SKAP1 Is a Novel Biomarker and Therapeutic Target for Gastric Cancer: Evidence from Expression, Functional, and Bioinformatic Analyses
Abstract
1. Introduction
2. Results
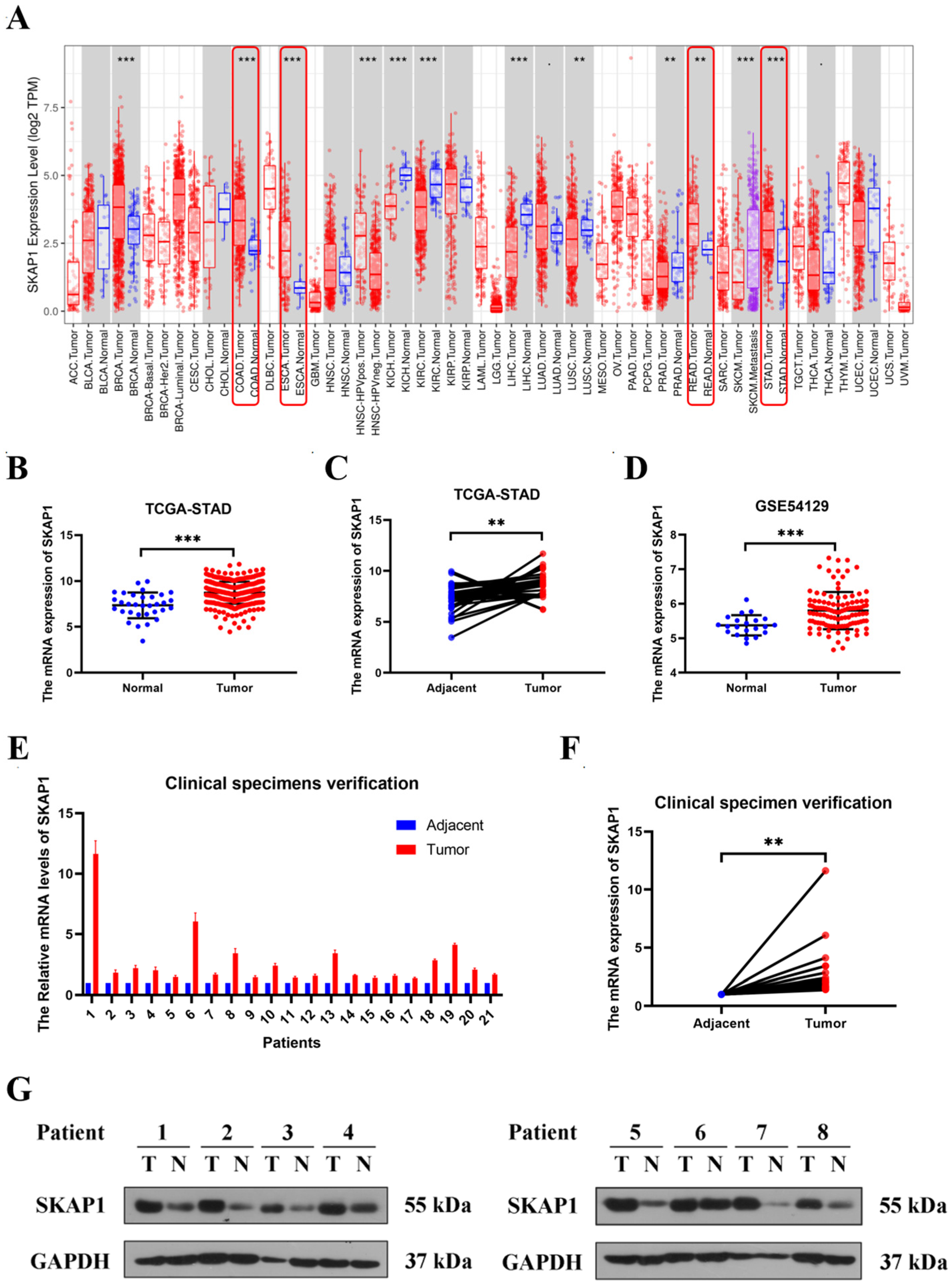
2.1. High Expression of SKAP1 in GC Tissue
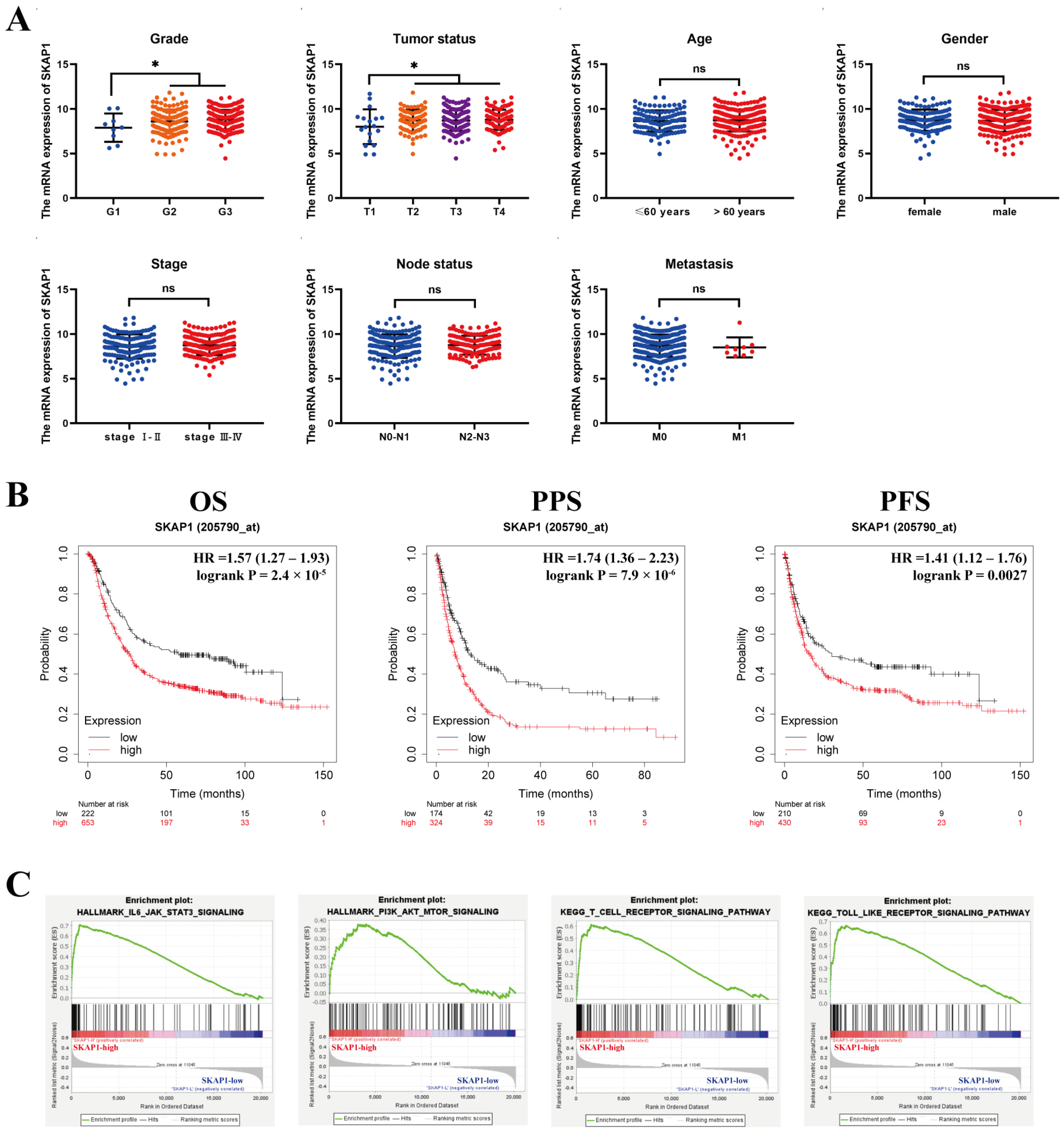
2.2. Relationship between SKAP1 and Clinicopathological Parameters and Prognosis of GC Patients
2.3. Bioinformatics Prediction of the Possible Mechanism by Which SKAP1 Regulates GC Progression
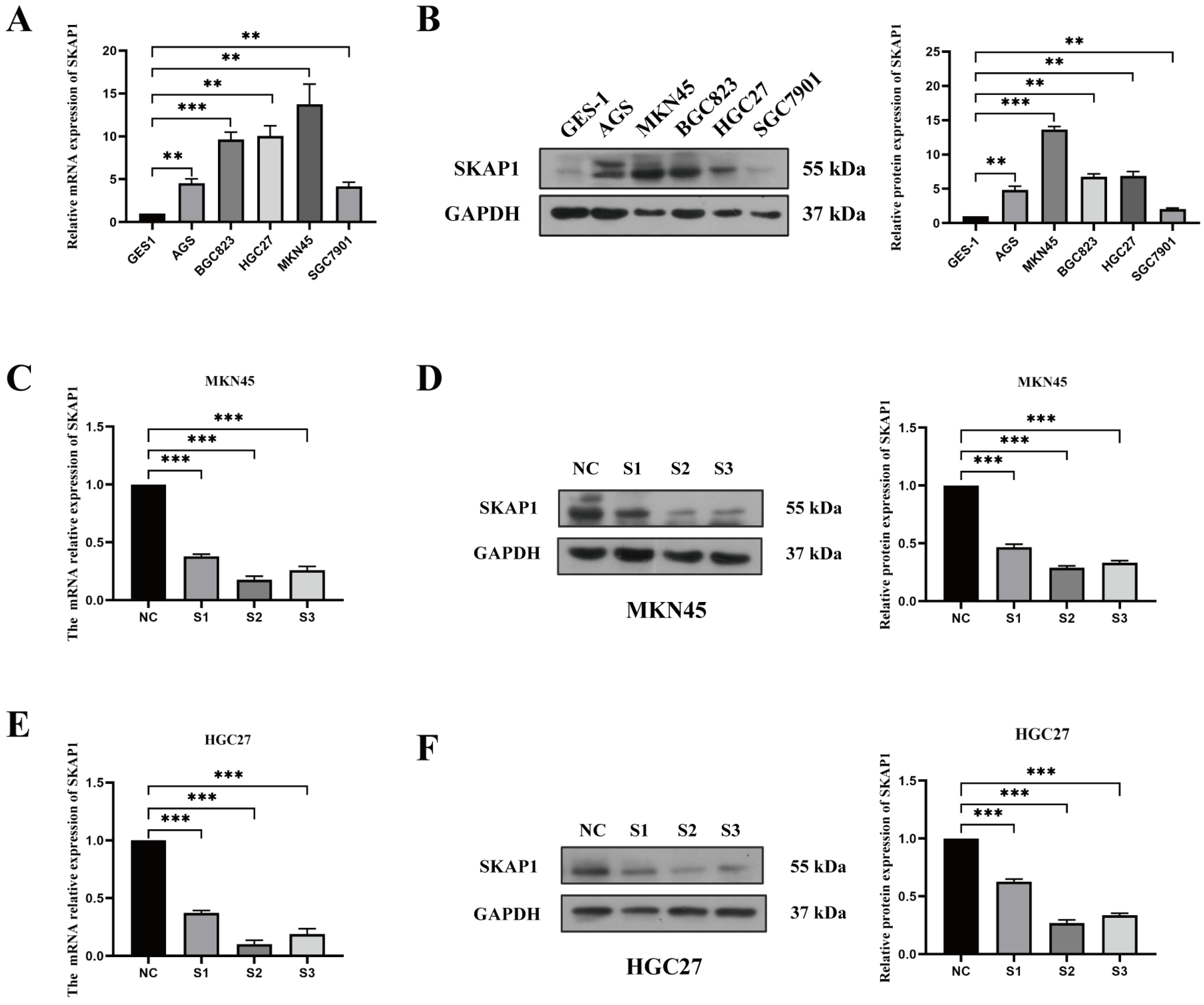
2.4. Effect of Silencing SKAP1 on GC Cell Biology
2.5. SKAP1 Regulates the JAK1/PI3K/AKT Signaling Axis in GC Cells
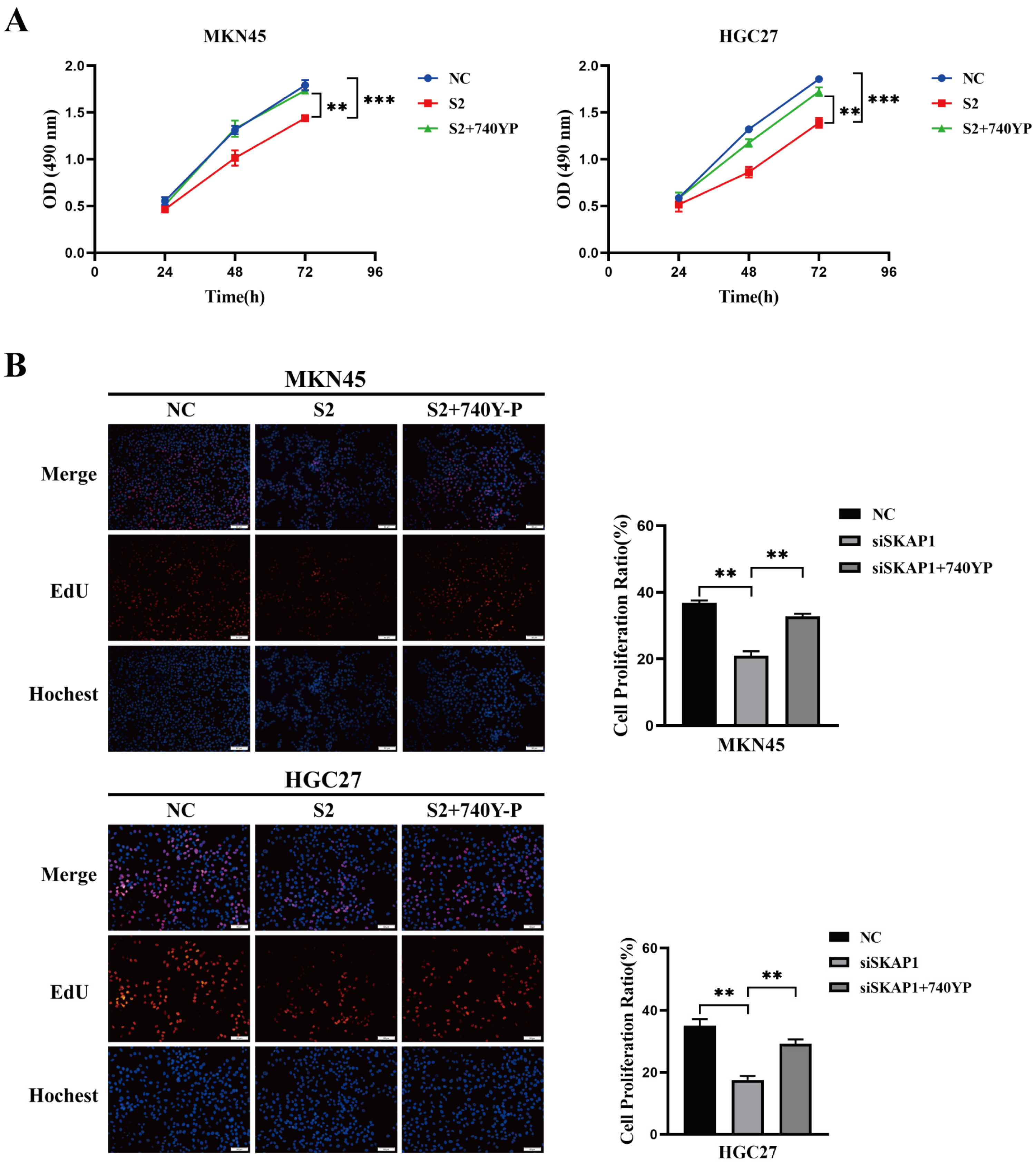
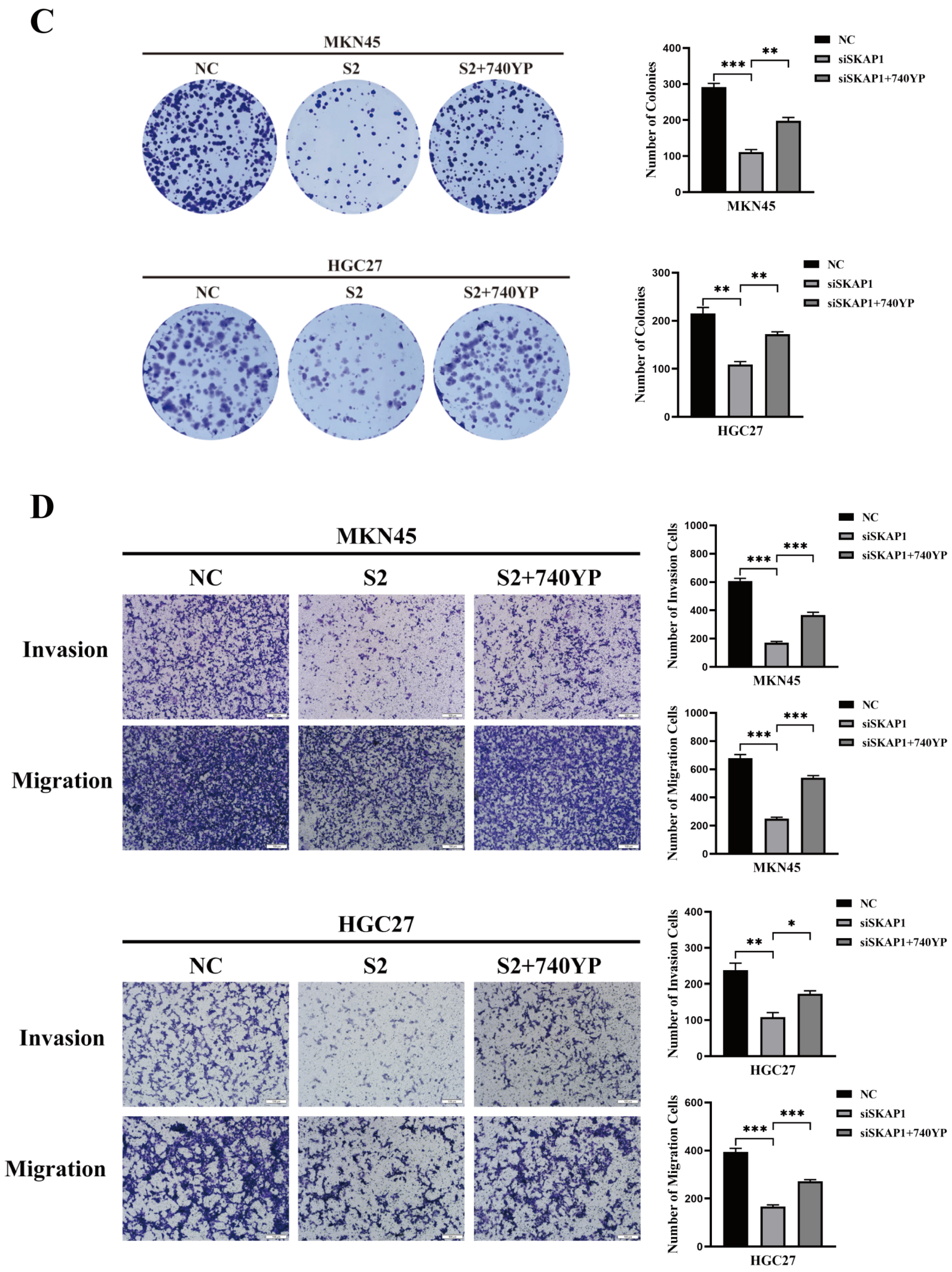
2.6. Effect of a PI3K Agonist on the SKAP1-Regulated Biological Behavior of GC Cells
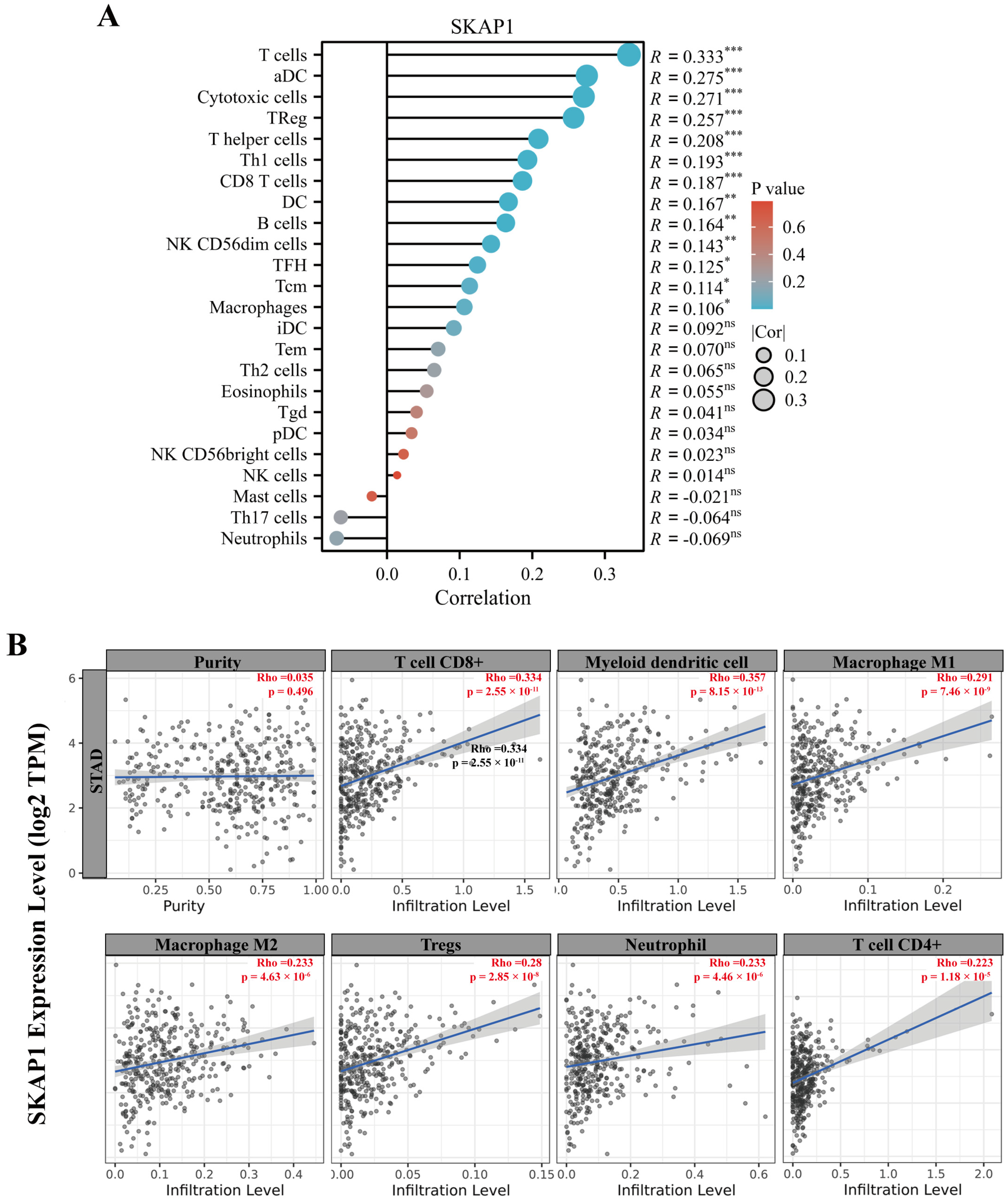
2.7. Bioinformatics Analysis of the Relationship between SKAP1 Expression and Immune Infiltration in GC
3. Discussion
4. Materials and Methods
4.1. Expression, Prognosis and Clinicopathological Data Analysis
4.2. Tissues and Cells
4.3. Real-Time Quantitative Reverse Transcription-PCR
4.4. Protein Extraction, Quantification, and Immunoblot Analysis
4.5. Cell Culture and Gene Transfection
4.6. MTS Cell Proliferation Assay
4.7. EdU Cell Proliferation Assay
4.8. Colony Formation Assay
4.9. Transwell Cell Invasion and Migration Assay
4.10. Flow Cytometry Apoptosis Assay
4.11. Relationship between SKAP1 Expression and Immune Infiltration in GC
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Chen, Y.; He, S. Family History of Gastric Cancer and Helicobacter pylori Treatment. N. Engl. J. Med. 2020, 382, 2171. [Google Scholar] [CrossRef] [PubMed]
- Fukase, K.; Kato, M.; Kikuchi, S.; Inoue, K.; Uemura, N.; Okamoto, S.; Terao, S.; Amagai, K.; Hayashi, S.; Asaka, M. Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: An open-label, randomised controlled trial. Lancet 2008, 372, 392–397. [Google Scholar] [CrossRef]
- Jamali, L.; Tofigh, R.; Tutunchi, S.; Panahi, G.; Borhani, F.; Akhavan, S.; Nourmohammadi, P.; Ghaderian, S.M.H.; Rasouli, M.; Mirzaei, H. Circulating microRNAs as diagnostic and therapeutic biomarkers in gastric and esophageal cancers. J. Cell Physiol. 2018, 233, 8538–8550. [Google Scholar] [CrossRef] [PubMed]
- Le Petit, G.F. Medazepam pKa determined by spectrophotometric and solubility methods. J. Pharm. Sci. 1976, 65, 1094–1095. [Google Scholar] [CrossRef]
- Wang, H.; Rudd, C.E. SKAP-55, SKAP-55-related and ADAP adaptors modulate integrin-mediated immune-cell adhesion. Trends Cell Biol. 2008, 18, 486–493. [Google Scholar] [CrossRef]
- Marie-Cardine, A.; Bruyns, E.; Eckerskorn, C.; Kirchgessner, H.; Meuer, S.C.; Schraven, B. Molecular cloning of SKAP55, a novel protein that associates with the protein tyrosine kinase p59fyn in human T-lymphocytes. J. Biol. Chem. 1997, 272, 16077–16080. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Z.; Zheng, Y.; Zhu, Y.; Wei, Z.; Xu, H.; Tang, Q.; Kong, X.; Hu, L. SKAP2, a novel target of HSF4b, associates with NCK2/F-actin at membrane ruffles and regulates actin reorganization in lens cell. J. Cell. Mol. Med. 2011, 15, 783–795. [Google Scholar] [CrossRef]
- Marie-Cardine, A.; Verhagen, A.M.; Eckerskorn, C.; Schraven, B. SKAP-HOM, a novel adaptor protein homologous to the FYN-associated protein SKAP55. FEBS Lett. 1998, 435, 55–60. [Google Scholar] [CrossRef]
- Liu, J.; Kang, H.; Raab, M.; da Silva, A.J.; Kraeft, S.K.; Rudd, C.E. FYB (FYN binding protein) serves as a binding partner for lymphoid protein and FYN kinase substrate SKAP55 and a SKAP55-related protein in T cells. Proc. Natl. Acad. Sci. USA 1998, 95, 8779–8784. [Google Scholar] [CrossRef]
- Kuranami, S.; Yokobori, T.; Mogi, A.; Altan, B.; Yajima, T.; Onozato, R.; Azuma, Y.; Iijima, M.; Kosaka, T.; Kuwano, H. Src kinase-associated phosphoprotein2 expression is associated with poor prognosis in non-small cell lung cancer. Anticancer Res. 2015, 35, 2411–2415. [Google Scholar] [PubMed]
- Tanaka, M.; Shimamura, S.; Kuriyama, S.; Maeda, D.; Goto, A.; Aiba, N. SKAP2 Promotes Podosome Formation to Facilitate Tumor-Associated Macrophage Infiltration and Metastatic Progression. Cancer Res 2016, 76, 358–369. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, R.; van Kester, M.S.; Dijkman, R.; Vermeer, M.H.; Mulder, A.A.; Szuhai, K.; Knijnenburg, J.; Boer, J.M.; Willemze, R.; Tensen, C.P. Oncogenomic analysis of mycosis fungoides reveals major differences with Sezary syndrome. Blood 2009, 113, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Schulte, I.; Batty, E.M.; Pole, J.C.; Blood, K.A.; Mo, S.; Cooke, S.L.; Ng, C.; Howe, K.L.; Chin, S.F.; Brenton, J.D.; et al. Structural analysis of the genome of breast cancer cell line ZR-75-30 identifies twelve expressed fusion genes. BMC Genom. 2012, 13, 719. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, A.R.; Sharma, G.; Chakraborty, C.; Kim, J. PLK-1: Angel or devil for cell cycle progression. Biochim. Biophys. Acta 2016, 1865, 190–203. [Google Scholar] [CrossRef]
- van de Weerdt, B.C.; Medema, R.H. Polo-like kinases: A team in control of the division. Cell Cycle (Georget. Tex.) 2006, 5, 853–864. [Google Scholar] [CrossRef]
- Ramani, P.; Nash, R.; Sowa-Avugrah, E.; Rogers, C. High levels of polo-like kinase 1 and phosphorylated translationally controlled tumor protein indicate poor prognosis in neuroblastomas. J. Neuro-Oncol. 2015, 125, 103–111. [Google Scholar] [CrossRef]
- Tut, T.G.; Lim, S.H.; Dissanayake, I.U.; Descallar, J.; Chua, W.; Ng, W.; de Souza, P.; Shin, J.S.; Lee, C.S. Upregulated Polo-Like Kinase 1 Expression Correlates with Inferior Survival Outcomes in Rectal Cancer. PLoS ONE 2015, 10, e0129313. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, G.; Ma, Z.; Li, L.; Liu, M.; Qin, L.; Yu, Z.; Zhao, L.; Liu, Y.; Zhang, X.; et al. Hyperactivation of HER2-SHCBP1-PLK1 axis promotes tumor cell mitosis and impairs trastuzumab sensitivity to gastric cancer. Nat. Commun. 2021, 12, 2812. [Google Scholar] [CrossRef]
- Raab, M.; Strebhardt, K.; Rudd, C.E. Immune adaptor SKAP1 acts a scaffold for Polo-like kinase 1 (PLK1) for the optimal cell cycling of T-cells. Sci. Rep. 2019, 9, 10462. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Wei, J.; Wu, Y.; Zhao, K.; Wu, Y.; Wu, S.; Yang, X.; Xing, C. THUMPD3-AS1 facilitates cell growth and aggressiveness by the miR-218-5p/SKAP1 axis in colorectal cancer. Cell Biochem. Biophys. 2022, 80, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Canson, D.M.; O’Mara, T.A.; Spurdle, A.B.; Glubb, D.M. Splicing annotation of endometrial cancer GWAS risk loci reveals potentially causal variants and supports a role for NF1 and SKAP1 as susceptibility genes. HGG Adv. 2023, 4, 100185. [Google Scholar] [CrossRef] [PubMed]
- Ménasché, G.; Kliche, S.; Chen, E.J.; Stradal, T.E.; Schraven, B.; Koretzky, G. RIAM links the ADAP/SKAP-55 signaling module to Rap1, facilitating T-cell-receptor-mediated integrin activation. Mol. Cell. Biol. 2007, 27, 4070–4081. [Google Scholar] [CrossRef] [PubMed]
- Raab, M.; Wang, H.; Lu, Y.; Smith, X.; Wu, Z.; Strebhardt, K.; Ladbury, J.E.; Rudd, C.E. T cell receptor “inside-out” pathway via signaling module SKAP1-RapL regulates T cell motility and interactions in lymph nodes. Immunity 2010, 32, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Ophir, M.J.; Liu, B.C.; Bunnell, S.C. The N terminus of SKAP55 enables T cell adhesion to TCR and integrin ligands via distinct mechanisms. J. Cell Biol. 2013, 203, 1021–1041. [Google Scholar] [CrossRef]
- Khan, R.B.; Goult, B.T. Adhesions Assemble!—Autoinhibition as a Major Regulatory Mechanism of Integrin-Mediated Adhesion. Front. Mol. Biosci. 2019, 6, 144. [Google Scholar] [CrossRef]
- Yan, B.; Calderwood, D.A.; Yaspan, B.; Ginsberg, M.H. Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J. Biol. Chem. 2001, 276, 28164–28170. [Google Scholar] [CrossRef]
- Almajali, B.; Johan, M.F.; Al-Wajeeh, A.S.; Wan Taib, W.R.; Ismail, I.; Alhawamdeh, M.; Al-Tawarah, N.M.; Ibrahim, W.N.; Al-Rawashde, F.A.; Al-Jamal, H.A.N. Gene Expression Profiling and Protein Analysis Reveal Suppression of the C-Myc Oncogene and Inhibition JAK/STAT and PI3K/AKT/mTOR Signaling by Thymoquinone in Acute Myeloid Leukemia Cells. Pharmaceuticals 2022, 15, 307. [Google Scholar] [CrossRef]
- Matsuoka, T.; Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 2018, 24, 2818–2832. [Google Scholar] [CrossRef]
- Smith, X.; Taylor, A.; Rudd, C.E. T-cell immune adaptor SKAP1 regulates the induction of collagen-induced arthritis in mice. Immunol. Lett. 2016, 176, 122–127. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Wu, P.; Hu, Y.Z. PI3K/Akt/mTOR pathway inhibitors in cancer: A perspective on clinical progress. Curr. Med. Chem. 2010, 17, 4326–4341. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Podlaha, O.; Riester, M.; De, S.; Michor, F. Evolution of the cancer genome. Trends Genet. TIG 2012, 28, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Demaria, O.; Cornen, S.; Daëron, M.; Morel, Y.; Medzhitov, R.; Vivier, E. Harnessing innate immunity in cancer therapy. Nature 2019, 574, 45–56. [Google Scholar] [CrossRef]
- Woo, S.R.; Corrales, L.; Gajewski, T.F. Innate immune recognition of cancer. Annu. Rev. Immunol. 2015, 33, 445–474. [Google Scholar] [CrossRef]
- Corrales, L.; Matson, V.; Flood, B.; Spranger, S.; Gajewski, T.F. Innate immune signaling and regulation in cancer immunotherapy. Cell Res. 2017, 27, 96–108. [Google Scholar] [CrossRef]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef]
- Alderton, G.K.; Bordon, Y. Tumour immunotherapy--leukocytes take up the fight. Nat. Rev. Immunol. 2012, 12, 237. [Google Scholar] [CrossRef]
- Yuen, G.J.; Demissie, E.; Pillai, S. B lymphocytes and cancer: A love-hate relationship. Trends Cancer 2016, 2, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Yu, Q.; Li, Y.; Zhang, M.; Peng, Z.; Wang, S.; Quan, Z.; Gao, D. SKAP1 Is a Novel Biomarker and Therapeutic Target for Gastric Cancer: Evidence from Expression, Functional, and Bioinformatic Analyses. Int. J. Mol. Sci. 2023, 24, 11870. https://doi.org/10.3390/ijms241411870
Zhu L, Yu Q, Li Y, Zhang M, Peng Z, Wang S, Quan Z, Gao D. SKAP1 Is a Novel Biomarker and Therapeutic Target for Gastric Cancer: Evidence from Expression, Functional, and Bioinformatic Analyses. International Journal of Molecular Sciences. 2023; 24(14):11870. https://doi.org/10.3390/ijms241411870
Chicago/Turabian StyleZhu, Lingqin, Qiongfang Yu, Yuanheng Li, Meng Zhang, Zhiwei Peng, Song Wang, Ziyi Quan, and Dian Gao. 2023. "SKAP1 Is a Novel Biomarker and Therapeutic Target for Gastric Cancer: Evidence from Expression, Functional, and Bioinformatic Analyses" International Journal of Molecular Sciences 24, no. 14: 11870. https://doi.org/10.3390/ijms241411870
APA StyleZhu, L., Yu, Q., Li, Y., Zhang, M., Peng, Z., Wang, S., Quan, Z., & Gao, D. (2023). SKAP1 Is a Novel Biomarker and Therapeutic Target for Gastric Cancer: Evidence from Expression, Functional, and Bioinformatic Analyses. International Journal of Molecular Sciences, 24(14), 11870. https://doi.org/10.3390/ijms241411870





