Oligo-Carrageenan Kappa Increases Expression of Genes Encoding Proteins Involved in Photosynthesis, C, N, and S Assimilation, and Growth in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
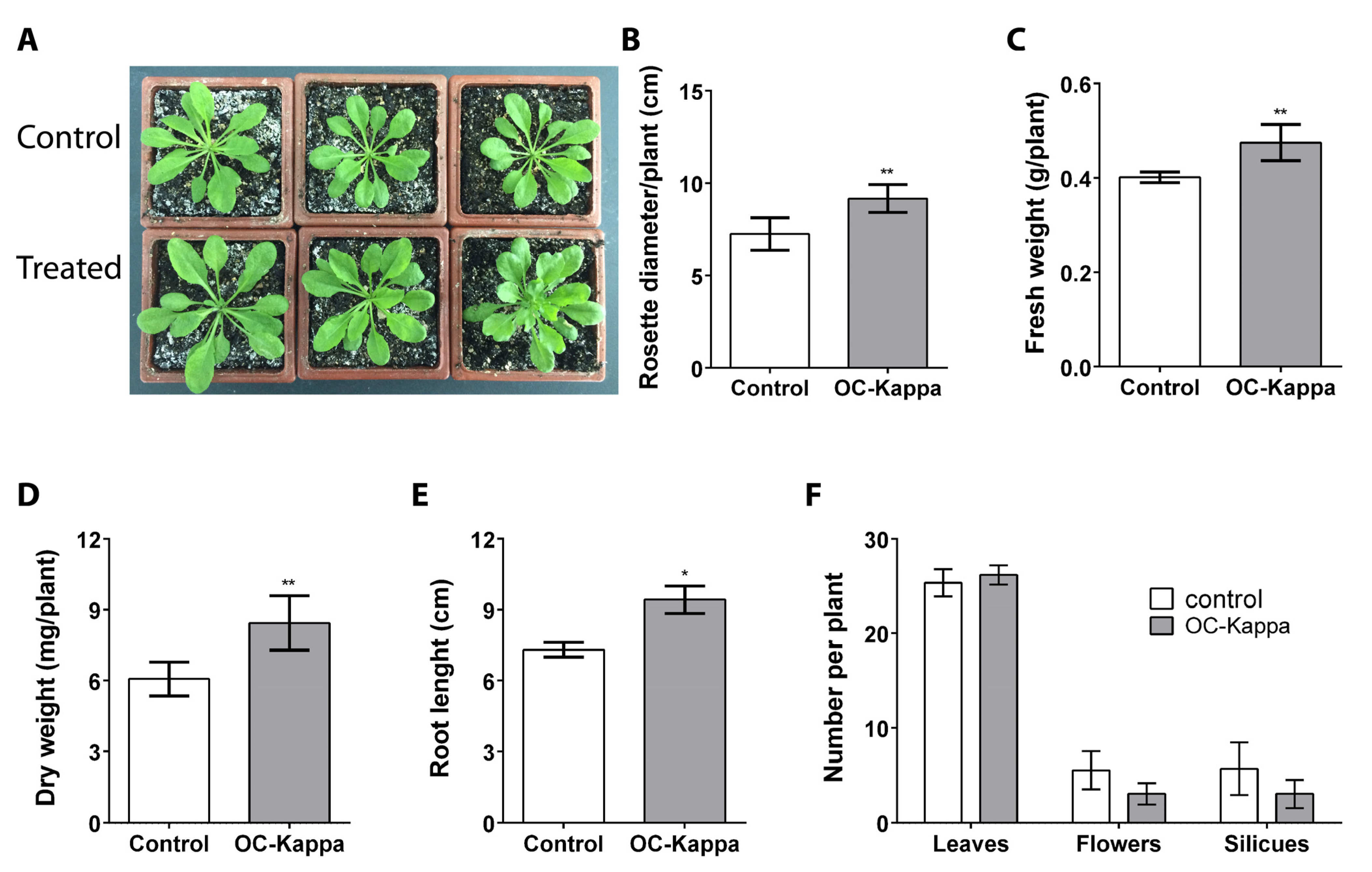
2.1. OC Kappa Induces an Increase in A. thaliana Growth
2.2. Identification of Transcripts Differentially Expressed in Response to OC Kappa
2.3. Classification of Differentially Expressed Transcripts in Response to OC Kappa
2.4. Identification of Transcripts Encoding Proteins Involved in Plant Growth and Development
2.5. Identification of Transcripts Encoding Proteins Involved in Photosynthesis
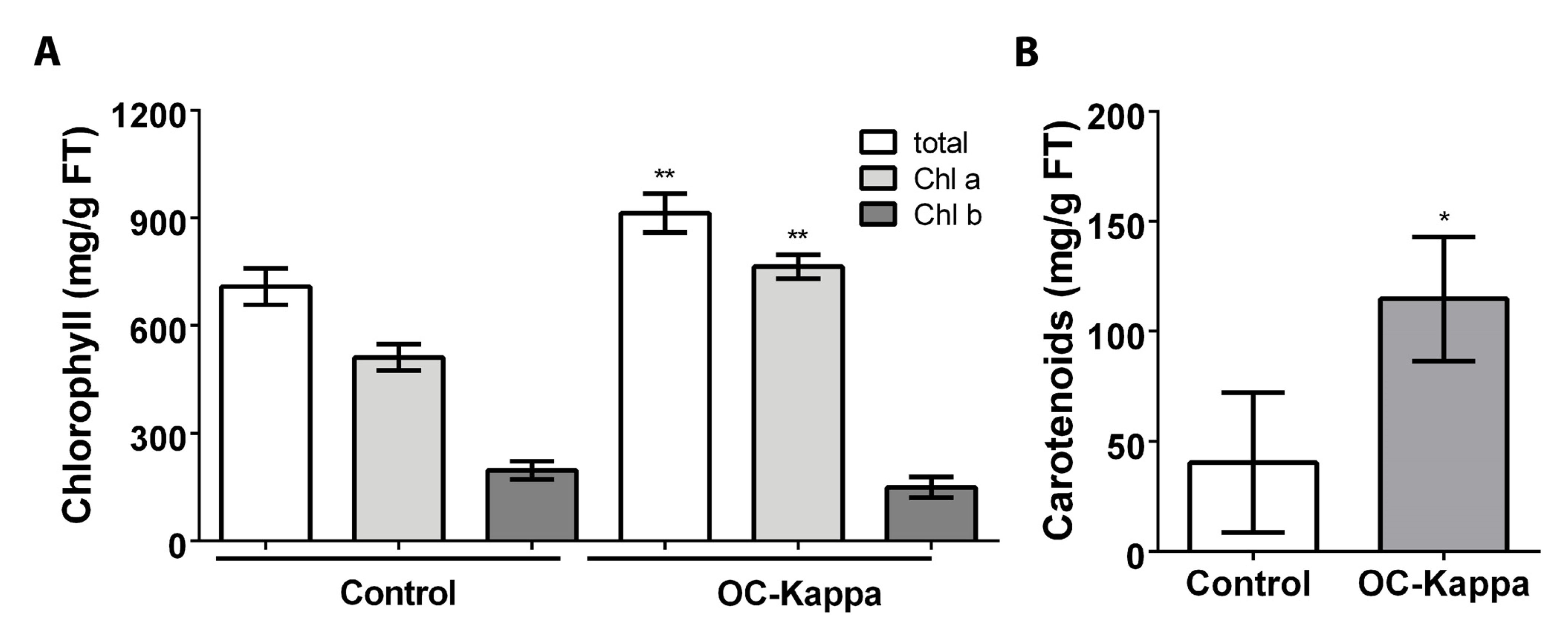
2.6. Quantification of Chlorophylls and Carotenoids in Plants Treated with OC Kappa
2.7. Identification of Transcripts Encoding Enzymes Involved in C, N, and S Assimilation
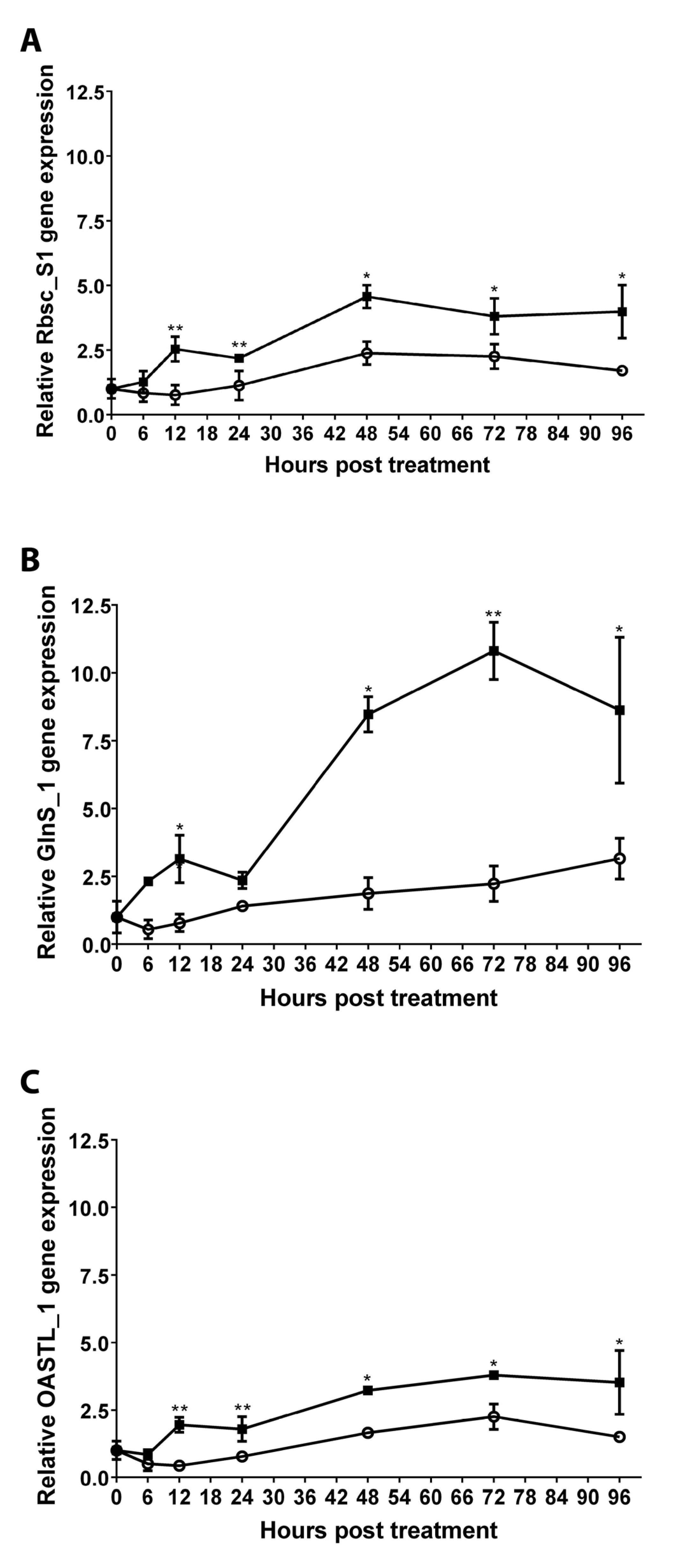
2.8. Quantification of Transcripts Encoding Enzymes Involved in C, N, and S Assimilation
3. Discussion
4. Materials and Methods
4.1. Plant Cultivation and Preparation of OC Kappa
4.2. Treatment with OC Kappa
4.3. Total RNA Extraction, Preparation of cDNA Libraries, and Sequencing
4.4. Annotation and Detection of Differentially Expressed Transcripts
4.5. Quantification of Chlorophylls
4.6. Quantification of Carotenoids
4.7. Quantification of Transcripts of Enzymes Involved in C, N, and S Assimilation by qRT-PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vassilev, N.; Vassileva, M.; Lopez, A.; Martos, V.; Reyes, A.; Maksimovic, I.; Eichler-Löbermann, B.; Malusà, E. Unexploited Potential of Some Biotechnological Techniques for Biofertilizer Production and Formulation. Appl. Microbiol. Biotechnol. 2015, 99, 4983–4996. [Google Scholar] [CrossRef] [PubMed]
- Igawa, T.K.; de Toledo, P.M.; Anjos, L.J.S. Climate Change Could Reduce and Spatially Reconfigure Cocoa Cultivation in the Brazilian Amazon by 2050. PLoS ONE 2022, 17, e0262729. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.P.; Tegeder, M. Implications of Nitrogen Phloem Loading for Carbon Metabolism and Transport during Arabidopsis Development. J. Integr. Plant Biol. 2017, 59, 409–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anoman, A.D.; Flores-Tornero, M.; Benstein, R.M.; Blau, S.; Rosa-Téllez, S.; Bräutigam, A.; Fernie, A.R.; Muñoz-Bertomeu, J.; Schilasky, S.; Meyer, A.J.; et al. Deficiency in the Phosphorylated Pathway of Serine Biosynthesis Perturbs Sulfur Assimilation. Plant Physiol. 2019, 180, 153–170. [Google Scholar] [CrossRef]
- Ristova, D.; Kopriva, S. Sulfur Signaling and Starvation Response in Arabidopsis. IScience 2022, 25, 104242. [Google Scholar] [CrossRef]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and Its Role in Plant Development: Structure, Signalling, Regulation and Response Mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Meng, Z.; Wang, B.; Chen, M. Research Progress on the Roles of Cytokinin in Plant Response to Stress. Int. J. Mol. Sci. 2020, 21, 6574. [Google Scholar] [CrossRef]
- Hedden, P. The Current Status of Research on Gibberellin Biosynthesis. Plant Cell Physiol. 2020, 61, 1832–1849. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A.; Ali, Q.; Liu, F. Brassinosteroids (BRs) Role in Plant Development and Coping with Different Stresses. Int. J. Mol. Sci. 2022, 23, 1012. [Google Scholar] [CrossRef]
- Choudhury, S.; Islam, N.; Ali, M.A. Growth and Yield of Summer Tomato as Influenced by Plant Growth Regulators. Int. J. Sustain. Agric. 2013, 5, 25–28. [Google Scholar] [CrossRef]
- Laporte, D.; Vera, J.; Chandía, N.P.; Zúñiga, E.A.; Matsuhiro, B.; Moenne, A. Structurally Unrelated Algal Oligosaccharides Differentially Stimulate Growth and Defense against Tobacco Mosaic Virus in Tobacco Plants. J. Appl. Phycol. 2007, 19, 79–88. [Google Scholar] [CrossRef]
- Shukla, P.S.; Borza, T.; Critchley, A.T.; Prithiviraj, B. Carrageenans from Red Seaweeds as Promoters of Growth and Elicitors of Defense Response in Plants. Front. Mar. Sci. 2016, 3, 81. [Google Scholar] [CrossRef] [Green Version]
- Moenne, A.; González, A. Chitosan-, Alginate- Carrageenan-Derived Oligosaccharides Stimulate Defense against Biotic and Abiotic Stresses, and Growth in Plants: A Historical Perspective. Carbohydr. Res. 2021, 503, 108298. [Google Scholar] [CrossRef] [PubMed]
- Vera, J.; Castro, J.; Gonzalez, A.; Moenne, A. Seaweed Polysaccharides and Derived Oligosaccharides Stimulate Defense Responses and Protection against Pathogens in Plants. Mar. Drugs 2011, 9, 2514–2525. [Google Scholar] [CrossRef]
- Ahmad, B.; Jahan, A.; Sadiq, Y.; Shabbir, A.; Jaleel, H.; Khan, M.M.A. Radiation-Mediated Molecular Weight Reduction and Structural Modification in Carrageenan Potentiates Improved Photosynthesis and Secondary Metabolism in Peppermint (Mentha piperita L.). Int. J. Biol. Macromol. 2019, 124, 1069–1079. [Google Scholar] [CrossRef]
- San, P.T.; Khanh, C.M.; Khanh, H.H.N.; Khoa, T.A.; Hoang, N.; Nhung, L.T.; Trinh, N.T.K.; Nguyen, T.D. K-Oligocarrageenan Promoting Growth of Hybrid Maize: Influence of Molecular Weight. Molecules 2020, 25, 3825. [Google Scholar] [CrossRef]
- Moenne, A. Composition and Method to Stimulate Plant Growth and Defense against Pathogens in Plants. US Pat. Off. Appl. 2009, 12, 666–700. [Google Scholar]
- Castro, J.; Vera, J.; González, A.; Moenne, A. Oligo-Carrageenans Stimulate Growth by Enhancing Photosynthesis, Basal Metabolism, and Cell Cycle in Tobacco Plants (Var. Burley). J. Plant Growth Regul. 2012, 31, 173–185. [Google Scholar] [CrossRef]
- González, A.; Moenne, F.; Gómez, M.; Sáez, C.A.; Contreras, R.A.; Moenne, A. Oligo-Carrageenan Kappa Increases NADPH, Ascorbate and Glutathione Syntheses and TRR/TRX Activities Enhancing Photosynthesis, Basal Metabolism, and Growth in Eucalyptus Trees. Front. Plant Sci. 2014, 5, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, A.; Contreras, R.A.; Zúñiga, G.; Moenne, A. Oligo-Carrageenan Kappa-Induced Reducing Redox Status and Activation of TRR/TRX System Increase the Level of Indole-3-Acetic Acid, Gibberellin A 3 and Trans-Zeatin in Eucalyptus Globulus Trees. Molecules 2014, 19, 12690–12698. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.H.; Huang, X.Z.; Xiao, S.L.; Fu, X.D. Evolutionarily Conserved DELLA-Mediated Gibberellin Signaling in Plants. J. Integr. Plant Biol. 2008, 50, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Teramura, H.; Murakoshi, S.; Nasuno, K.; Nishida, N.; Ito, T.; Yoshida, M.; Kamiya, Y.; Yamaguchi, S.; Takahashi, Y. DELLAs Function as Coactivators of GAI-ASSOCIATED FACTOR1 in Regulation of Gibberellin Homeostasis and Signaling in Arabidopsis. Plant Cell 2014, 26, 2920–2938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agusti, J.; Lichtenberger, R.; Schwarz, M.; Nehlin, L.; Greb, T. Characterization of Transcriptome Remodeling during Cambium Formation Identifies MOL1 and RUL1 as Opposing Regulators of Secondary Growth. PLoS Genet. 2011, 7, e1001312. [Google Scholar] [CrossRef] [Green Version]
- Gursanscky, N.R.; Jouannet, V.; Grünwald, K.; Sanchez, P.; Laaber-Schwarz, M.; Greb, T. MOL1 Is Required for Cambium Homeostasis in Arabidopsis. Plant J. 2016, 86, 210–220. [Google Scholar] [CrossRef] [Green Version]
- Mishler-Elmore, J.W.; Zhou, Y.; Sukul, A.; Oblak, M.; Tan, L.; Faik, A.; Held, M.A. Extensins: Self-Assembly, Crosslinking, and the Role of Peroxidases. Front. Plant Sci. 2021, 14, 664738. [Google Scholar] [CrossRef]
- Hromadová, D.; Soukup, A.; Tylová, E. Arabinogalactan Proteins in Plant Roots—An Update on Possible Functions. Front. Plant Sci. 2021, 17, 674010. [Google Scholar] [CrossRef]
- İşkil, R.; Surgun-Acar, Y. Expression Analysis of Cell Wall Assembly and Remodelling-Related Genes in Arabidopsis Roots Subjected to Boron Stress and Brassinosteroid at Different Developmental Stages. Acta Bot. Bras. 2018, 32, 546–554. [Google Scholar] [CrossRef] [Green Version]
- Babu, Y.; Bayer, M. Plant Polygalacturonases Involved in Cell Elongation and Separation—The Same but Different? Plants 2014, 3, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Nibbering, P.; Petersen, B.L.; Motawia, M.S.; Jørgensen, B.; Ulvskov, P.; Niittylä, T. Golgi-Localized Exo-B1,3-Galactosidases Involved in Cell Expansion and Root Growth in Arabidopsis. J. Biol. Chem. 2020, 295, 10581–10592. [Google Scholar] [CrossRef]
- Pauly, M.; Keegstra, K. Biosynthesis of the Plant Cell Wall Matrix Polysaccharide Xyloglucan∗. Annu. Rev. Plant Biol. 2016, 67, 235–259. [Google Scholar] [CrossRef]
- Buttery, B.R.; Buzzell, R.L. The relantionship between chloropyll content and rate of photosynthesis in soybeans. Can. J. Plant Sci. 1977, 57, 1–5. [Google Scholar] [CrossRef]
- Thye, K.L.; Wan Abdullah, W.M.A.N.; Balia Yusof, Z.N.; Wee, C.Y.; Ong-Abdullah, J.; Loh, J.Y.; Cheng, W.H.; Lamasudin, D.U.; Lai, K.S. λ-Carrageenan Promotes Plant Growth in Banana via Enhancement of Cellular Metabolism, Nutrient Uptake, and Cellular Homeostasis. Sci. Rep. 2022, 12, 19639. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Zaffagnini, M.; Morisse, S.; Sparla, F.; Pérez-Pérez, M.E.; Francia, F.; Danon, A.; Marchand, C.H.; Fermani, S.; Trost, P.; et al. Redox Regulation of the Calvin-Benson Cycle: Something Old, Something New. Front. Plant Sci. 2013, 25, 470. [Google Scholar] [CrossRef] [Green Version]
- Kang, Z.; Qin, T.; Zhao, Z. Thioredoxins and Thioredoxin Reductase in Chloroplasts: A Review. Gene 2019, 706, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) Database and Informatics Resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of Total Carotenoids and Chlorophylls a and b of Leaf Extracts in Different Solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef] [Green Version]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]

| Process | ID | Proteins | Log FC |
|---|---|---|---|
| Growth | AT5G03120 | MOK16.3 | 1.8 |
| AT3G19850 | MPN9.9 BTB/POZ | 2.1 | |
| AT3G61750 | Cyt b561 | 1.4 | |
| AT4G23820 | PECL | 1.2 | |
| AT3G28040 | MMG15.21 | 1.1 | |
| AT5G07080 | ACT | 3.2 | |
| Cell growth | AT4G03070 | AOP1 | 1.8 |
| AT5G24870 | UBQT | 1.3 | |
| AT5G65380 | DTX27 | 1.7 | |
| Unidimensional cell growth | AT5G44020 | HAD | 1.8 |
| AT2G37640 | EXPA3 | 1.6 | |
| AT1G69530 | EXPA1 | 2.4 | |
| AT3G29030 | EXPA5 | 1.2 | |
| AT2G40610 | EXPA8 | 6.5 | |
| AT1G75750 | GASA1 | 2.1 | |
| AT2G18300 | HBI1 | 2.8 | |
| AT1G75780 | TUBB1 | 1.3 | |
| Pollen tube growth | AT2G43750 | OASB | 1.6 |
| AT5G04950 | NAS1 | 2.4 | |
| AT1G09240 | NAS3 | 2.8 | |
| AT5G46220 | TOD1 | 4.3 | |
| Meristem growth | AT1G27470 | WD-40 | 1.1 |
| Cell wall modification | AT1G74670 | GASA6 | 2.9 |
| AT1G48100 | PGLR4 | 2.8 | |
| developmental growth | AT3G16175 | TES | 3.9 |
| AT1G72880 | SurE | 1.2 | |
| AT1G04040 | HAD | 2.8 | |
| AT3G03820 | SAUR29 | 9.7 | |
| AT5G18670 | BAM9 | 2.4 | |
| AT5G65890 | ACR1 | 1.8 | |
| AT1G58520 | RXW8 | 2.5 | |
| AT4G30140 | GDSL | 2.2 | |
| AT5G24580 | HIPPO9 | 2.9 | |
| AT3G16150 | MSL1.19 | 6.3 | |
| AT3G24480 | LRX4 | 1.2 | |
| AT3G27020 | YSL6 | 1.4 | |
| AT5G48900 | PLY20 | 1.2 | |
| AT1G69160 | BIG1E | 2.2 | |
| AT3G25717 | DVL6 | 2.4 | |
| AT2G32450 | T32F6.3 | 1.1 | |
| AT5G54130 | CBEEP | 2.2 | |
| AT4G14740 | DL3410C | 1.2 | |
| AT2G28470 | BGAL8 | 1.5 | |
| AT1G09250 | BHLH149 | 1.3 | |
| Regulation of growth | AT1G26945 | PRE6 | 4.2 |
| AT5G39860 | PRE1 | 9.2 | |
| AT2G17500 | PILS5 | 1.6 | |
| AT2G26670 | HO1 | 1.4 | |
| AT1G35670 | CPK11 | 1.9 | |
| AT5G64770 | GLV2 | 3.3 | |
| AT1G72430 | SAUR78 | 2.0 | |
| Regulation of cell growth | AT3G51800 | EBP1 | 1.5 |
| Regulation of | AT5G15430 | CBD | 6.7 |
| developmental growth | AT4G25780 | CAP | 2.0 |
| Response to growth | AT1G10370 | GSTU17 | 4.9 |
| hormone |
| Process | ID | Proteins | Log FC |
|---|---|---|---|
| PSII | AT1G03600 | Psb27-H1 | 1.5 |
| AT4G28660 | Psb28 | 1.7 | |
| AT2G20890 | Psb29 | 1.1 | |
| ATCG00070 | PsbK | 1.4 | |
| AT5G66570 | PsbO1 | 1.6 | |
| AT3G50820 | PsbO2 | 2.1 | |
| AT1G06680 | PsbP1 | 2.4 | |
| AT4G15510 | PsbP1 | 2.1 | |
| AT4G21280 | PsbQ1 | 2.9 | |
| AT4G05180 | PsbQ2 | 2.5 | |
| AT1G79040 | PsbR | 1.8 | |
| AT1G44575 | PsbS | 2.1 | |
| AT3G21055 | PsbT | 1.9 | |
| AT2G30570 | PsbW | 2.2 | |
| AT2G06520 | PsbX | 2.2 | |
| AT2G06520 | PsbX | 2.2 | |
| AT1G67740 | PsbY | 3.2 | |
| PSI | AT4G02770 | PsaD1 | 2.5 |
| AT1G03130 | PsaD2 | 4.0 | |
| AT4G28750 | PsaE-1 | 2.6 | |
| AT2G20260 | PsaE-2 | 1.6 | |
| AT1G31330 | PsaF | 2.7 | |
| AT1G55670 | PsaG | 2.5 | |
| AT1G52230 | PsaH | 3.3 | |
| AT1G30380 | PsaK | 2.1 | |
| AT4G12800 | PsaL | 2.3 | |
| AT1G49975 | PsaN | 1.2 | |
| AT5G64040 | PsaN | 3.2 | |
| AT1G08380 | PsaO | 3.1 | |
| AT4G22890 | PGRL1A | 1.4 | |
| LHCII | AT1G29920 | LhcB 1.1 | 4.2 |
| AT1G29910 | LhcB 1.2 | 3.3 | |
| AT1G29930 | LhcB 1.3 | 3.2 | |
| AT2G34430 | LhcB 1.5 | 1.2 | |
| AT2G34420 | LhcB 1.5 | 2.7 | |
| AT2G05100 | LhcB 2.1 | 3.0 | |
| AT2G05070 | LhcB 2.2 | 3.7 | |
| AT3G27690 | LhcB 2.4 | 3.9 | |
| AT5G54270 | LhcB 3 | 1.8 | |
| AT5G01530 | LhcB 4.1 | 8.0 | |
| AT3G08940 | LhcB 4.2 | 7.7 | |
| AT4G10340 | LhcB 5 | 7.3 | |
| AT1G15820 | LhcB 6 | 4.4 | |
| AT1G76570 | LhcB 7 | 5.3 | |
| LHCI | AT3G54890 | LHCA 1 | 7.4 |
| AT3G61470 | LHCA 2 | 1.9 | |
| AT1G61520 | LHCA 3 | 3.7 | |
| AT4G17600 | LIL3.1 | 1.5 | |
| AT3G47470 | LHCA 4 | 1.6 | |
| AT1G45474 | LHCA 5 | 2.0 | |
| AT1G19150 | LHCA 6 | 1.7 | |
| Electron transport chain | AT1G15980 | PnsB1 | 1.6 |
| AT1G76100 | PETE | 1.4 | |
| AT5G08720 | PPP1 | 1.9 | |
| Chlorophyll synthesis | AT1G74470 | CHLP | 2.5 |
| AT3G56940 | CRD1 | 3.4 | |
| AT4G14690 | ELIP2 | 7.1 | |
| Biosynthesis, repair and | AT1G16720 | Hcf173 | 2.2 |
| reassembly of PSII | AT4G35250 | Hcf244 | 2.3 |
| AT1G75690 | Lqy1 | 2.2 | |
| AT5G02120 | OHP1 | 2.5 | |
| ATP synthase | AT4G00895 | ATP synthase subunit delta, ATPaseF1 | 1.2 |
| AT4G09650 | ATP synthase subunit delta, ATPaseF1 | 2.9 |
| Process | ID | Proteins | Log FC |
|---|---|---|---|
| Calvin–Benson cycle | AT1G10960 | FD1 | 3.0 |
| AT4G14890 | FDC1 | 1.8 | |
| AT5G66190 | FNR1 | 1.2 | |
| AT4G38970 | FBA2 | 2.0 | |
| AT2G21330 | FBA1 | 2.6 | |
| AT1G12900 | G3PDHA2 | 2.5 | |
| AT1G42970 | G3PDHB | 1.1 | |
| AT3G26650 | G3PDHA1 | 1.4 | |
| AT4G22890 | PGRL1A | 1.4 | |
| AT2G01290 | R5PI2 | 2.5 | |
| AT3G04790 | R5PI3 | 2.0 | |
| AT1G67090 | RBCS-1A | 1.5 | |
| AT5G38430 | RBCS-1B | 2.5 | |
| AT5G38410 | RBCS-3B | 2.7 | |
| AT5G38420 | RBCS-2B | 2.7 | |
| AT2G39730 | RBCA | 3.0 | |
| AT3G04550 | RAF1.2 | 1.3 | |
| AT5G19855 | RBCX2 | 1.4 | |
| AT3G02730 | TRXF1 | 1.2 | |
| AT1G03680 | TRXM1 | 1.5 | |
| AT3G06730 | ClTRX | 1.6 | |
| AT1G76080 | CDSP32 | 1.7 | |
| AT5G16400 | TRXF2 | 2.2 | |
| AT5G17630 | XPT | 1.6 | |
| AT5G46110 | TPT | 1.9 | |
| AT3G01550 | PPT2 | 4.1 | |
| Nitrogen assimilation | AT5G56860 | GATA21 | 1.1 |
| AT1G66200 | GLN1-2 | 1.8 | |
| AT5G35630 | GLN2 | 1.6 | |
| AT1G52190 | NPF1.2 | 1.3 | |
| AT2G26690 | NPF6.2 | 2.8 | |
| AT1G12110 | NPF6.3 | 2.5 | |
| AT3G21670 | NPF6.4 | 5.9 | |
| AT2G15620 | NIR1 | 1.4 | |
| Sulfur uptake | AT1G62180 | APR2 | 2.9 |
| AT4G21990 | APR3 | 1.8 | |
| AT5G67520 | APK4 | 3.4 | |
| AT2G43750 | OASB | 1.6 | |
| AT5G28030 | DES1 | 1.9 | |
| AT5G28020 | CYSD2 | 3.2 | |
| AT2G25680 | SULTR2 | 2.5 | |
| AT3G51895 | SULTR3 | 1.7 | |
| AT1G61740 | TAUE2 | 1.0 | |
| AT2G36630 | TAUE2 | 2.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méndez, T.; Fuentes, A.; Cofre, D.; Moenne, A.; Laporte, D. Oligo-Carrageenan Kappa Increases Expression of Genes Encoding Proteins Involved in Photosynthesis, C, N, and S Assimilation, and Growth in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 11894. https://doi.org/10.3390/ijms241511894
Méndez T, Fuentes A, Cofre D, Moenne A, Laporte D. Oligo-Carrageenan Kappa Increases Expression of Genes Encoding Proteins Involved in Photosynthesis, C, N, and S Assimilation, and Growth in Arabidopsis thaliana. International Journal of Molecular Sciences. 2023; 24(15):11894. https://doi.org/10.3390/ijms241511894
Chicago/Turabian StyleMéndez, Tamara, Alejandra Fuentes, Diego Cofre, Alejandra Moenne, and Daniel Laporte. 2023. "Oligo-Carrageenan Kappa Increases Expression of Genes Encoding Proteins Involved in Photosynthesis, C, N, and S Assimilation, and Growth in Arabidopsis thaliana" International Journal of Molecular Sciences 24, no. 15: 11894. https://doi.org/10.3390/ijms241511894





