The Evaluation of Rac1 Signaling as a Potential Therapeutic Target of Alzheimer’s Disease
Abstract
1. Introduction
2. Results
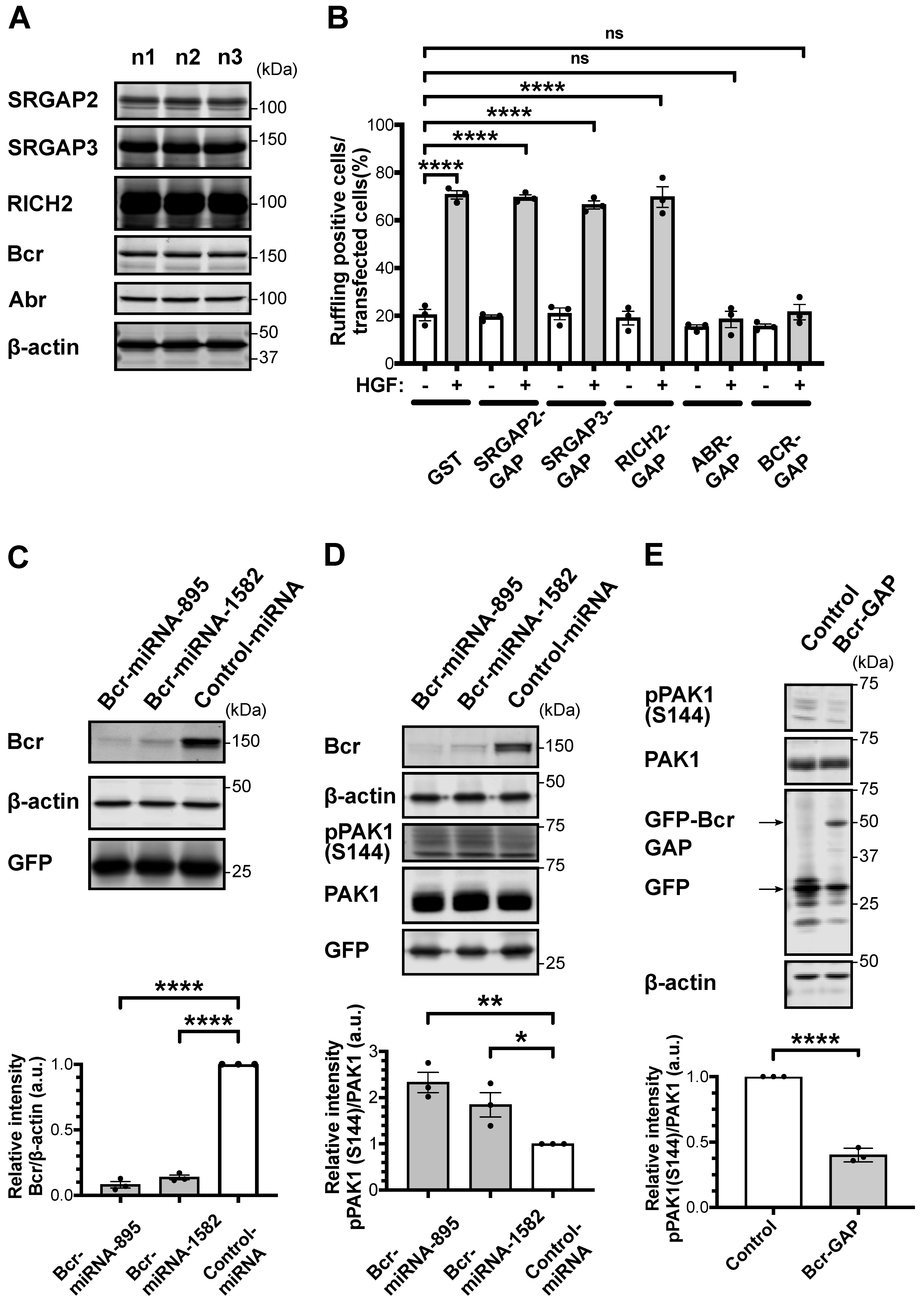
2.1. RacGAP Bcr Knockdown in Striatal/Accumbal Neurons Enhances Rac1-PAK Signaling
2.2. Bcr Knockdown in the NAc Enhances Aversive Learning
3. Discussion
4. Materials and Methods
4.1. Antibodies
4.2. Cell Culture and Transfection
4.3. Animal
4.4. Plasmid Construction
4.5. Primary Neuron Culture
4.6. Cell Ruffling Assay
4.7. Ex Vivo Sample Preparation
4.8. In Vivo Sample Preparation
4.9. SDS-PAGE Immunoblotting Assay
4.10. AAV Preparation
4.11. AAV Injection
4.12. Passive Avoidance Test
4.13. Immunohistochemical Analysis
4.14. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Etienne-Manneville, S.; Hall, A. Rho GTPases in Cell Biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Bosco, E.E.; Mulloy, J.C.; Zheng, Y. Rac1 GTPase: A “Rac” of All Trades. Cell. Mol. Life Sci. 2009, 66, 370–374. [Google Scholar] [CrossRef]
- Van Aelst, L.; D’Souza-Schorey, C. Rho GTPases and Signaling Networks. Genes Dev. 1997, 11, 2295–2322. [Google Scholar] [CrossRef]
- Hayashi-Takagi, A.; Yagishita, S.; Nakamura, M.; Shirai, F.; Wu, Y.I.; Loshbaugh, A.L.; Kuhlman, B.; Hahn, K.M.; Kasai, H. Labelling and Optical Erasure of Synaptic Memory Traces in the Motor Cortex. Nature 2015, 525, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Bolognin, S.; Lorenzetto, E.; Diana, G.; Buffelli, M. The Potential Role of Rho GTPases in Alzheimer’s Disease Pathogenesis. Mol. Neurobiol. 2014, 50, 406–422. [Google Scholar] [CrossRef]
- Dobrigna, M.; Poëa-Guyon, S.; Rousseau, V.; Vincent, A.; Toutain, A.; Barnier, J.-V. The Molecular Basis of P21-Activated Kinase-Associated Neurodevelopmental Disorders: From Genotype to Phenotype. Front. Neurosci. 2023, 17, 1123784. [Google Scholar] [CrossRef]
- Ruiz-Velasco, R.; Lanning, C.C.; Williams, C.L. The Activation of Rac1 by M3 Muscarinic Acetylcholine Receptors Involves the Translocation of Rac1 and IQGAP1 to Cell Junctions and Changes in the Composition of Protein Complexes Containing Rac1, IQGAP1, and Actin. J. Biol. Chem. 2002, 277, 33081–33091. [Google Scholar] [CrossRef]
- Ridley, A.J.; Paterson, H.F.; Johnston, C.L.; Diekmann, D.; Hall, A. The Small GTP-Binding Protein Rac Regulates Growth Factor-Induced Membrane Ruffling. Cell 1992, 70, 401–410. [Google Scholar] [CrossRef]
- Vetter, I.R.; Wittinghofer, A. The Guanine Nucleotide-Binding Switch in Three Dimensions. Science 2001, 294, 1299–1304. [Google Scholar] [CrossRef]
- Scala, M.; Nishikawa, M.; Ito, H.; Tabata, H.; Khan, T.; Accogli, A.; Davids, L.; Ruiz, A.; Chiurazzi, P.; Cericola, G.; et al. Variant-Specific Changes in RAC3 Function Disrupt Corticogenesis in Neurodevelopmental Phenotypes. Brain 2022, 145, 3308–3327. [Google Scholar] [CrossRef] [PubMed]
- Manser, E.; Leung, T.; Salihuddin, H.; Zhao, Z.; Lim, L. A Brain Serine/Threonine Protein Kinase Activated by Cdc42 and Rac1. Nature 1994, 367, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Knaus, U.G.; Morris, S.; Dong, H.-J.; Chernoff, J.; Bokoch, G.M. Regulation of Human Leukocyte P21-Activated Kinases Through G Protein—Coupled Receptors. Science 1995, 269, 221–223. [Google Scholar] [CrossRef]
- Wang, F.; Herzmark, P.; Weiner, O.D.; Srinivasan, S.; Servant, G.; Bourne, H.R. Lipid Products of PI(3)Ks Maintain Persistent Cell Polarity and Directed Motility in Neutrophils. Nat. Cell Biol. 2002, 4, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, R.; Downward, J. SnapShot: Class I PI3K Isoform Signaling. Cell 2013, 154, 940.e1. [Google Scholar] [CrossRef] [PubMed]
- Yamahashi, Y.; Lin, Y.-H.; Mouri, A.; Iwanaga, S.; Kawashima, K.; Tokumoto, Y.; Watanabe, Y.; Faruk, M.O.; Zhang, X.; Tsuboi, D.; et al. Phosphoproteomic of the Acetylcholine Pathway Enables Discovery of the PKC-β-PIX-Rac1-PAK Cascade as a Stimulatory Signal for Aversive Learning. Mol. Psychiatry 2022, 27, 3479–3492. [Google Scholar] [CrossRef] [PubMed]
- Buccafusco, J.J. Methods of Behavior Analysis in Neuroscience; CRC Press/Taylor&Francis: Boca Raton, FL, USA, 2009; ISBN 9781420052343. [Google Scholar]
- Kreider-Letterman, G.; Carr, N.M.; Garcia-Mata, R. Fixing the GAP: The Role of RhoGAPs in Cancer. Eur. J. Cell Biol. 2022, 101, 151209. [Google Scholar] [CrossRef]
- Oh, D.; Han, S.; Seo, J.; Lee, J.-R.; Choi, J.; Groffen, J.; Kim, K.; Cho, Y.S.; Choi, H.-S.; Shin, H.; et al. Regulation of Synaptic Rac1 Activity, Long-Term Potentiation Maintenance, and Learning and Memory by BCR and ABR Rac GTPase-Activating Proteins. J. Neurosci. 2010, 30, 14134–14144. [Google Scholar] [CrossRef]
- Charrier, C.; Joshi, K.; Coutinho-Budd, J.; Kim, J.-E.; Lambert, N.; de Marchena, J.; Jin, W.-L.; Vanderhaeghen, P.; Ghosh, A.; Sassa, T.; et al. Inhibition of SRGAP2 Function by Its Human-Specific Paralogs Induces Neoteny during Spine Maturation. Cell 2012, 149, 923–935. [Google Scholar] [CrossRef]
- Raynaud, F.; Janossy, A.; Dahl, J.; Bertaso, F.; Perroy, J.; Varrault, A.; Vidal, M.; Worley, P.F.; Boeckers, T.M.; Bockaert, J.; et al. Shank3-Rich2 Interaction Regulates AMPA Receptor Recycling and Synaptic Long-Term Potentiation. J. Neurosci. 2013, 33, 9699–9715. [Google Scholar] [CrossRef]
- Iwata, R.; Matsukawa, H.; Yasuda, K.; Mizuno, H.; Itohara, S.; Iwasato, T. Developmental RacGAP A2-Chimaerin Signaling Is a Determinant of the Morphological Features of Dendritic Spines in Adulthood. J. Neurosci. 2015, 35, 13728–13744. [Google Scholar] [CrossRef]
- Bacon, C.; Endris, V.; Rappold, G.A. The Cellular Function of SrGAP3 and Its Role in Neuronal Morphogenesis. Mech. Dev. 2013, 130, 391–395. [Google Scholar] [CrossRef]
- Iwata, R.; Ohi, K.; Kobayashi, Y.; Masuda, A.; Iwama, M.; Yasuda, Y.; Yamamori, H.; Tanaka, M.; Hashimoto, R.; Itohara, S.; et al. RacGAP A2-Chimaerin Function in Development Adjusts Cognitive Ability in Adulthood. Cell Rep. 2014, 8, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Tcherkezian, J.; Lamarche-Vane, N. Current Knowledge of the Large RhoGAP Family of Proteins. Biol. Cell 2007, 99, 67–86. [Google Scholar] [CrossRef] [PubMed]
- Fukata, Y.; Oshiro, N.; Kinoshita, N.; Kawano, Y.; Matsuoka, Y.; Bennett, V.; Matsuura, Y.; Kaibuchi, K. Phosphorylation of Adducin by Rho-Kinase Plays a Crucial Role in Cell Motility. J. Cell Biol. 1999, 145, 347–361. [Google Scholar] [CrossRef]
- Fukata, M.; Nakagawa, M.; Itoh, N.; Kawajiri, A.; Yamaga, M.; Kuroda, S.; Kaibuchi, K. Involvement of IQGAP1, an Effector of Rac1 and Cdc42 GTPases, in Cell-Cell Dissociation during Cell Scattering. Mol. Cell Biol. 2001, 21, 2165–2183. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J.; Self, A.J.; Kasmi, F.; Paterson, H.F.; Hall, A.; Marshall, C.J.; Ellis, C. Rho Family GTPase Activating Proteins P190, Bcr and RhoGAP Show Distinct Specificities in Vitro and in Vivo. EMBO J. 1993, 12, 5151–5160. [Google Scholar] [CrossRef]
- Tan, E.C.; Leung, T.; Manser, E.; Lim, L. The Human Active Breakpoint Cluster Region-Related Gene Encodes a Brain Protein with Homology to Guanine Nucleotide Exchange Proteins and GTPase-Activating Proteins. J. Biol. Chem. 1993, 268, 27291–27298. [Google Scholar] [CrossRef]
- Chong, C.; Tan, L.; Lim, L.; Manser, E. The Mechanism of PAK Activation. J. Biol. Chem. 2001, 276, 17347–17353. [Google Scholar] [CrossRef]
- ZHAO, Z.; MANSER, E. PAK and Other Rho-Associated Kinases—Effectors with Surprisingly Diverse Mechanisms of Regulation. Biochem. J. 2005, 386, 201–214. [Google Scholar] [CrossRef]
- Lei, M.; Lu, W.; Meng, W.; Parrini, M.-C.; Eck, M.J.; Mayer, B.J.; Harrison, S.C. Structure of PAK1 in an Autoinhibited Conformation Reveals a Multistage Activation Switch. Cell 2000, 102, 387–397. [Google Scholar] [CrossRef]
- Zhao, L.; Ma, Q.-L.; Calon, F.; Harris-White, M.E.; Yang, F.; Lim, G.P.; Morihara, T.; Ubeda, O.J.; Ambegaokar, S.; Hansen, J.E.; et al. Role of P21-Activated Kinase Pathway Defects in the Cognitive Deficits of Alzheimer Disease. Nat. Neurosci. 2006, 9, 234–242. [Google Scholar] [CrossRef]
- Lauterborn, J.C.; Cox, C.D.; Chan, S.W.; Vanderklish, P.W.; Lynch, G.; Gall, C.M. Synaptic Actin Stabilization Protein Loss in Down Syndrome and Alzheimer Disease. Brain Pathol. 2020, 30, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Newey, S.E.; Velamoor, V.; Govek, E.-E.; Van Aelst, L. Rho GTPases, Dendritic Structure, and Mental Retardation. J. Neurobiol. 2005, 64, 58–74. [Google Scholar] [CrossRef]
- Stankiewicz, T.R.; Linseman, D.A. Rho Family GTPases: Key Players in Neuronal Development, Neuronal Survival, and Neurodegeneration. Front. Cell. Neurosci. 2014, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Lionarons, D.A.; Hancock, D.C.; Rana, S.; East, P.; Moore, C.; Murillo, M.M.; Carvalho, J.; Spencer-Dene, B.; Herbert, E.; Stamp, G.; et al. RAC1P29S Induces a Mesenchymal Phenotypic Switch via Serum Response Factor to Promote Melanoma Development and Therapy Resistance. Cancer Cell 2019, 36, 68–83.e9. [Google Scholar] [CrossRef] [PubMed]
- Katayama, K.; Zheng, Y.; Inoue, N. Rac Is Required for the Survival of Cortical Neurons. Exp. Neurol. 2023, 361, 114316. [Google Scholar] [CrossRef] [PubMed]
- Bradley, S.J.; Bourgognon, J.-M.; Sanger, H.E.; Verity, N.; Mogg, A.J.; White, D.J.; Butcher, A.J.; Moreno, J.A.; Molloy, C.; Macedo-Hatch, T.; et al. M1 Muscarinic Allosteric Modulators Slow Prion Neurodegeneration and Restore Memory Loss. J. Clin. Investig. 2016, 127, 487–499. [Google Scholar] [CrossRef]
- Abd-Elrahman, K.S.; Sarasija, S.; Colson, T.L.; Ferguson, S.S.G. A Positive Allosteric Modulator for the Muscarinic Receptor (M1 MAChR) Improves Pathology and Cognitive Deficits in Female APPswe/PSEN1ΔE9 Mice. Br. J. Pharmacol. 2022, 179, 1769–1783. [Google Scholar] [CrossRef]
- Brown, A.J.H.; Bradley, S.J.; Marshall, F.H.; Brown, G.A.; Bennett, K.A.; Brown, J.; Cansfield, J.E.; Cross, D.M.; de Graaf, C.; Hudson, B.D.; et al. From Structure to Clinic: Design of a Muscarinic M1 Receptor Agonist with the Potential to Treat Alzheimer’s Disease. Cell 2021, 184, 5886–5901.e22. [Google Scholar] [CrossRef]
- Cowan, C.W.; Shao, Y.R.; Sahin, M.; Shamah, S.M.; Lin, M.Z.; Greer, P.L.; Gao, S.; Griffith, E.C.; Brugge, J.S.; Greenberg, M.E. Vav Family GEFs Link Activated Ephs to Endocytosis and Axon Guidance. Neuron 2005, 46, 205–217. [Google Scholar] [CrossRef]
- Ögren, S.O.; Stiedl, O. Passive Avoidance. In Encyclopedia of Psychopharmacology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1220–1228. [Google Scholar]
- Nagai, T.; Nakamuta, S.; Kuroda, K.; Nakauchi, S.; Nishioka, T.; Takano, T.; Zhang, X.; Tsuboi, D.; Funahashi, Y.; Nakano, T.; et al. Phosphoproteomics of the Dopamine Pathway Enables Discovery of Rap1 Activation as a Reward Signal In Vivo. Neuron 2016, 89, 550–565. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Yamahashi, Y.; Riedl, M.; Amano, M.; Kaibuchi, K. The Evaluation of Rac1 Signaling as a Potential Therapeutic Target of Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 11880. https://doi.org/10.3390/ijms241511880
Wang H, Yamahashi Y, Riedl M, Amano M, Kaibuchi K. The Evaluation of Rac1 Signaling as a Potential Therapeutic Target of Alzheimer’s Disease. International Journal of Molecular Sciences. 2023; 24(15):11880. https://doi.org/10.3390/ijms241511880
Chicago/Turabian StyleWang, Huanhuan, Yukie Yamahashi, Marcel Riedl, Mutsuki Amano, and Kozo Kaibuchi. 2023. "The Evaluation of Rac1 Signaling as a Potential Therapeutic Target of Alzheimer’s Disease" International Journal of Molecular Sciences 24, no. 15: 11880. https://doi.org/10.3390/ijms241511880
APA StyleWang, H., Yamahashi, Y., Riedl, M., Amano, M., & Kaibuchi, K. (2023). The Evaluation of Rac1 Signaling as a Potential Therapeutic Target of Alzheimer’s Disease. International Journal of Molecular Sciences, 24(15), 11880. https://doi.org/10.3390/ijms241511880




