Alternative Splicing during Fiber Development in G. hirsutum
Abstract
:1. Introduction
2. Results
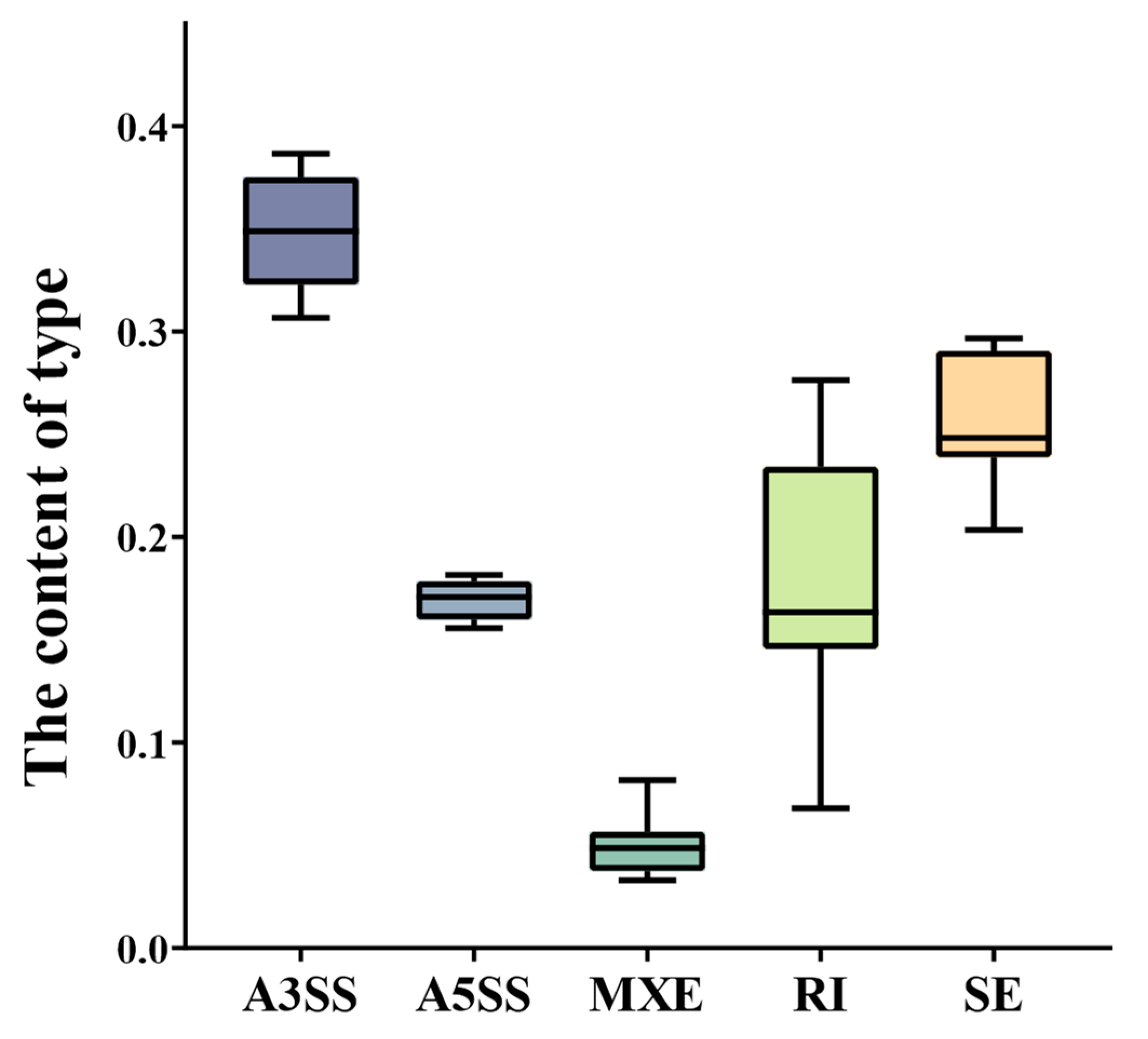
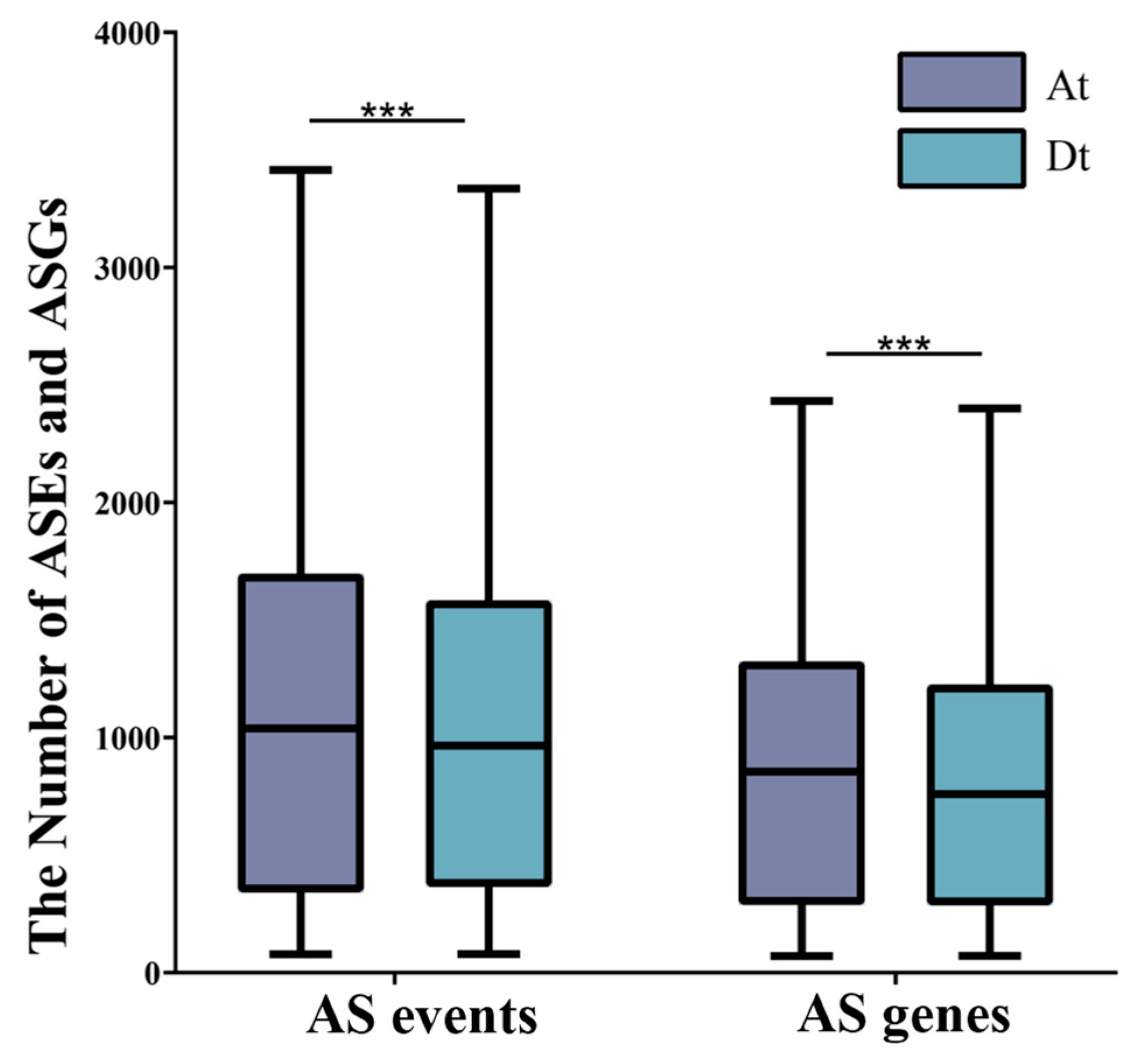
2.1. Statistical Analysis of AS Genes and AS Events
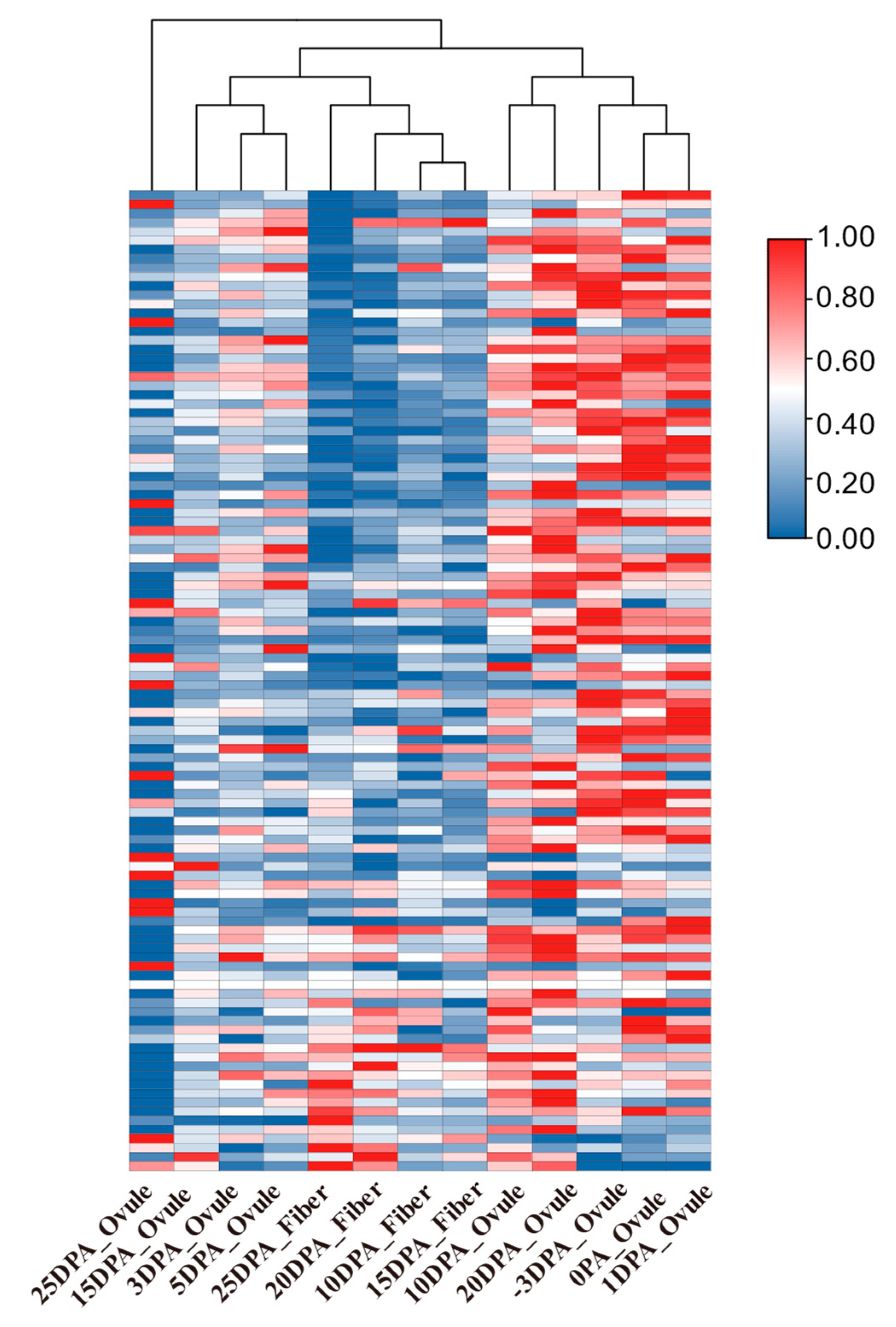
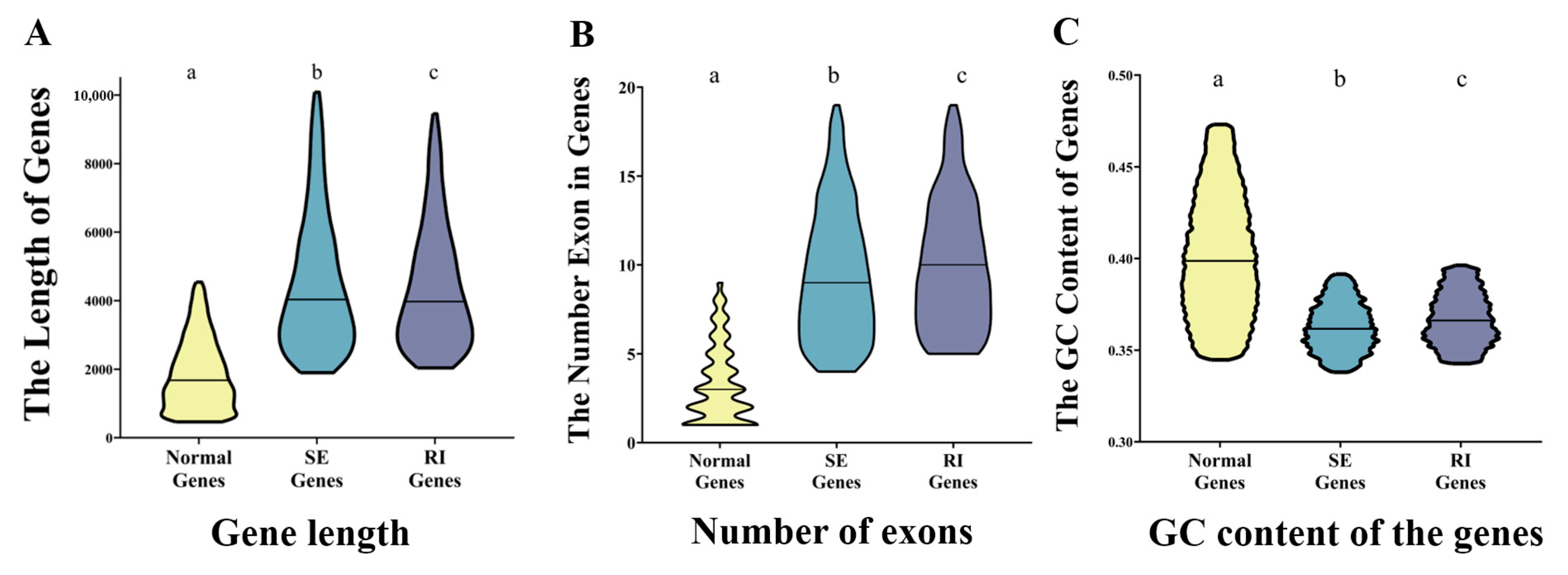
2.2. Identification of the Fiber-Development-Related AS Genes and AS Events
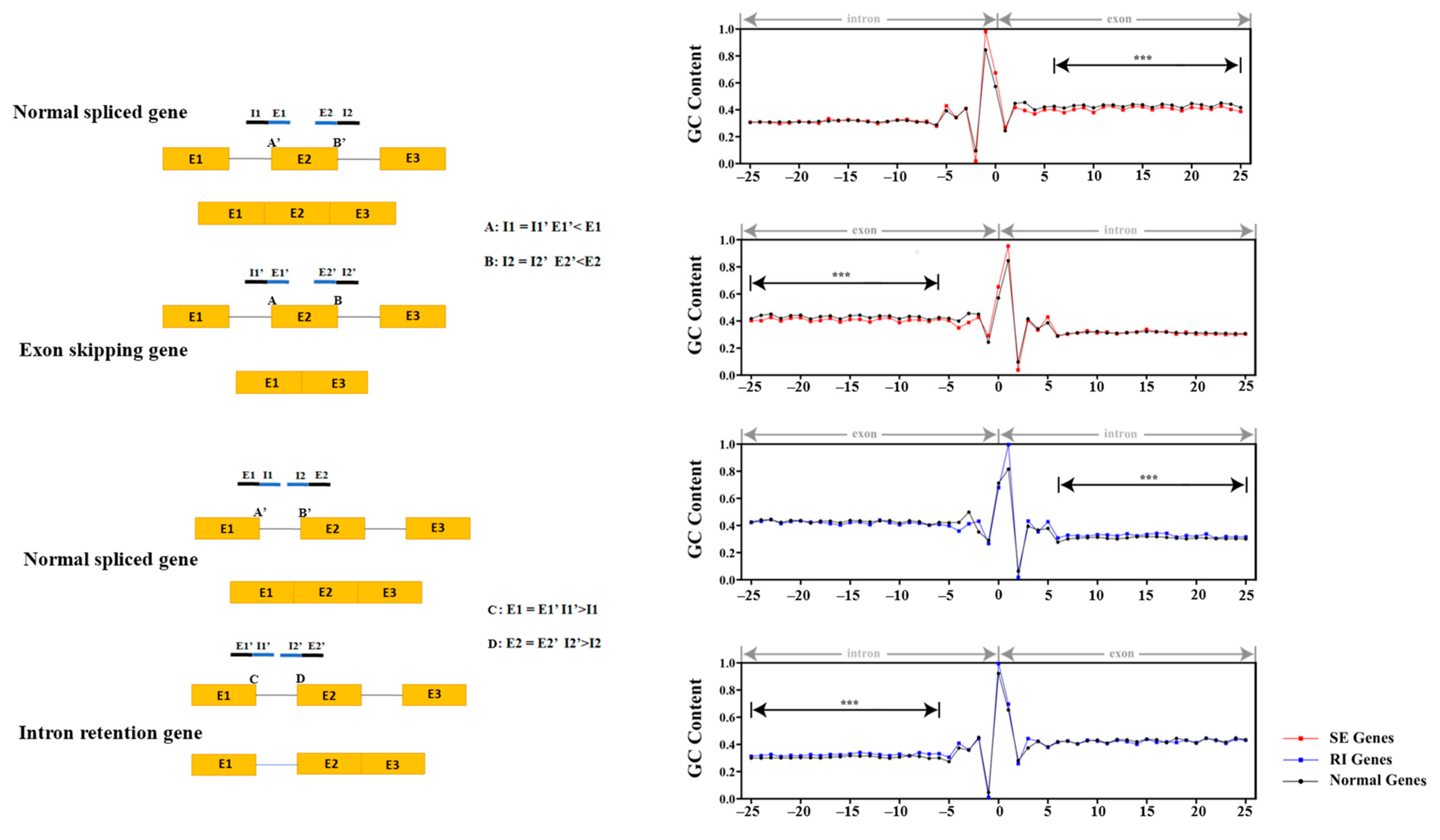
2.3. Regulation of AS in Cotton
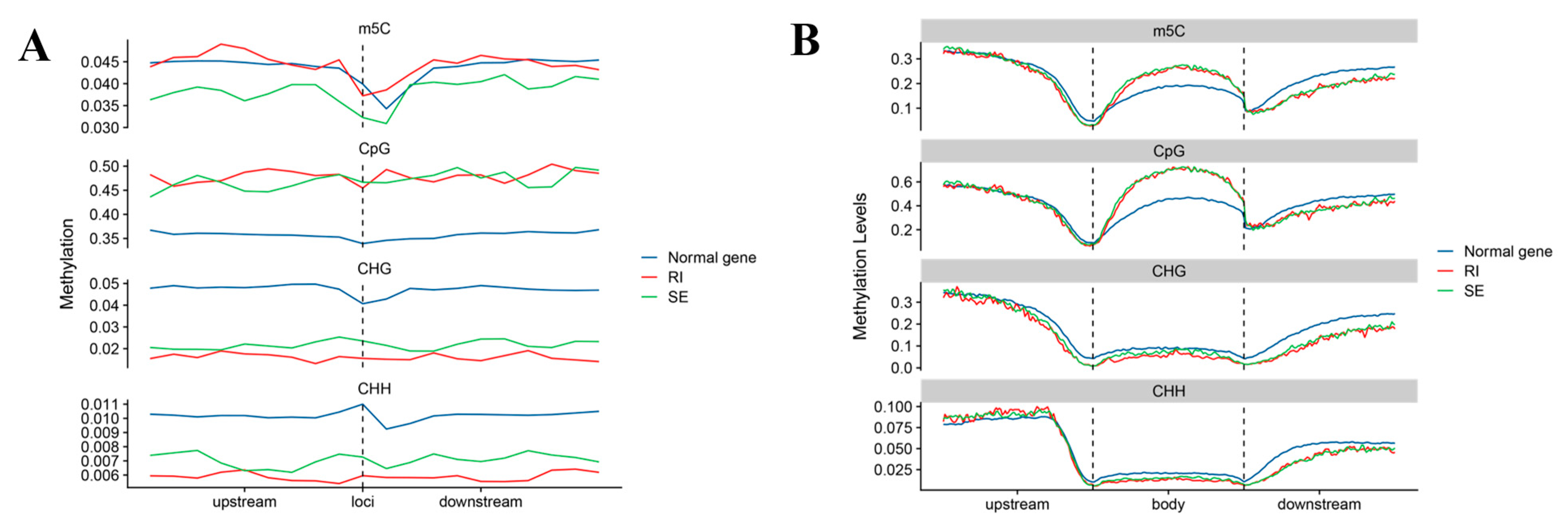
2.4. Different Types of AS Genes Methylation in Cotton
3. Discussion
4. Materials and Methods
4.1. Data Availability
4.2. Processing of the Raw Data and Alignment
4.3. Performing AS and Statistical Analysis
4.4. Gene Expression Analysis
4.5. AS Genes’ Sequence Analysis
4.6. Methylation’s Sequence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, G.; Huang, J.Q.; Chen, X.Y.; Zhu, Y.X. Recent Advances and Future Perspectives in Cotton Research. Annu. Rev. Plant Biol. 2021, 72, 437–462. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.W.; Yu, N.; Li, C.H.; Luo, B.; Gou, J.Y.; Wang, L.J.; Chen, X.Y. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 2004, 16, 2323–2334. [Google Scholar] [CrossRef] [Green Version]
- Haigler, C.; Betancur, L.; Stiff, M.; Tuttle, J. Cotton fiber: A powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 2012, 3, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Mei, H.; Lu, H.; Chen, R.; Hu, Y.; Zhang, T. Transcriptome Time-Course Analysis in the Whole Period of Cotton Fiber Development. Front. Plant Sci. 2022, 13, 864529. [Google Scholar] [CrossRef]
- Lam, P.Y.; Wang, L.; Lo, C.; Zhu, F.Y. Alternative Splicing and Its Roles in Plant Metabolism. Int. J. Mol. Sci. 2022, 23, 7355. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants--coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Jia, Z.; Pu, Q.; Tian, Y.; Zhu, F.; Liu, Y. ABA Mediates Plant Development and Abiotic Stress via Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 3796. [Google Scholar] [CrossRef]
- Liu, J.; Chen, S.; Liu, M.; Chen, Y.; Fan, W.; Lee, S.; Xiao, H.; Kudrna, D.; Li, Z.; Chen, X.; et al. Full-Length Transcriptome Sequencing Reveals Alternative Splicing and lncRNA Regulation during Nodule Development in Glycine max. Int. J. Mol. Sci. 2022, 23, 7371. [Google Scholar] [CrossRef]
- Liu, X.X.; Guo, Q.H.; Xu, W.B.; Liu, P.; Yan, K. Rapid Regulation of Alternative Splicing in Response to Environmental Stresses. Front. Plant Sci. 2022, 13, 832177. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, X.H.; Guo, Q.H.; Liu, P.; Li, Y.; Wu, C.A.; Yang, G.D.; Huang, J.G.; Zhang, S.Z.; Zheng, C.C.; et al. Salt responsive alternative splicing of a RING finger E3 ligase modulates the salt stress tolerance by fine-tuning the balance of COP9 signalosome subunit 5A. PLoS Genet. 2021, 17, e1009898. [Google Scholar] [CrossRef]
- Rosenkranz, R.R.E.; Ullrich, S.; Löchli, K.; Simm, S.; Fragkostefanakis, S. Relevance and Regulation of Alternative Splicing in Plant Heat Stress Response: Current Understanding and Future Directions. Front. Plant Sci. 2022, 13, 911277. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, K.; Bartsch, M.; Ding, J.; Soppe, W.J. Seed Dormancy in Arabidopsis Requires Self-Binding Ability of DOG1 Protein and the Presence of Multiple Isoforms Generated by Alternative Splicing. PLoS Genet. 2015, 11, e1005737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzyk, P.; Brown, J.W.; Nimmo, H.G. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 2012, 24, 961–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Chen, H.; Deng, X.W.; Irish, V.F.; Wei, N. Phytochrome B Induces Intron Retention and Translational Inhibition of PHYTOCHROME-INTERACTING FACTOR3. Plant Physiol. 2020, 182, 159–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutz, U.; Posé, D.; Pfeifer, M.; Gundlach, H.; Hagmann, J.; Wang, C.; Weigel, D.; Mayer, K.F.; Schmid, M.; Schwechheimer, C. Modulation of Ambient Temperature-Dependent Flowering in Arabidopsis thaliana by Natural Variation of FLOWERING LOCUS M. PLoS Genet. 2015, 11, e1005588. [Google Scholar] [CrossRef]
- Zhan, X.; Qian, B.; Cao, F.; Wu, W.; Yang, L.; Guan, Q.; Gu, X.; Wang, P.; Okusolubo, T.A.; Dunn, S.L.; et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 2015, 6, 8139. [Google Scholar] [CrossRef] [Green Version]
- Cucinotta, M.; Cavalleri, A.; Guazzotti, A.; Astori, C.; Manrique, S.; Bombarely, A.; Oliveto, S.; Biffo, S.; Weijers, D.; Kater, M.M.; et al. Alternative Splicing Generates a MONOPTEROS Isoform Required for Ovule Development. Curr. Biol. 2021, 31, 892–899.e893. [Google Scholar] [CrossRef]
- Liu, L.; Tang, Z.; Liu, F.; Mao, F.; Yujuan, G.; Wang, Z.; Zhao, X. Normal, novel or none: Versatile regulation from alternative splicing. Plant Signal Behav. 2021, 16, 1917170. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Feng, M.; Yang, G.; Sun, L.; Qin, Z.; Cao, J.; Wen, J.; Li, H.; Zhou, Y.; Chen, X.; et al. Changes in Alternative Splicing in Response to Domestication and Polyploidization in Wheat. Plant Physiol. 2020, 184, 1955–1968. [Google Scholar] [CrossRef]
- Gao, P.; Quilichini, T.D.; Zhai, C.; Qin, L.; Nilsen, K.T.; Li, Q.; Sharpe, A.G.; Kochian, L.V.; Zou, J.; Reddy, A.S.N.; et al. Alternative splicing dynamics and evolutionary divergence during embryogenesis in wheat species. Plant Biotechnol. J. 2021, 19, 1624–1643. [Google Scholar] [CrossRef]
- Feng, Y.; Wei, R.; Liu, A.; Fan, S.; Che, J.; Zhang, Z.; Tian, B.; Yuan, Y.; Shi, G.; Shang, H. Genome-wide identification, evolution, expression, and alternative splicing profiles of peroxiredoxin genes in cotton. PeerJ 2021, 9, e10685. [Google Scholar] [CrossRef]
- Lightfoot, D.J.; Malone, K.M.; Timmis, J.N.; Orford, S.J. Evidence for alternative splicing of MADS-box transcripts in developing cotton fibre cells. Mol. Genet. Genom. 2008, 279, 75–85. [Google Scholar] [CrossRef]
- Feng, S.; Xu, M.; Liu, F.; Cui, C.; Zhou, B. Reconstruction of the full-length transcriptome atlas using PacBio Iso-Seq provides insight into the alternative splicing in Gossypium australe. BMC Plant Biol. 2019, 19, 365. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Xiao, G.; Zhu, Y.X. Single-nucleotide resolution mapping of the Gossypium raimondii transcriptome reveals a new mechanism for alternative splicing of introns. Mol. Plant 2014, 7, 829–840. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Li, W.; Zhang, F.; Guo, W. RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. BMC Genom. 2018, 19, 73. [Google Scholar] [CrossRef]
- Wang, M.; Wang, P.; Liang, F.; Ye, Z.; Li, J.; Shen, C.; Pei, L.; Wang, F.; Hu, J.; Tu, L.; et al. A global survey of alternative splicing in allopolyploid cotton: Landscape, complexity and regulation. New Phytol. 2018, 217, 163–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thatcher, S.R.; Zhou, W.; Leonard, A.; Wang, B.B.; Beatty, M.; Zastrow-Hayes, G.; Zhao, X.; Baumgarten, A.; Li, B. Genome-wide analysis of alternative splicing in Zea mays: Landscape and genetic regulation. Plant Cell 2014, 26, 3472–3487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chodavarapu, R.K.; Feng, S.; Bernatavichute, Y.V.; Chen, P.Y.; Stroud, H.; Yu, Y.; Hetzel, J.A.; Kuo, F.; Kim, J.; Cokus, S.J.; et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010, 466, 388–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, S.; Park, J.W.; Lu Z-x Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differ-ential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Cortés, A.J.; Skeen, P.; Blair, M.W.; Chacón-Sánchez, M.I. Does the Genomic Landscape of Species Divergence in Phaseolus Beans Coerce Parallel Signatures of Adaptation and Domestication? Front. Plant Sci. 2018, 9, 1816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortés, A.J.; Blair, M.W. Genotyping by Sequencing and Genome–Environment Associations in Wild Common Bean Predict Widespread Divergent Adaptation to Drought. Front. Plant Sci. 2018, 9, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.; Ahi, E.P. The importance of alternative splicing in adaptive evolution. Mol. Ecol. 2022, 31, 1928–1938. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, Y.; Yao, J.; Huang, H.; Qian, B.; Liu, X.; Chen, Y.; Pang, J.; Zhan, X.; Zhu, J.K.; et al. Modulation of plant development and chilling stress responses by alternative splicing events under control of the spliceosome protein SmEb in Arabidopsis. Plant Cell Env. 2022, 45, 2762–2779. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tian, Y.; Chi, W.; Zhang, H.; Yu, J.; Chen, G.; Wu, W.; Jiang, X.; Wang, S.; Lin, Z.; et al. Alternative splicing of OsGS1;1 affects nitrogen-use efficiency, grain development, and amylose content in rice. Plant J. 2022, 110, 1751–1762. [Google Scholar] [CrossRef]
- Zhu, J.; Yan, X.; Liu, S.; Xia, X.; An, Y.; Xu, Q.; Zhao, S.; Liu, L.; Guo, R.; Zhang, Z.; et al. Alternative splicing of CsJAZ1 negatively regulates flavan-3-ol biosynthesis in tea plants. Plant J. 2022, 110, 243–261. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, W.; Wei, J.; Gao, Z.; Guan, J.; Cui, Z.; Wang, X. SKIP Regulates ABA Signaling through Alternative Splicing in Arabidopsis. Plant Cell Physiol. 2022, 63, 494–507. [Google Scholar] [CrossRef]
- Yan, X.; Bai, D.; Song, H.; Lin, K.; Pang, E. Alternative splicing during fruit development among fleshy fruits. BMC Genom. 2021, 22, 762. [Google Scholar] [CrossRef]
- Wang, Z.; Ji, H.; Yuan, B.; Wang, S.; Su, C.; Yao, B.; Zhao, H.; Li, X. ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 2015, 6, 8138. [Google Scholar] [CrossRef] [Green Version]
- Penfield, S.; Josse, E.M.; Halliday, K.J. A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol. Biol. 2010, 73, 89–95. [Google Scholar] [CrossRef]
- Meyer, K.; Koester, T.; Staiger, D. Pre-mRNA Splicing in Plants: In Vivo Functions of RNA-Binding Proteins Implicated in the Splicing Process. Biomolecules 2015, 5, 1717–1740. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, L.; Tan, M.; Wang, L.; Zhao, W.; You, J.; Wang, L.; Yan, X.; Wang, W. The pattern of alternative splicing and DNA methylation alteration and their interaction in linseed (Linum usitatissimum L.) response to repeated drought stresses. Biol. Res. 2023, 56, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xu, J.; Pu, X.; Lv, H.; Liu, Y.; Ma, H.; Wu, F.; Wang, Q.; Feng, X.; Liu, T.; et al. Maize DNA Methylation in Response to Drought Stress Is Involved in Target Gene Expression and Alternative Splicing. Int. J. Mol. Sci. 2021, 22, 8285. [Google Scholar] [CrossRef]
- Yu, J.; Jung, S.; Cheng, C.-H.; Ficklin, S.P.; Lee, T.; Zheng, P.; Jones, D.; Percy, R.G.; Main, D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2013, 42, D1229–D1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differ-entiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq–A Python framework to work with high-throughput sequencing data. bioRxiv 2014, 2824. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Song, Q.; Guan, X.; Chen, Z.J. Dynamic Roles for Small RNAs and DNA Methylation during Ovule and Fiber Development in Allotetraploid Cotton. PLoS Genet. 2015, 11, e1005724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Lim, J.-Q.; Sung, W.-K.; Li, G. An integrated package for bisulfite DNA methylation data analysis with Indel-sensitive mapping. BMC Bioinform. 2019, 20, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
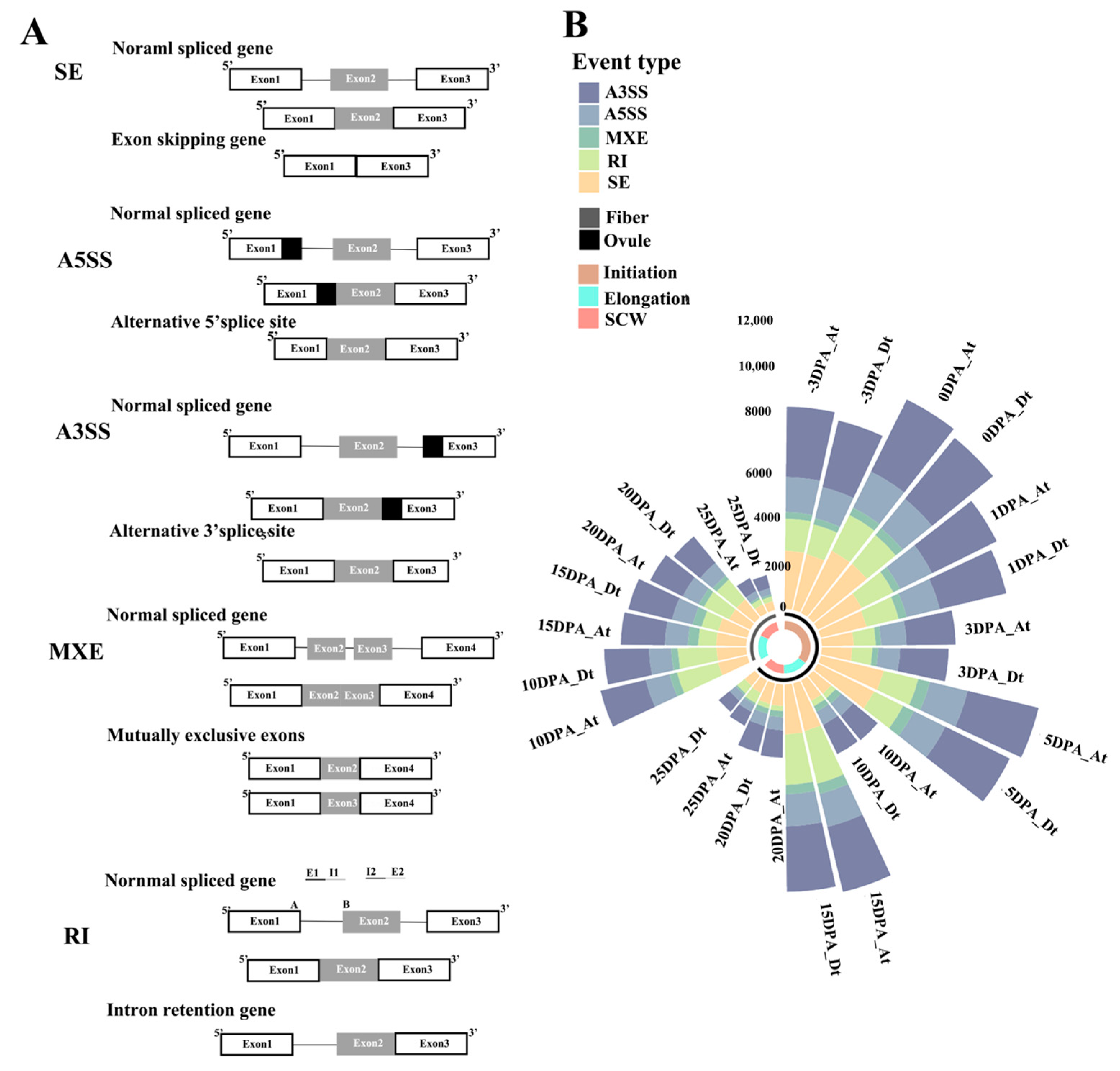


| A3SS | A5SS | MXE | RI | SE | SUM | |
|---|---|---|---|---|---|---|
| −3 DPA_Ovule | 4369 | 2343 | 438 | 1960 | 3072 | 8437 |
| 0 DPA_Ovule | 4832 | 2656 | 563 | 2339 | 3642 | 9360 |
| 1 DPA_Ovule | 4362 | 2386 | 527 | 2133 | 2614 | 8309 |
| 3 DPA_Ovule | 3253 | 1664 | 394 | 1431 | 1733 | 6354 |
| 5 DPA_Ovule | 4599 | 2514 | 799 | 2176 | 3374 | 9237 |
| 10 DPA_Ovule | 2268 | 1114 | 475 | 453 | 1316 | 4605 |
| 15 DPA_Ovule | 4177 | 2206 | 489 | 3188 | 2645 | 8669 |
| 20 DPA_Ovule | 1977 | 949 | 417 | 371 | 1240 | 4091 |
| 25 DPA_Ovule | 1102 | 602 | 141 | 804 | 805 | 2894 |
| 10 DPA_Fiber | 3040 | 1636 | 494 | 2612 | 1676 | 6849 |
| 15 DPA_Fiber | 3053 | 1528 | 434 | 1215 | 1675 | 6067 |
| 20 DPA_Fiber | 2402 | 1213 | 392 | 1833 | 1321 | 5420 |
| 25 DPA_Fiber | 1053 | 508 | 164 | 411 | 599 | 2363 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Wen, S.; Yu, Z.; Luo, K.; Rong, J.; Ding, M. Alternative Splicing during Fiber Development in G. hirsutum. Int. J. Mol. Sci. 2023, 24, 11812. https://doi.org/10.3390/ijms241411812
Zheng J, Wen S, Yu Z, Luo K, Rong J, Ding M. Alternative Splicing during Fiber Development in G. hirsutum. International Journal of Molecular Sciences. 2023; 24(14):11812. https://doi.org/10.3390/ijms241411812
Chicago/Turabian StyleZheng, Jing, Shuhan Wen, Zhipeng Yu, Keyan Luo, Junkang Rong, and Mingquan Ding. 2023. "Alternative Splicing during Fiber Development in G. hirsutum" International Journal of Molecular Sciences 24, no. 14: 11812. https://doi.org/10.3390/ijms241411812




