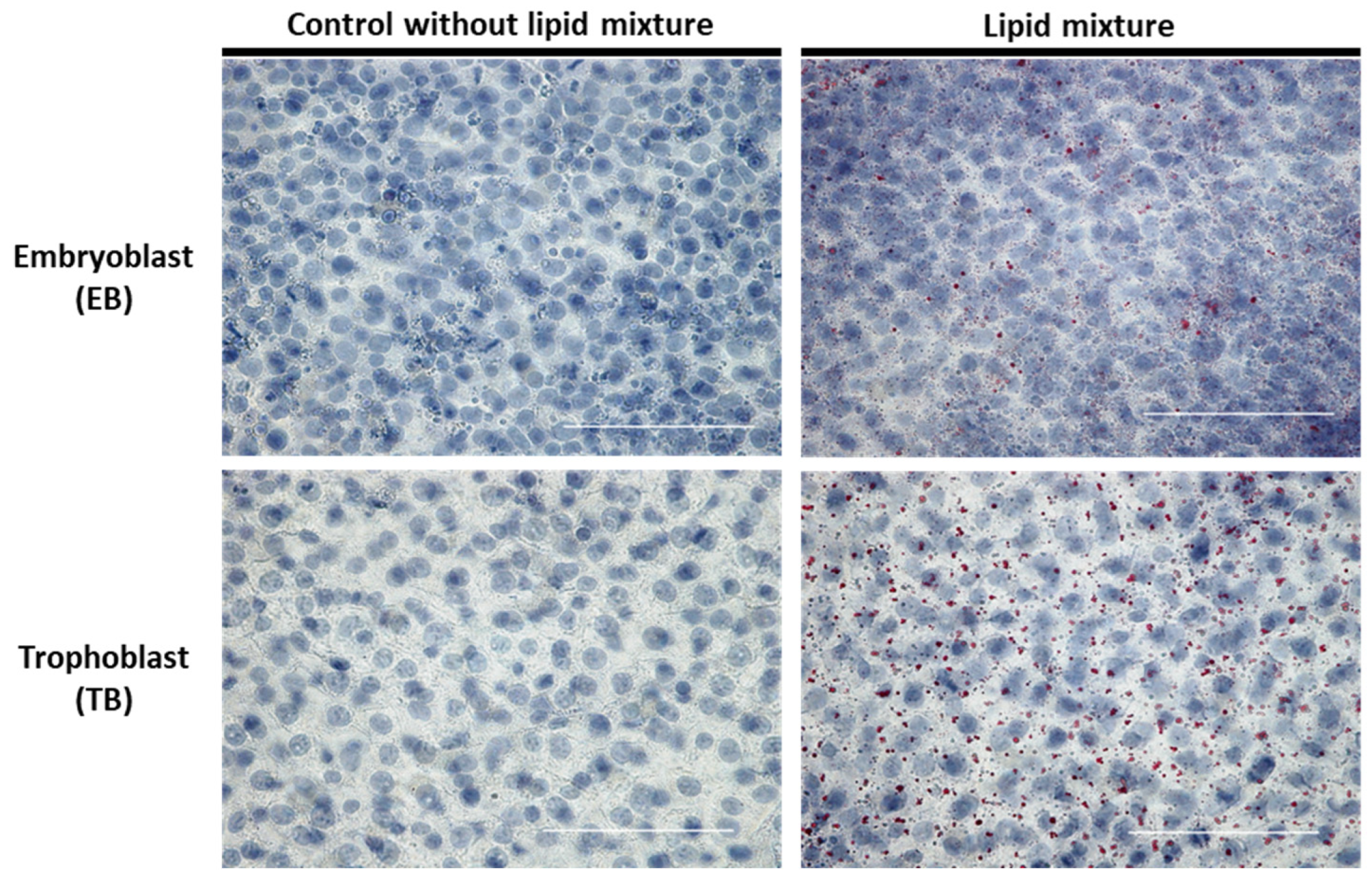
2.5. Lipid-Dependent Regulation of Intracellular Lipid Accumulation in In Vitro-Cultured Blastocysts
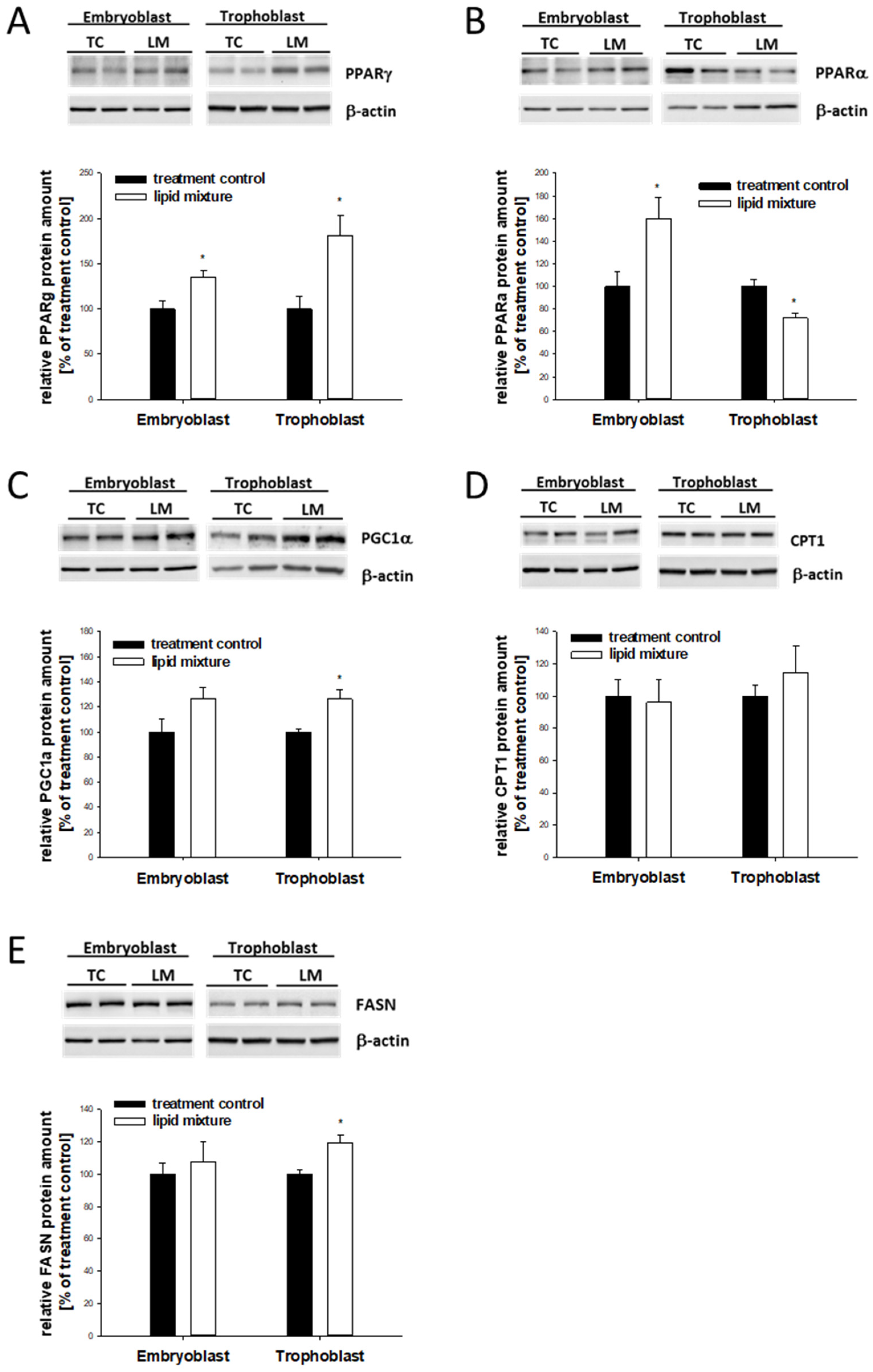
2.6. Lipid-Dependent Expression of Lipogenic Marker Genes in In Vitro-Cultured Blastocysts
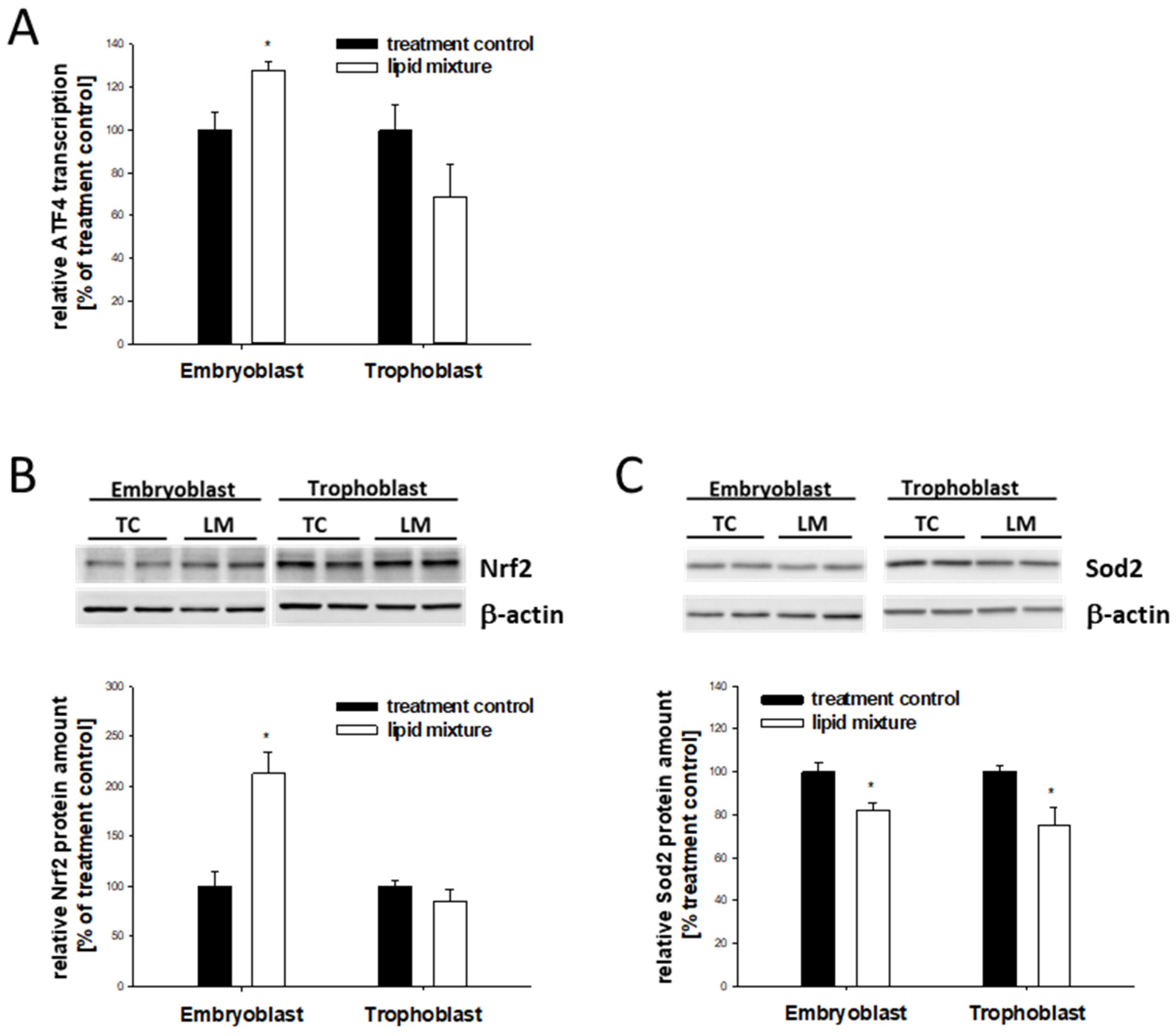
2.7. Lipid-Dependent Expression of Stress Markers in In Vitro-Cultured Blastocysts
3. Discussion
4. Materials and Methods
4.1. Alloxan Treatment
4.2. Embryo Recovery and In Vitro Culture
4.3. Staining of Rabbit Blastocysts
4.4. RNA Isolation and cDNA Synthesis
4.5. Polymerase Chain Reaction (RT-PCR)
4.6. Measurement of mRNA Levels Using Quantitative PCR (qPCR)
4.7. Protein Sample Preparation
4.8. Immunoblotting
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cockburn, K.; Rossant, J. Making the blastocyst: Lessons from the mouse. J. Clin. Investig. 2010, 120, 995–1003. [Google Scholar] [CrossRef]
- Leese, H.J. Metabolism of the preimplantation embryo: 40 years on. Reproduction 2012, 143, 417–427. [Google Scholar] [CrossRef]
- Zhao, Z.; Reece, E.A. New concepts in diabetic embryopathy. Clin. Lab. Med. 2013, 33, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vera, I.; Bonet, B.; Viana, M.; Quintanar, A.; Martín, M.D.; Blanco, P.; Donnay, S.; Albi, M. Changes in plasma lipids and increased low-density lipoprotein susceptibility to oxidation in pregnancies complicated by gestational diabetes: Consequences of obesity. Metab. Clin. Exp. 2007, 56, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Herrera, E. Metabolic adaptations in pregnancy and their implications for the availability of substrates to the fetus. Eur. J. Clin. Nutr. 2000, 54 (Suppl. S1), S47–S51. [Google Scholar] [CrossRef] [PubMed]
- Nolan, C.J.; Riley, S.F.; Sheedy, M.T.; Walstab, J.E.; Beischer, N.A. Maternal serum triglyceride, glucose tolerance, and neonatal birth weight ratio in pregnancy. Diabetes Care 1995, 18, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Wyman, A.; Pinto, A.B.; Sheridan, R.; Moley, K.H. One-cell zygote transfer from diabetic to nondiabetic mouse results in congenital malformations and growth retardation in offspring. Endocrinology 2008, 149, 466–469. [Google Scholar] [CrossRef]
- Rousseau-Ralliard, D.; Couturier-Tarrade, A.; Thieme, R.; Brat, R.; Rolland, A.; Boileau, P.; Aubrière, M.-C.; Daniel, N.; Dahirel, M.; Derisoud, E.; et al. A short periconceptional exposure to maternal type-1 diabetes is sufficient to disrupt the feto-placental phenotype in a rabbit model. Mol. Cell. Endocrinol. 2019, 480, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Ibayashi, M.; Aizawa, R.; Mitsui, J.; Tsukamoto, S. Homeostatic regulation of lipid droplet content in mammalian oocytes and embryos. Reproduction 2021, 162, R99–R109. [Google Scholar] [CrossRef]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta 2011, 1812, 1007–1022. [Google Scholar] [CrossRef]
- Tontonoz, P.; Spiegelman, B.M. Fat and beyond: The diverse biology of PPARgamma. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Rakhshandehroo, M.; Knoch, B.; Müller, M.; Kersten, S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010, 2010, 612089. [Google Scholar] [CrossRef]
- Patel, D.D.; Knight, B.L.; Wiggins, D.; Humphreys, S.M.; Gibbons, G.F. Disturbances in the normal regulation of SREBP-sensitive genes in PPAR alpha-deficient mice. J. Lipid Res. 2001, 42, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Knight, B.L.; Hebbachi, A.; Hauton, D.; Brown, A.-M.; Wiggins, D.; Patel, D.D.; Gibbons, G.F. A role for PPARalpha in the control of SREBP activity and lipid synthesis in the liver. Biochem. J. 2005, 389, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Mauvoisin, D.; Mounier, C. Hormonal and nutritional regulation of SCD1 gene expression. Biochimie 2011, 93, 78–86. [Google Scholar] [CrossRef]
- Liang, H.; Ward, W.F. PGC-1alpha: A key regulator of energy metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef]
- Kim, T.; Yang, Q. Peroxisome-proliferator-activated receptors regulate redox signaling in the cardiovascular system. World J. Cardiol. 2013, 5, 164–174. [Google Scholar] [CrossRef]
- Ding, G.; Fu, M.; Qin, Q.; Lewis, W.; Kim, H.W.; Fukai, T.; Bacanamwo, M.; Chen, Y.E.; Schneider, M.D.; Mangelsdorf, D.J.; et al. Cardiac peroxisome proliferator-activated receptor gamma is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc. Res. 2007, 76, 269–279. [Google Scholar] [CrossRef]
- Guellich, A.; Damy, T.; Lecarpentier, Y.; Conti, M.; Claes, V.; Samuel, J.L.; Quillard, J.; Hébert, J.L.; Pineau, T.; Coirault, C. Role of oxidative stress in cardiac dysfunction of PPARalpha−/− mice. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H93–H102. [Google Scholar] [CrossRef]
- Girnun, G.D.; Domann, F.E.; Moore, S.A.; Robbins, M.E. Identification of a functional peroxisome proliferator-activated receptor response element in the rat catalase promoter. Mol. Endocrinol. 2002, 16, 2793–2801. [Google Scholar] [CrossRef]
- Polvani, S.; Tarocchi, M.; Galli, A. PPARγ and Oxidative Stress: Con(β) Catenating NRF2 and FOXO. PPAR Res. 2012, 2012, 641087. [Google Scholar] [CrossRef]
- Gureev, A.P.; Shaforostova, E.A.; Popov, V.N. Regulation of Mitochondrial Biogenesis as a Way for Active Longevity: Interaction Between the Nrf2 and PGC-1α Signaling Pathways. Front. Genet. 2019, 10, 435. [Google Scholar] [CrossRef]
- Papp, E.; Nardai, G.; Söti, C.; Csermely, P. Molecular chaperones, stress proteins and redox homeostasis. Biofactors 2003, 17, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Bellmann, K.; Jäättelä, M.; Wissing, D.; Burkart, V.; Kolb, H. Heat shock protein hsp70 overexpression confers resistance against nitric oxide. FEBS Lett. 1996, 391, 185–188. [Google Scholar] [CrossRef]
- Jäättelä, M.; Wissing, D.; Kokholm, K.; Kallunki, T.; Egeblad, M. Hsp70 exerts its anti-apoptotic function downstream of caspase-3-like proteases. EMBO J. 1998, 17, 6124–6134. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.; Martinus, R.D. Hyperglycaemia and oxidative stress upregulate HSP60 & HSP70 expression in HeLa cells. Springerplus 2013, 2, 431. [Google Scholar] [CrossRef]
- Zhao, Z. Endoplasmic reticulum stress in maternal diabetes-induced cardiac malformations during critical cardiogenesis period. Birth Defects Res. B Dev. Reprod. Toxicol. 2012, 95, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Eckert, R.L.; Reece, E.A. Reduction in embryonic malformations and alleviation of endoplasmic reticulum stress by nitric oxide synthase inhibition in diabetic embryopathy. Reprod. Sci. 2012, 19, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhao, Z.; Eckert, R.L.; Reece, E.A. The essential role of protein kinase Cδ in diabetes-induced neural tube defects. J. Matern. Fetal Neonatal Med. 2012, 25, 2020–2024. [Google Scholar] [CrossRef]
- Cho, H.; Wu, M.; Zhang, L.; Thompson, R.; Nath, A.; Chan, C. Signaling dynamics of palmitate-induced ER stress responses mediated by ATF4 in HepG2 cells. BMC Syst. Biol. 2013, 7, 9. [Google Scholar] [CrossRef]
- Schindler, M.; Fischer, S.; Thieme, R.; Fischer, B.; Santos, A.N. cAMP-responsive element binding protein: A vital link in embryonic hormonal adaptation. Endocrinology 2013, 154, 2208–2221. [Google Scholar] [CrossRef]
- Houghton, F.D. Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation 2006, 74, 11–18. [Google Scholar] [CrossRef]
- Hewitson, L.C.; Leese, H.J. Energy metabolism of the trophectoderm and inner cell mass of the mouse blastocyst. J. Exp. Zool. 1993, 267, 337–343. [Google Scholar] [CrossRef]
- Gürke, J.; Schindler, M.; Pendzialek, S.M.; Thieme, R.; Grybel, K.J.; Heller, R.; Spengler, K.; Fleming, T.P.; Fischer, B.; Navarrete Santos, A. Maternal diabetes promotes mTORC1 downstream signalling in rabbit preimplantation embryos. Reproduction 2016, 151, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Schindler, M.; Pendzialek, M.; Navarrete Santos, A.; Plösch, T.; Seyring, S.; Gürke, J.; Haucke, E.; Knelangen, J.M.; Fischer, B.; Santos, A.N. Maternal diabetes leads to unphysiological high lipid accumulation in rabbit preimplantation embryos. Endocrinology 2014, 155, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Schindler, M.; Dannenberger, D.; Nuernberg, G.; Pendzialek, M.; Grybel, K.; Seeling, T.; Santos, A.N. Embryonic fatty acid metabolism in diabetic pregnancy: The difference between embryoblasts and trophoblasts. Mol. Hum. Reprod. 2020, 26, 837–849. [Google Scholar] [CrossRef]
- Kilby, M.D.; Neary, R.H.; Mackness, M.I.; Durrington, P.N. Fetal and maternal lipoprotein metabolism in human pregnancy complicated by type I diabetes mellitus. J. Clin. Endocrinol. Metab. 1998, 83, 1736–1741. [Google Scholar] [CrossRef]
- Merzouk, H.; Madani, S.; Korso, N.; Bouchenak, M.; Prost, J.; Belleville, J. Maternal and fetal serum lipid and lipoprotein concentrations and compositions in type 1 diabetic pregnancy: Relationship with maternal glycemic control. J. Lab. Clin. Med. 2000, 136, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Mau, K.H.T.; Karimlou, D.; Barneda, D.; Brochard, V.; Royer, C.; Leeke, B.; de Souza, R.A.; Pailles, M.; Percharde, M.; Srinivas, S.; et al. Dynamic enlargement and mobilization of lipid droplets in pluripotent cells coordinate morphogenesis during mouse peri-implantation development. Nat. Commun. 2022, 13, 3861. [Google Scholar] [CrossRef]
- Königsdorf, C.A.I.; Navarrete Santos, A.; Schmidt, J.-S.; Fischer, S.; Fischer, B. Expression profile of fatty acid metabolism genes in preimplantation blastocysts of obese and non-obese mice. Obes. Facts 2012, 5, 575–586. [Google Scholar] [CrossRef]
- Mishima, T.; Miner, J.H.; Morizane, M.; Stahl, A.; Sadovsky, Y. The expression and function of fatty acid transport protein-2 and -4 in the murine placenta. PLoS ONE 2011, 6, e25865. [Google Scholar] [CrossRef] [PubMed]
- Scifres, C.M.; Chen, B.; Nelson, D.M.; Sadovsky, Y. Fatty acid binding protein 4 regulates intracellular lipid accumulation in human trophoblasts. J. Clin. Endocrinol. Metab. 2011, 96, E1083-91. [Google Scholar] [CrossRef]
- Storch, J.; McDermott, L. Structural and functional analysis of fatty acid-binding proteins. J. Lipid Res. 2009, 50, S126–S131. [Google Scholar] [CrossRef] [PubMed]
- Ayers, S.D.; Nedrow, K.L.; Gillilan, R.E.; Noy, N. Continuous nucleocytoplasmic shuttling underlies transcriptional activation of PPARgamma by FABP4. Biochemistry 2007, 46, 6744–6752. [Google Scholar] [CrossRef] [PubMed]
- Schaiff, W.T.; Knapp, F.F.R.; Barak, Y.; Biron-Shental, T.; Nelson, D.M.; Sadovsky, Y. Ligand-activated peroxisome proliferator activated receptor gamma alters placental morphology and placental fatty acid uptake in mice. Endocrinology 2007, 148, 3625–3634. [Google Scholar] [CrossRef]
- Lefebvre, P.; Chinetti, G.; Fruchart, J.-C.; Staels, B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Investig. 2006, 116, 571–580. [Google Scholar] [CrossRef]
- Schindler, M.; Pendzialek, M.; Grybel, K.J.; Seeling, T.; Gürke, J.; Fischer, B.; Navarrete Santos, A. Adiponectin stimulates lipid metabolism via AMPK in rabbit blastocysts. Hum. Reprod. 2017, 32, 1382–1392. [Google Scholar] [CrossRef]
- Hewitson, L.C.; Martin, K.L.; Leese, H.J. Effects of metabolic inhibitors on mouse preimplantation embryo development and the energy metabolism of isolated inner cell masses. Mol. Reprod. Dev. 1996, 43, 323–330. [Google Scholar] [CrossRef]
- Dunning, K.R.; Cashman, K.; Russell, D.L.; Thompson, J.G.; Norman, R.J.; Robker, R.L. Beta-oxidation is essential for mouse oocyte developmental competence and early embryo development. Biol. Reprod. 2010, 83, 909–918. [Google Scholar] [CrossRef]
- Valsangkar, D.; Downs, S.M. A requirement for fatty acid oxidation in the hormone-induced meiotic maturation of mouse oocytes. Biol. Reprod. 2013, 89, 43. [Google Scholar] [CrossRef]
- Paczkowski, M.; Schoolcraft, W.B.; Krisher, R.L. Fatty acid metabolism during maturation affects glucose uptake and is essential to oocyte competence. Reproduction 2014, 148, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Chirala, S.S.; Chang, H.; Matzuk, M.; Abu-Elheiga, L.; Mao, J.; Mahon, K.; Finegold, M.; Wakil, S.J. Fatty acid synthesis is essential in embryonic development: Fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc. Natl. Acad. Sci. USA 2003, 100, 6358–6363. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef]
- Daniel, Z.C.T.R.; Richards, S.E.; Salter, A.M.; Buttery, P.J. Insulin and dexamethasone regulate stearoyl-CoA desaturase mRNA levels and fatty acid synthesis in ovine adipose tissue explants. J. Anim. Sci. 2004, 82, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Claycombe, K.J.; Jones, B.H.; Standridge, M.K.; Guo, Y.; Chun, J.T.; Taylor, J.W.; Moustaïd-Moussa, N. Insulin increases fatty acid synthase gene transcription in human adipocytes. Am. J. Physiol. 1998, 274, R1253–R1259. [Google Scholar] [CrossRef]
- Schaffer, J.E. Lipotoxicity: When tissues overeat. Curr. Opin. Lipidol. 2003, 14, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Borradaile, N.M.; Han, X.; Harp, J.D.; Gale, S.E.; Ory, D.S.; Schaffer, J.E. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J. Lipid Res. 2006, 47, 2726–2737. [Google Scholar] [CrossRef]
- Wu, L.L.; Russell, D.L.; Wong, S.L.; Chen, M.; Tsai, T.-S.; St John, J.C.; Norman, R.J.; Febbraio, M.A.; Carroll, J.; Robker, R.L. Mitochondrial dysfunction in oocytes of obese mothers: Transmission to offspring and reversal by pharmacological endoplasmic reticulum stress inhibitors. Development 2015, 142, 681–691. [Google Scholar] [CrossRef]
- Takahashi, M.; Keicho, K.; Takahashi, H.; Ogawa, H.; Schulte, R.M.; Okano, A. Effect of oxidative stress on development and DNA damage in in-vitro cultured bovine embryos by comet assay. Theriogenology 2000, 54, 137–145. [Google Scholar] [CrossRef]
- Duvnjak, M.; Lerotić, I.; Barsić, N.; Tomasić, V.; Virović Jukić, L.; Velagić, V. Pathogenesis and management issues for non-alcoholic fatty liver disease. World J. Gastroenterol. 2007, 13, 4539–4550. [Google Scholar] [CrossRef]
- Sturmey, R.G.; Hawkhead, J.A.; Barker, E.A.; Leese, H.J. DNA damage and metabolic activity in the preimplantation embryo. Hum. Reprod. 2009, 24, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Sun, Q.; Su, Q.; Dang, S.; Chen, G. Taurine promotes cognitive function in prenatally stressed juvenile rats via activating the Akt-CREB-PGC1α pathway. Redox Biol. 2016, 10, 179–190. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Bhattarai, K.R.; Riaz, T.A.; Kim, H.R.; Chae, H.J. The aftermath of the interplay between the endoplasmic reticulum stress response and redox signaling. Exp. Mol. Med. 2021, 53, 151–167. [Google Scholar] [CrossRef]
- Ramin, N.; Thieme, R.; Fischer, S.; Schindler, M.; Schmidt, T.; Fischer, B.; Navarrete Santos, A. Maternal diabetes impairs gastrulation and insulin and IGF-I receptor expression in rabbit blastocysts. Endocrinology 2010, 151, 4158–4167. [Google Scholar] [CrossRef] [PubMed]
- Moley, K.H. Hyperglycemia and apoptosis: Mechanisms for congenital malformations and pregnancy loss in diabetic women. Trends Endocrinol. Metab. 2001, 12, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, R.J. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999, 13, 1211–1233. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, L.; Tabas, I. Role of endoplasmic reticulum stress in metabolic disease and other disorders. Annu. Rev. Med. 2012, 63, 317–328. [Google Scholar] [CrossRef]
- Xiao, G.; Zhang, T.; Yu, S.; Lee, S.; Calabuig-Navarro, V.; Yamauchi, J.; Ringquist, S.; Dong, H.H. ATF4 protein deficiency protects against high fructose-induced hypertriglyceridemia in mice. J. Biol. Chem. 2013, 288, 25350–25361. [Google Scholar] [CrossRef]
- Maurer, R.R. Advances in rabbit embryo culture. In Methods in Mammalian Reproduction; Daniels, J.C., Ed.; Academic Press: New York, NY, USA, 1978; Volume 1978, pp. 259–272. [Google Scholar]
- Fischer, B.; Chavatte-Palmer, P.; Viebahn, C.; Navarrete Santos, A.; Duranthon, V. Rabbit as a reproductive model for human health. Reproduction 2012, 144, 1–10. [Google Scholar] [CrossRef]
- Beyaz, S.; Mana, M.D.; Roper, J.; Kedrin, D.; Saadatpour, A.; Hong, S.-J.; Bauer-Rowe, K.E.; Xifaras, M.E.; Akkad, A.; Arias, E.; et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016, 531, 53–58. [Google Scholar] [CrossRef]
- Zaytouni, T.; Tsai, P.-Y.; Hitchcock, D.S.; DuBois, C.D.; Freinkman, E.; Lin, L.; Morales-Oyarvide, V.; Lenehan, P.J.; Wolpin, B.M.; Mino-Kenudson, M.; et al. Critical role for arginase 2 in obesity-associated pancreatic cancer. Nat. Commun. 2017, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Tonack, S.; Fischer, B.; Navarrete Santos, A. Expression of the insulin-responsive glucose transporter isoform 4 in blastocysts of C57/BL6 mice. Anat. Embryol. 2004, 208, 225–230. [Google Scholar] [CrossRef]
- Seeling, T.; Haucke, E.; Navarrete Santos, A.; Grybel, K.J.; Gürke, J.; Pendzialek, S.M.; Schindler, M.; Simm, A.; Navarrete Santos, A. Glyoxalase 1 expression is downregulated in preimplantation blastocysts of diabetic rabbits. Reprod. Domest. Anim. 2019, 54 (Suppl. S3), 4–11. [Google Scholar] [CrossRef] [PubMed]
- Thieme, R.; Schindler, M.; Ramin, N.; Fischer, S.; Mühleck, B.; Fischer, B.; Navarrete Santos, A. Insulin growth factor adjustment in preimplantation rabbit blastocysts and uterine tissues in response to maternal type 1 diabetes. Mol. Cell. Endocrinol. 2012, 358, 96–103. [Google Scholar] [CrossRef]
- de Nivelle, J.; Thoma, J.; Toto Nienguesso, A.; Seeling, T.; Jung, J.-S.; Navarrete Santos, A.; Schindler, M. Rabbit as an Aging Model in Reproduction: Advanced Maternal Age Alters GLO1 Expression in the Endometrium at the Time of Implantation. Appl. Sci. 2020, 10, 7732. [Google Scholar] [CrossRef]
- Pendzialek, S.M.; Schindler, M.; Plösch, T.; Gürke, J.; Haucke, E.; Hecht, S.; Fischer, B.; Santos, A.N. Cholesterol metabolism in rabbit blastocysts under maternal diabetes. Reprod. Fertil. Dev. 2017, 29, 1921–1931. [Google Scholar] [CrossRef] [PubMed]
| Relative mRNA Amount (in % of Normoinsulinaemic EB) | ||||
|---|---|---|---|---|
| Embryoblast | Trophoblast | |||
| Gene | Normoinsulinemic | Diabetic | Normoinsulinemic | Diabetic |
| PPARα | 100.0 ± 16.9 | 116.0 ± 11.8 | 44.4 ± 8.6 b | 29.1 ± 6.3 b |
| PPARγ | 100.0 ± 15.1 | 177.9 ± 35.4 a | 753.1 ± 58.9 b | 528.2 ± 135.8 b |
| CPT1 | 100.0 ± 16.2 | 125.1 ± 35.4 | 222.4 ± 58.1 | 90.8 ± 15.9 |
| CD36 | 100.0 ± 24.3 | 60.3 ± 11.3 | 286.6 ± 29.1 b | 295.4 ± 41.6 b |
| Relative mRNA Amount [5 of Nontreated Control] | ||||
|---|---|---|---|---|
| Gene | Embryoblast | Trophoblast | ||
| mRNA | Treatment Control | Lipid Mixture | Treatment Control | Lipid Mixture |
| PPARα | 100.0 ± 7.8 | 112.1 ± 11.0 | 100.0 ± 10.3 | 70.6 ± 6.6 * |
| PPARγ | 100.0 ± 8.6 | 169.9 ± 20.4 * | 100.0 ± 10.5 | 101.3 ± 11.7 |
| CPT1 | 100.0 ± 9.9 | 136.9 ± 19.7 | 100.0 ± 13.8 | 59.1 ± 6.6 * |
| FASN | 100.0 ± 10.9 | 167.9 ± 49.2 | 100.0 ± 12.4 | 49.7 ± 6.4 * |
| FABP4 | 100.0 ± 7.1 | 170.1 ± 23.4 * | 100.0 ± 7.8 | 200.3 ± 24.7 * |
| FATP4 | 100.0 ± 11.5 | 141.0 ± 11.9 * | 100.0 ± 13.8 | 85.2 ± 9.5 |
| CD36 | 100.0 ± 16.6 | 238.5 ± 43.9 * | 100.0 ± 9.3 | 104.4 ± 11.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schindler, M.; Geisler, S.M.; Seeling, T.; Navarrete Santos, A. Ectopic Lipid Accumulation Correlates with Cellular Stress in Rabbit Blastocysts from Diabetic Mothers. Int. J. Mol. Sci. 2023, 24, 11776. https://doi.org/10.3390/ijms241411776
Schindler M, Geisler SM, Seeling T, Navarrete Santos A. Ectopic Lipid Accumulation Correlates with Cellular Stress in Rabbit Blastocysts from Diabetic Mothers. International Journal of Molecular Sciences. 2023; 24(14):11776. https://doi.org/10.3390/ijms241411776
Chicago/Turabian StyleSchindler, Maria, Sophia Mareike Geisler, Tom Seeling, and Anne Navarrete Santos. 2023. "Ectopic Lipid Accumulation Correlates with Cellular Stress in Rabbit Blastocysts from Diabetic Mothers" International Journal of Molecular Sciences 24, no. 14: 11776. https://doi.org/10.3390/ijms241411776
APA StyleSchindler, M., Geisler, S. M., Seeling, T., & Navarrete Santos, A. (2023). Ectopic Lipid Accumulation Correlates with Cellular Stress in Rabbit Blastocysts from Diabetic Mothers. International Journal of Molecular Sciences, 24(14), 11776. https://doi.org/10.3390/ijms241411776





