Chimerism Testing by Next Generation Sequencing for Detection of Engraftment and Early Disease Relapse in Allogeneic Hematopoietic Cell Transplantation and an Overview of NGS Chimerism Studies
Abstract
:1. Introduction
2. Results
2.1. Accuracy and Proficiency Testing
2.2. Comparison of NGS Chimerism with STR-PCR
2.3. Performance Specification of NGS Chimerism
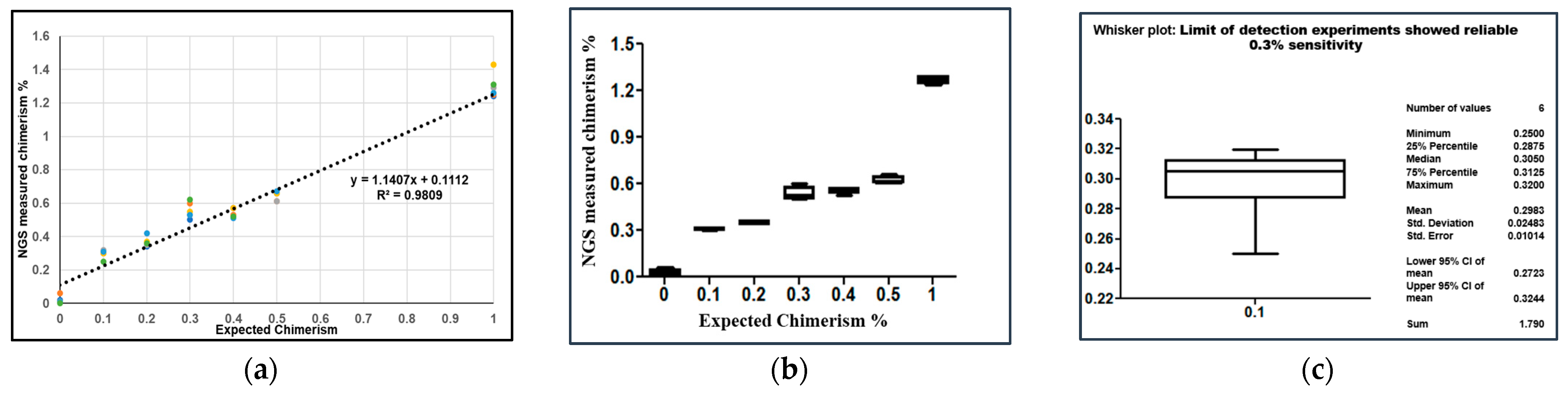
2.4. Analytical Sensitivity of NGS
2.5. Precision and Reproducibility Testing
2.6. Inter-Tech Variability
3. Discussion
4. Materials and Methods
4.1. Sample Selection
4.2. DNA Extraction
4.3. Artificial Mixed Chimeric Specimen Preparation
4.4. NGS, Panel and Chimerism Analysis
4.5. Accuracy, Performance Specification, and Analytical Sensitivity
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bader, P.; Niemeyer, C.; Willasch, A.; Kreyenberg, H.; Strahm, B.; Kremens, B.; Gruhn, B.; Dilloo, D.; Vormoor, J.; Lang, P.; et al. Children with myelodysplastic syndrome (MDS) and increasing mixed chimaerism after allogeneic stem cell transplantation have a poor outcome which can be improved by preemptive immunotherapy. Br. J. Haematol. 2005, 128, 649–658. [Google Scholar] [CrossRef]
- Jacque, N.; Nguyen, S.; Golmard, J.-L.; Uzunov, M.; Garnier, A.; Leblond, V.; Vernant, J.-P.; Bories, D.; Dhédin, N. Chimerism analysis in peripheral blood using indel quantitative real-time PCR is a useful tool to predict post-transplant relapse in acute leukemia. Bone Marrow Transplant. 2015, 50, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Sellmann, L.; Rabe, K.; Bünting, I.; Dammann, E.; Göhring, G.; Ganser, A.; Stadler, M.; Weissinger, E.M.; Hambach, L. Diagnostic value of highlysensitive chimerism analysis after allogeneic stem cell transplantation. Bone Marrow Transplant. 2018, 53, 1457–1465. [Google Scholar] [CrossRef]
- Valero-Garcia, J.; González-Espinosa, M.D.; Barrios, M.; Carmona-Antoñanzas, G.; García-Planells, J.; Ruiz-Lafora, C.; Fuentes-Gálvez, A.; Jiménez-Velasco, A. Earlier relapse detection after allogeneic haematopoietic stem cell transplantation by chimerism assays: Digital PCR versus quantitative real-time PCR of insertion/deletion polymorphisms. PLoS ONE 2019, 14, e0212708. [Google Scholar]
- Bejanyan, N.; Weisdorf, D.J.; Logan, B.R.; Wang, H.-L.; Devine, S.M.; de Lima, M.; Bunjes, D.W.; Zhang, M.-J. Survival of patients with acute myeloid leukemia relapsing after allogeneic hematopoietic cell transplantation: A Center for International Blood and Marrow Transplant research study. Biol. Blood Marrow Transplant. 2015, 21, 454–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bader, P.; Niethammer, D.; Willasch, A.; Kreyenberg, H.; Klingebiel, T. How and when should we monitor chimerism after allogeneic stem cell transplantation? Bone Marrow Transplant. 2005, 35, 107–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, C.; Labopin, M.; Schaap, N.; Veelken, H.; Schleuning, M.; Stadler, M.; Finke, J.; Hurst, E.; Baron, F.; Ringden, O.; et al. Prophylactic donor lymphocyte infusion after allogeneic stem cell transplantation in acute leukaemia—A matched pair analysis by the acute leukaemia working party of ebmt. Br. J. Haematol. 2019, 184, 782–787. [Google Scholar] [CrossRef]
- Clark, J.R.; Scott, S.D.; Jack, A.L.; Lee, H.; Mason, J.; Carter, G.I.; Pearce, L.; Jackson, T.; Clouston, H.; Sproul, A.; et al. United Kingdom National External Quality Assessment Service for Leucocyte Immunophenotyping Chimerism Working Group: Monitoring of chimerism following allogeneic haematopoietic stem cell transplantation (HSCT): Technical recommendations for the use of short tandem repeat (STR) based techniques, on behalf of the United Kingdom National External Quality Assessment Service for Leucocyte Immunophenotyping Chimerism Working Group. Br. J. Haematol. 2015, 168, 26–37. [Google Scholar]
- Lawler, M.; Humphries, P.; McCann, S.R. Evaluation of mixed chimerism by in vitro amplification of dinucleotide repeat sequences using the polymerase chain reaction. Blood 1991, 77, 2504–2514. [Google Scholar] [CrossRef] [Green Version]
- Thiede, C.; Bornhäuser, M.; Oelschlägel, U.; Brendel, C.; Leo, R.; Daxberger, H.; Mohr, B.; Florek, M.; Kroschinsky, F.; Geissler, G.; et al. Sequential monitoring of chimerism and detection of minimal residual disease after allogeneic blood stem cell transplantation (BSCT) using multiplex PCR amplification of short tandem repeat-markers. Leukemia 2001, 15, 293–302. [Google Scholar] [CrossRef] [Green Version]
- Alizadeh, M.; Bernard, M.; Danic, B.; Dauriac, C.; Birebent, B.; Lapart, C.; Lamy, T.; Le Prisé, P.Y.; Beauplet, A.; Bories, D.; et al. Quantitative assessment of hematopoietic chimerism after bone marrow transplantation by real-time quantitative polymerase chain reaction. Blood 2002, 99, 4618–4625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Velasco, A.; Barrios, M.; Román-Gómez, J.; Navarro, G.; Buño, I.; Castillejo, J.A.; Rodríguez, A.I.; García-Gemar, G.; Torres, A.; Heiniger, A.I. Reliable quantification of hematopoietic chimerism after allogeneic transplantation for acute leukemia using amplification by real-time PCR of null alleles and insertion/deletion polymorphisms. Leukemia 2005, 19, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Lion, T.; Watzinger, F.; Preuner, S.; Kreyenberg, H.; Tilanus, M.; de Weger, R.; van Loon, J.; de Vries, L.; Cavé, H.; Acquaviva, C.; et al. The EuroChimerism concept for a standardized approach to chimerism analysis after allogeneic stem cell transplantation. Leukemia 2012, 26, 1821–1828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Hwang, I.S.; Shin, S.; Choi, J.R.; Lee, S.-T. SNP-based next-generation sequencing reveals low-level mixed chimerism after allogeneic hematopoietic stem cell transplantation. Ann. Hematol. 2018, 97, 1731–1734. [Google Scholar] [CrossRef]
- Kim, S.Y.; Jeong, M.H.; Park, N.; Ra, E.; Park, H.; Seo, S.H.; Kim, J.Y.; Seong, M.-W.; Park, S.S. Chimerism monitoring after allogeneic hematopoietic stem cell transplantation using quantitative real-time PCR of biallelic insertion/deletion polymorphisms. J. Mol. Diagn. 2014, 16, 679–688. [Google Scholar] [CrossRef]
- Aloisio, M.; Licastro, D.; Caenazzo, L.; Torboli, V.; D’eustacchio, A.; Severini, G.M.; Athanasakis, E. A technical application of quantitative next-generation sequencing for chimerism evaluation. Mol. Med. Rep. 2016, 14, 2967–2974. [Google Scholar] [CrossRef] [Green Version]
- Kristt, D.; Israeli, M.; Narinski, R.; Or, H.; Yaniv, I.; Stein, J.; Klein, T. Hematopoietic chimerism monitoring based on strs: Quantitative platform performance on sequential samples. J. Biomol. Tech. 2005, 16, 380–391. [Google Scholar]
- Cusick, M.F.; Clark, L.; Tu, T.; Goforth, J.; Zhang, X.; LaRue, B.; Gutierrez, R.; Jindra, P.T. Performance characteristics of chimerism testing by next generation sequencing. Hum. Immunol. 2022, 83, 61–69. [Google Scholar] [CrossRef]
- Pettersson, L.; Vezzi, F.; Vonlanthen, S.; Alwegren, K.; Hedrum, A.; Hauzenberger, D. Development and performance of a next generation sequencing (NGS) assay for monitoring of mixed chimerism. Clin. Chim. Acta 2021, 512, 40–48. [Google Scholar] [CrossRef]
- Vynck, M.; Nollet, F.; Sibbens, L.; Lievens, B.; Denys, A.; Cauwelier, B.; Devos, H. Performance Assessment of the Devyser. High-Throughput Sequencinge Based Assay for Chimerism Monitoring in Patients after Allogeneic Hematopoietic Stem Cell Transplantation. J. Mol. Diagn. 2021, 23, 1116–1126. [Google Scholar] [CrossRef]
- Chia, W.C.; Khoo, T.S.; Abdul Wahid, S.F.S.; Razak, N.F.A.; Alauddin, H.; Raja Sabudin, R.Z.A.; Othman, A.; Hassan, R.; Hussin, N.H. Multiplex str panel for assessment of chimerism following hematopoietic stem cell transplantation (hsct). Ann. Hematol. 2019, 98, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, Y.; Feng, Y.; Zhou, H.; Ma, X.; Wu, D.; Chen, S.; Sun, A. The clinical application of SNP-based next-generation sequencing (SNP-NGS) for evaluation of chimerism and microchimerism after HLA-mismatched stem cell microtransplantation. Int. J. Hematol. 2022, 116, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Kim, Y.J.; Park, S.S.; Han, E.; Kim, M.; Kim, Y. Simultaneous Monitoring of Mutation and Chimerism Using Next-Generation Sequencing in Myelodysplastic Syndrome. J. Clin. Med. 2019, 8, 2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedini, P.; Cherouat, N.; Basire, A.; Simon, S.; Budon, L.; Pourtein, M.; Grondin, S.; Moskovtchenko, P.; Chiaroni, J.; Michel, G.; et al. Evaluation of Next Generation Sequencing and Crystal Digital PCR for Chimerism Monitoring of Post-Allogeneic Hematopoietic Stem Cell Transplantation. Transplant. Cell. Ther. 2021, 27, 89. [Google Scholar] [CrossRef] [PubMed]
- Pedini, P.; Graiet, H.; Laget, L.; Filosa, L.; Chatron, J.; Cherouat, N.; Chiaroni, J.; Hubert, L.; Frassati, C.; Picard, C. Qualitative and quantitative comparison of cell-free DNA and cell-free fetal DNA isolation by four (semi-)automated extraction methods: Impact in two clinical applications: Chimerism quantification and noninvasive prenatal diagnosis. J. Transl. Med. 2021, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Picard, C.; Frassati, C.; Cherouat, N.; Maioli, S.; Moskovtchenko, P.; Cherel, M.; Chiaroni, J.; Pedini, P. New methods for the quantification of mixed chimerism in transplantation. Front. Immunol. 2023, 14, 1023116. [Google Scholar] [CrossRef]
- Thiede, C.; Bornhäuser, M.; Ehninger, G. Evaluation of str informativity for chimerism testing—comparative analysis of 27 str systems in 203 matched related donor recipient pairs. Leukemia 2004, 18, 248–254. [Google Scholar] [CrossRef]
- Han, J.; Sun, J.; Zhao, L.; Zhao, W.; Liu, Y.; Li, C. Validation study of a 15-plex rapid str amplification system for human identification. Forensic. Sci. Int. Genet. 2017, 28, 71–81. [Google Scholar] [CrossRef]
- Odriozola, A.; Riancho, J.A.; Colorado, M.; Zarrabeitia, M.T. Evaluation of the sensitivity of two recently developed str multiplexes for the analysis of chimerism after haematopoietic stem cell transplantation. Int. J. Immunogenet. 2013, 40, 88–92. [Google Scholar] [CrossRef]
- Tau, T.; Wally, A.; Fanie, T.P.; Ngono, G.L.; Mpoloka, S.W.; Davison, S.; D’Amato, M.E. Genetic variation and population structure of botswana populations as identified with ampflstr identifiler short tandem repeat (str) loci. Sci. Rep. 2017, 7, 6768. [Google Scholar] [CrossRef] [Green Version]
- Mattsson, M.; Uzunel, L.; Tammik, J.; Aschan, O. Ringden, Leukemia lineage specific chimerism analysis is a sensitive predictor of relapse in patients with acute myeloid leukemia and myelodysplastic syndrome after allogeneic stem cell transplantation. Leukemia 2001, 15, 1976–1985. [Google Scholar] [CrossRef]
- Blouin, A.G.; Askar, M. Chimerism analysis for clinicians: A review of the literature and worldwide practices. Bone Marrow Transplant. 2022, 57, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xie, B.; Yan, J. Application of next-generation sequencing technology in forensic science. Genom. Proteom. Bioinform. 2014, 12, 190–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liesveld, J.L.; Rothberg, P.G. Mixed chimerism in SCT: Conflict or peaceful coexistence? Bone Marrow Transplant. 2008, 42, 297–310. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Bian, Y.; Zhang, Z.; Zheng, H.; Wang, Z.; Zha, L.; Cai, J.; Gao, Y.; Ji, C.; Hou, Y.; et al. Parallel analysis of 124 universal SNPs for human identification by targeted semiconductor sequencing. Sci. Rep. 2015, 5, 18683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre-Ruiz, P.; Ariceta, B.; Viguria, M.C.; Zudaire, M.T.; Blasco-Iturri, Z.; Arnedo, P.; Aguilera-Diaz, A.; Jauregui, A.; Mañú, A.; Prosper, F.; et al. Assessment of Minimal Residual Disease by Next Generation Sequencing in Peripheral Blood as a Complementary Tool for Personalized Transplant Monitoring in Myeloid Neoplasms. J. Clin. Med. 2020, 9, 3818. [Google Scholar] [CrossRef]
- Tang, Z.; Zhang, T.; Yang, B.; Su, J.; Song, Q. spaCI: Deciphering spatial cellular communications via adaptive graph model. Brief. Bioinform. 2023, 24, 1–13. [Google Scholar] [CrossRef]
- Triozzi, P.L.; Stirling, E.R.; Song, Q.; Westwood, B.; Kooshki, M.; Forbes, M.E.; Holbrook, B.; Cook, K.L.; Alexander-Miller, M.A.; Miller, L.D.; et al. Circulating immune bioenergetic, metabolic, and genetic signatures predict melanoma patients response to anti-PD-1 immune checkpoint blockade. Clin. Cancer Res. 2022, 28, 1192–1202. [Google Scholar] [CrossRef]



| No. | Population | Sample Size | Number of Markers and Discription | Methodology | Sequencing Kit | Technology | NGS: Informative SNPs | NGS Sensitivity/Specificity | STR/NGS Analysis | STR: Informative SNPs | STR Sensitivity/Specificity | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Republic of Korea | 15 BM samples | 124 SNPs; among these, 90 autosomal SNPs | NGS vs. STR | HID-Ion AmpliSeq™ Identity Panel (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA) | Ion PGM™ System (Life Technologies) | 20.4 (13–32) | <1%/100% | 11 STRs: GenomeLab Human STR Primer Set (Beckman Coulter, Fullerton, CA, USA) | 5.7 (2–9) | 1–5% | [14] |
| 2 | Italy | 10 | 44-amplicon custom chimerism panel | NGS | Ion AmpliSeq custom chimerism (ACCh) panel | Ion Torrent | 16 | 0.04–1.0%/100% | AmpFlSTR Identifiler Plus PCR Amplification Kit (Thermo Fisher Scientific, Inc., CA, USA) | not available | 4%/100% | [16] |
| 3 | USA | not specified | N= 230: Primer A: Amelogenin, 27 autosomal STRs, 24 Y-STRs, 7 X-STRs, 94 informative SNPs. Primer Mix B, 78 aiSNPs | NGS vs. STR | NGS ForenSeq | MiSeq—FGx- Illumina | not available | 1%/100% | CE-STR: 16–21 STR loci | not available | 5%/99% | [18] |
| 4 | Belgium | 422 | 24 loci with a known biallelic insertion/deletion polymorphism | NGS vs. STR | Devyser next-generation sequencing chimerism assay | MiSeq or NextSeq550Dx instrument (Illumina, San Diego, CA) | 15 | 0.1%/100% | Power- Plex 16 HS PCR-CE assay (Promega assay) | not available | 2–5%/100% | [20] |
| 5 | Sweden | 651 samples | 24 indels | NGS vs. STR and RQ-PCR | Devyser Chimerism NGS kit by Devyser AB, Stockholm, Sweden | Illumina MiSeq | 9 (~40% informative) | 0.1%/100% | In-house STR marker and RQ-PCR by Alizadeh et al. [21] | 1 for STR and 2 for RQ-PCR | 2–5%/100% | [19] |
| 6 | People’s Republic of China | 48 | 48 SNPs | In house 48 primer sets | SNP-NGS | Illumina MiSeq | not available | 0.01–0.05%/100% | STR | not available | 1–10% | [22] |
| 7 | Republic of Korea | 53 | 121 SNPs | Agilent Technologies, Santa Clara, CA, USA | Customized target kit | Illumina HiSeq4000 platform | (9–37) | 0.5–1.0%/91.7% | AmpFlSTR Identifier PCR Amplification (Applied Biosystems, Warrington, UK) | 25.5 (9–41) | 1–5% | [23] |
| 8 | France | 91 | 24 indels | NGS vs. STR and cdPCR | Devyser Chimerism NGS kit by Devyser AB, Stockholm, Sweden | Illumina MiSeq | not available | 0.1%/100% | Crystal Digital PCR KMR kits (GenDX, Utrecht, The Netherlands) | not available | 0.1%/100% for cdPCR | [24] |
| 9 | France | 24 | 24 indels | NGS | Devyser Chimerism NGS kit by Devyser AB, Stockholm, Sweden | Illumina MiSeq | 8 | 0.1%/100% | ddPCR | not available | 0.10% | [25] |
| 10 | France | 24 | 24 indels | NGS | Devyser® panel (Devyser Chimerism NGS) | Illumina MiSeq | Not available | 0.1%/100% | AlloSeq HCT (CareDx) and NGStrack | not available | 0.3% for AlloSeq and 0.5% for NGStrac | [26] |
| 11 | Temple University Hospital, USA | 174 | 202 SNPs across all autosomal chromosomes | NGS vs. STR | AlloSeq HCT (CareDx) | Illumina MiSeq | 110 | 0.3%/100% | AmpFlSTR Identifiler Plus PCR Amplification kit (Thermo Fisher Scientific, Inc.) | 2–9 | 5%/99% | Current Paper |
| Disease Diagnosis | N = 46 | % |
|---|---|---|
| Acute Lymphocytic Leukemia (ALL) | 3 | 7.0 |
| Acute Myeloid Leukemia (AML) | 24 | 55.8 |
| Chronic Myelogenous Leukemia (CML) | 1 | 2.3 |
| Chronic Myelomonocytic Leukemia (CMML) | 2 | 4.7 |
| Hodgkins Lymphoma (HL) | 3 | 7.0 |
| Myelodysplastic Syndromes (MDS) | 7 | 16.3 |
| Neuroblastoma (NBL) | 1 | 2.3 |
| Severe combined immunodeficiency (SCID) | 1 | 2.3 |
| Telomere Biology Disorder (TBD) | 1 | 2.3 |
| Unknown | 3 | 7.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liacini, A.; Tripathi, G.; McCollick, A.; Gravante, C.; Abdelmessieh, P.; Shestovska, Y.; Mathew, L.; Geier, S. Chimerism Testing by Next Generation Sequencing for Detection of Engraftment and Early Disease Relapse in Allogeneic Hematopoietic Cell Transplantation and an Overview of NGS Chimerism Studies. Int. J. Mol. Sci. 2023, 24, 11814. https://doi.org/10.3390/ijms241411814
Liacini A, Tripathi G, McCollick A, Gravante C, Abdelmessieh P, Shestovska Y, Mathew L, Geier S. Chimerism Testing by Next Generation Sequencing for Detection of Engraftment and Early Disease Relapse in Allogeneic Hematopoietic Cell Transplantation and an Overview of NGS Chimerism Studies. International Journal of Molecular Sciences. 2023; 24(14):11814. https://doi.org/10.3390/ijms241411814
Chicago/Turabian StyleLiacini, Abdelhamid, Gaurav Tripathi, Amanda McCollick, Christopher Gravante, Peter Abdelmessieh, Yuliya Shestovska, Leena Mathew, and Steven Geier. 2023. "Chimerism Testing by Next Generation Sequencing for Detection of Engraftment and Early Disease Relapse in Allogeneic Hematopoietic Cell Transplantation and an Overview of NGS Chimerism Studies" International Journal of Molecular Sciences 24, no. 14: 11814. https://doi.org/10.3390/ijms241411814
APA StyleLiacini, A., Tripathi, G., McCollick, A., Gravante, C., Abdelmessieh, P., Shestovska, Y., Mathew, L., & Geier, S. (2023). Chimerism Testing by Next Generation Sequencing for Detection of Engraftment and Early Disease Relapse in Allogeneic Hematopoietic Cell Transplantation and an Overview of NGS Chimerism Studies. International Journal of Molecular Sciences, 24(14), 11814. https://doi.org/10.3390/ijms241411814





