Dysbindin Domain-Containing 1 in Prostate Cancer: New Insights into Bioinformatic Validation of Molecular and Immunological Features
Abstract
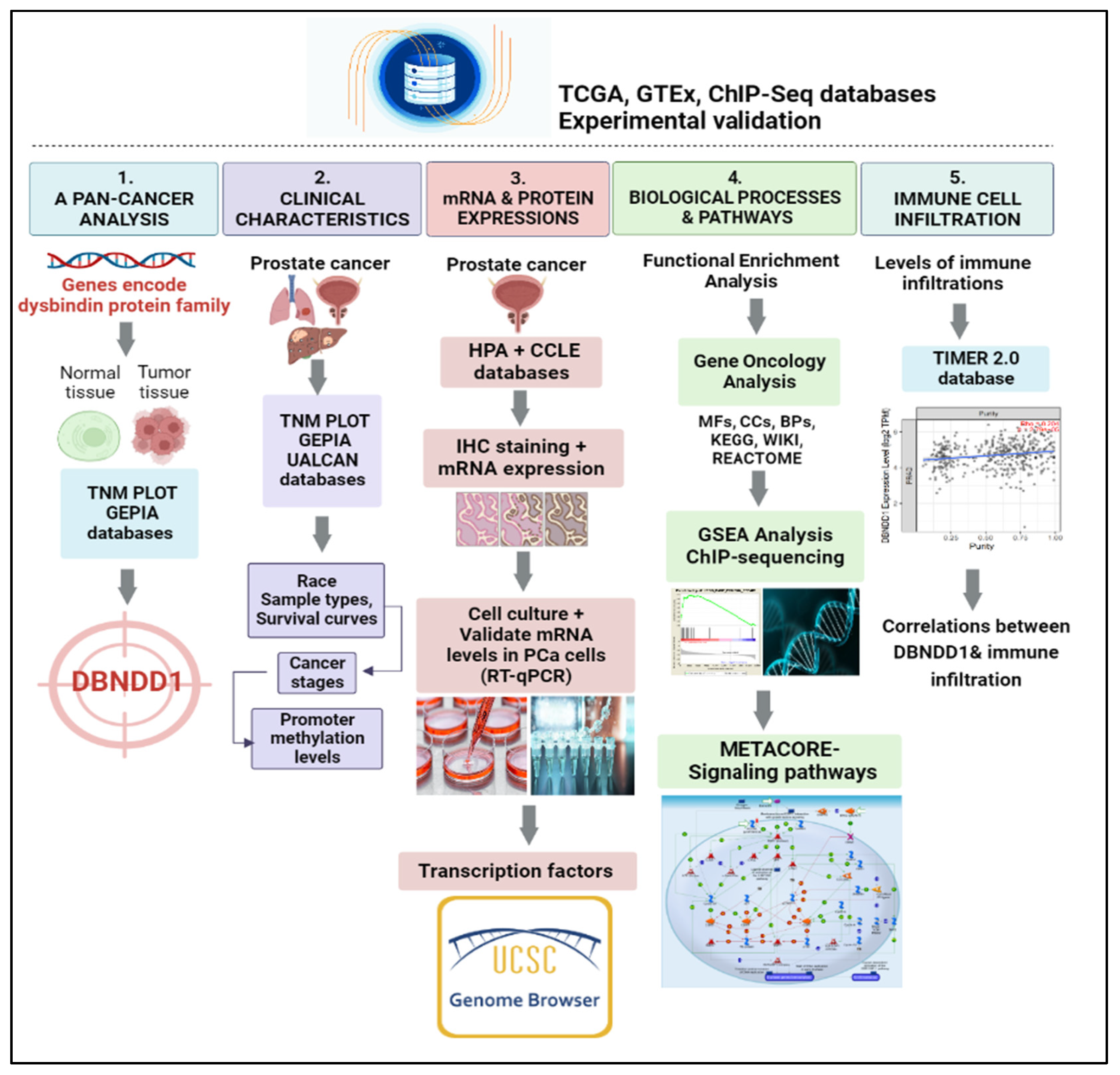
1. Introduction
2. Results
2.1. Expressions of Genes Encoding Dysbindin Protein Family Members in a Pan-Cancer Analysis
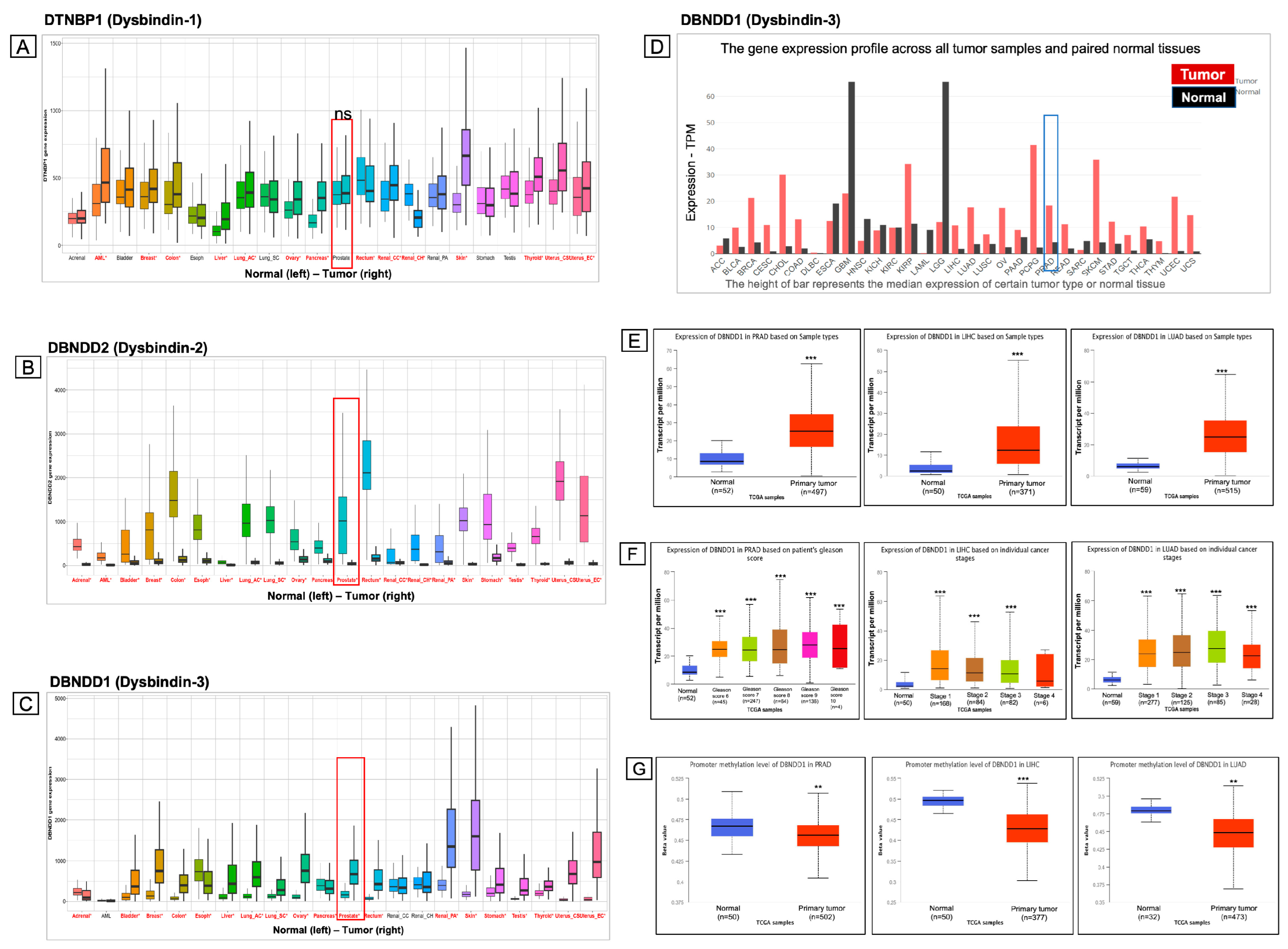
2.2. Survival Analysis of DBNDD1 Gene Expression
2.3. Differential Expression Analysis Identifies Upregulation of the DBNDD1 Gene in PCa
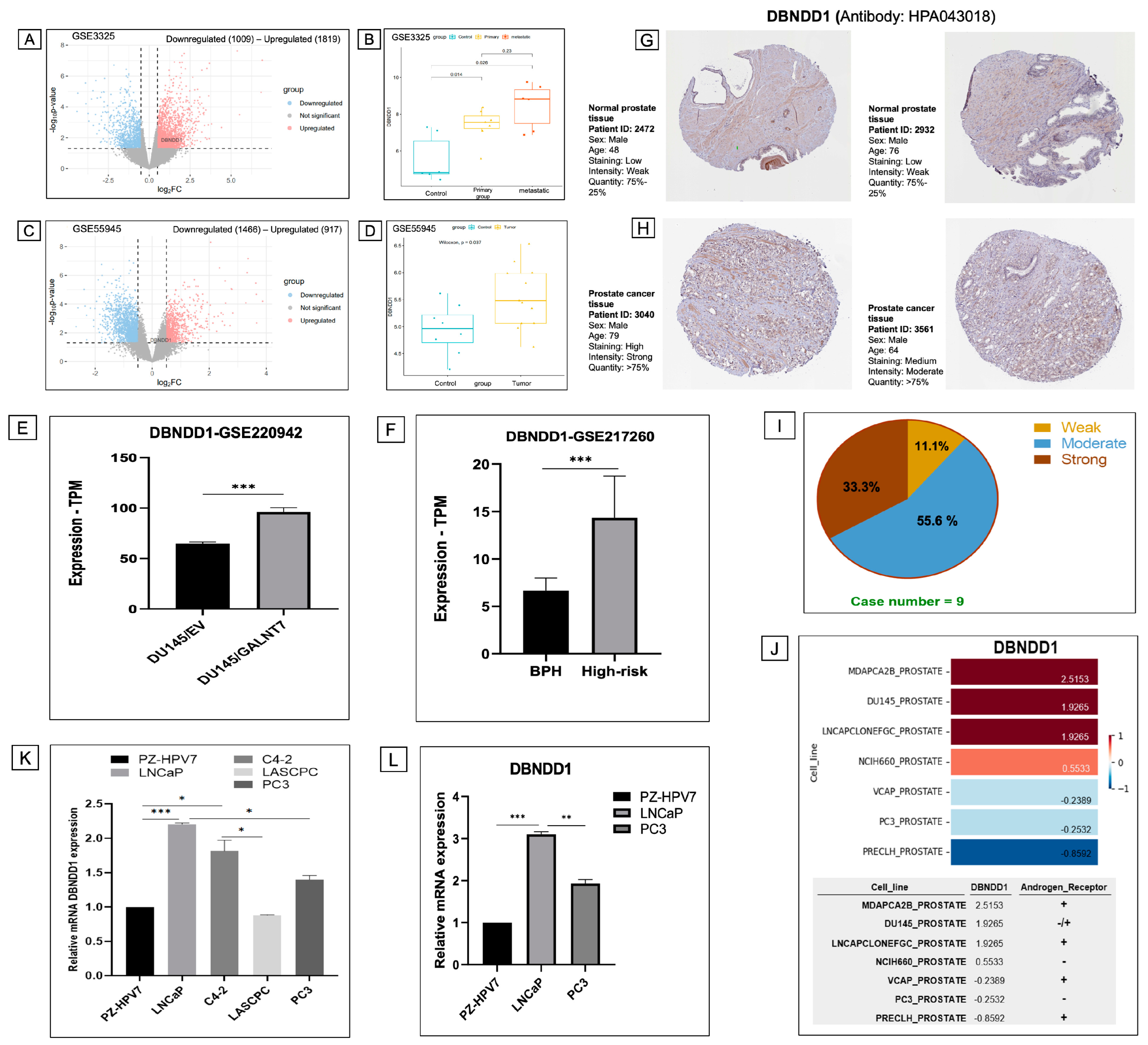
2.4. Upregulated DBNDD1 Expression Was Strongly Correlated with Cell Cycle Process

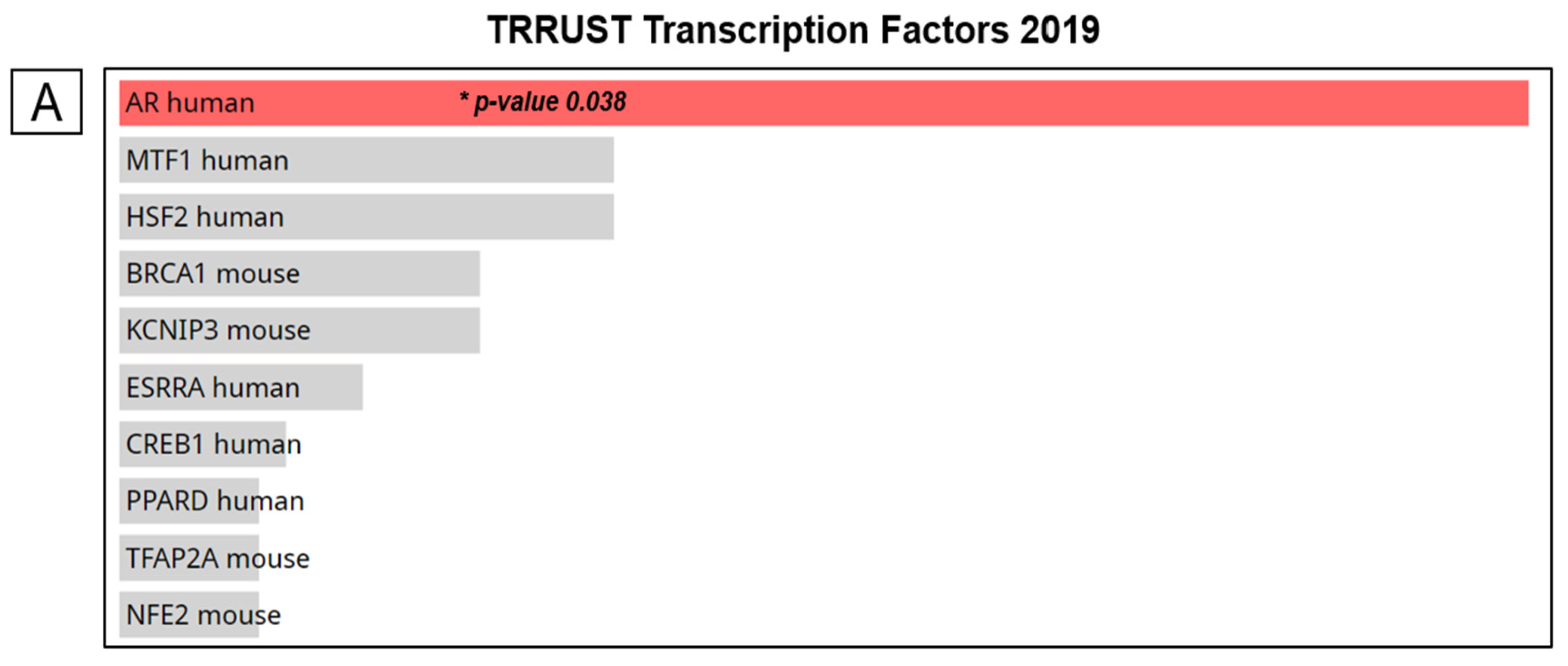
2.5. DBNDD1 Gene Expression and Transcription Factor-Binding Sites

2.6. High Expression of DBNDD1 Is Associated with E2F Transcription Factor Targets and Mitotic Spindle Checkpoint Signaling in PCa

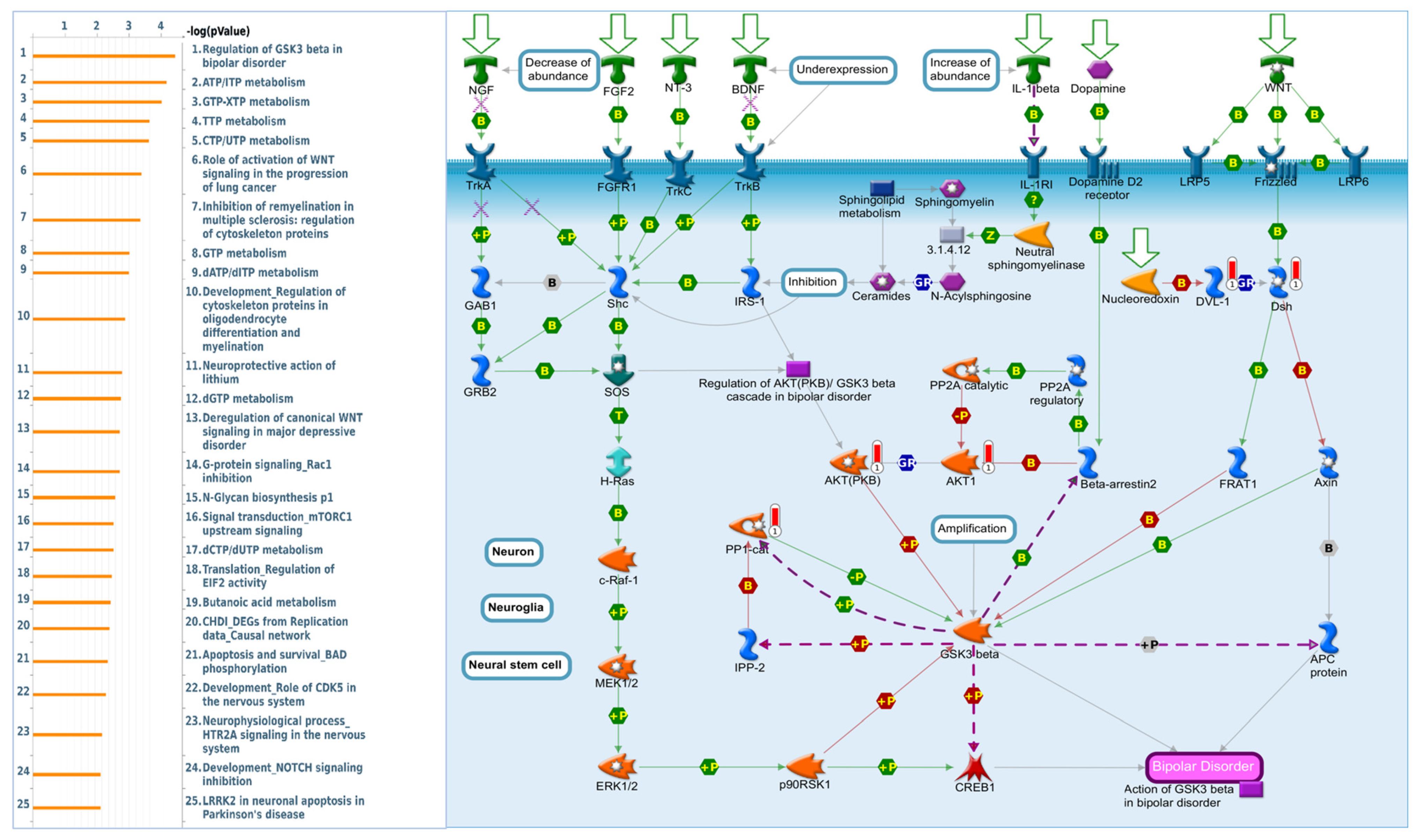
2.7. DBNDD1 Exhibits an Essential Role in the Regulation of Glycogen Synthase Kinase (GSK)-3β in Bipolar Disorder
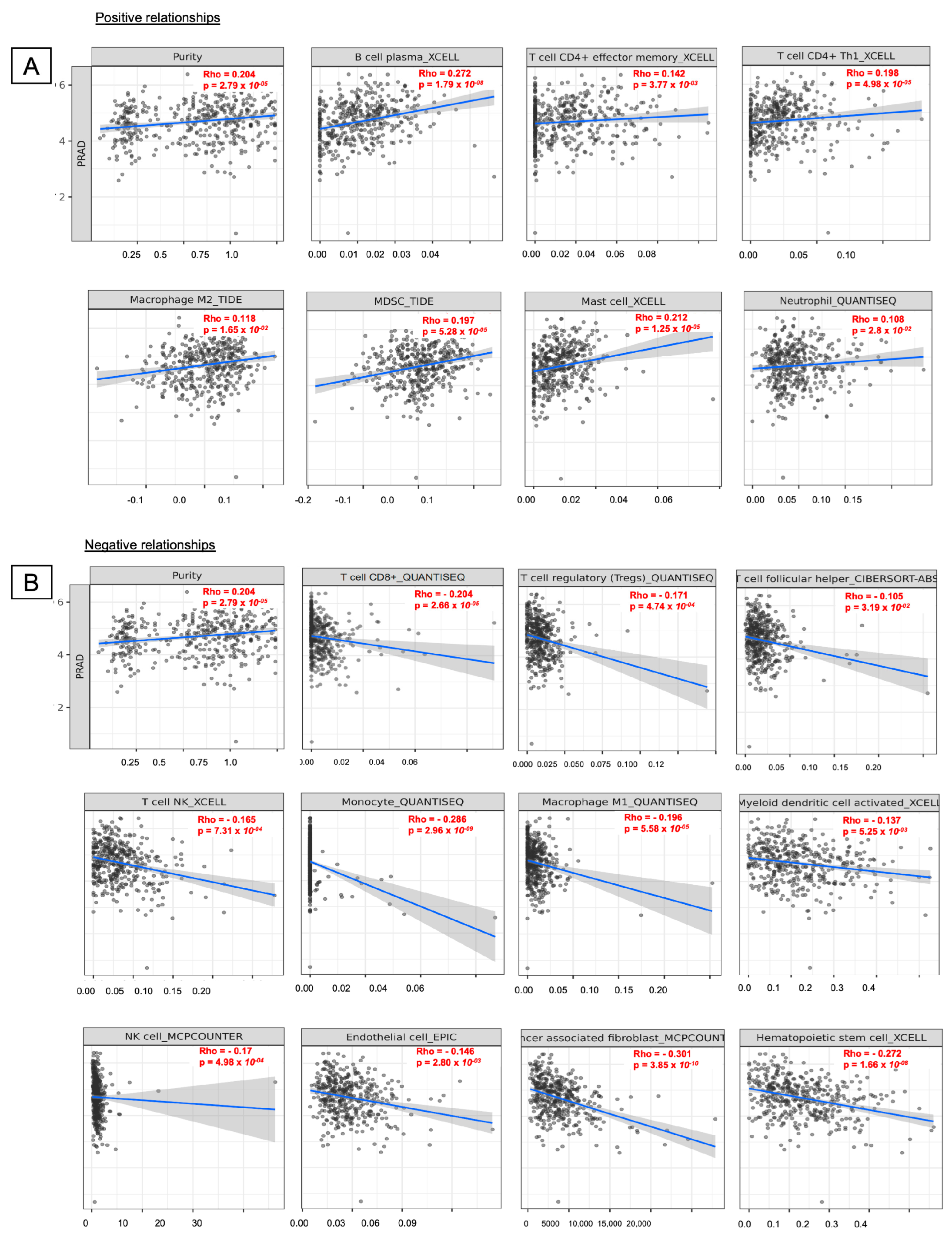
2.8. Levels of Immune Infiltration Were Associated with DBNDD1 Expression in PCa
3. Discussion
4. Materials and Methods
4.1. Differential Gene Expression Analysis
4.2. Enrichment Analysis
4.3. Identification of Correlations between Gene Expressions and Immune Cell Infiltration
4.4. Survival Curve Analysis
4.5. UALCAN
4.6. The TNM Plot
4.7. Database of Human Transcription Factor (TF) Targets
4.8. RNA-Seq Database in Gene Expression Omnibus (GEO)
4.9. Cell Culture
4.10. Reverse-Transcription (RT)-Quantitative Polymerase Chain Reaction (qPCR)
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Wang, Y.; Pei, L.; Tan, X.; Dong, C. Identification of Prostate Cancer Risk Genetics Biomarkers Based on Intergraded Bioinformatics Analysis. Front. Surg. 2022, 9, 856446. [Google Scholar] [CrossRef] [PubMed]
- Sekhoacha, M.; Riet, K.; Motloung, P.; Gumenku, L.; Adegoke, A.; Mashele, S. Prostate Cancer Review: Genetics, Diagnosis, Treatment Options, and Alternative Approaches. Molecules 2022, 27, 5730. [Google Scholar] [CrossRef]
- Marima, R.; Hull, R.; Mathabe, K.; Setlai, B.; Batra, J.; Sartor, O.; Mehrotra, R.; Dlamini, Z. Prostate cancer racial, socioeconomic, geographic disparities: Targeting the genomic landscape and splicing events in search for diagnostic, prognostic and therapeutic targets. Am. J. Cancer Res. 2021, 11, 1012–1030. [Google Scholar] [PubMed]
- Valcarcel-Jimenez, L.; Macchia, A.; Martín-Martín, N.; Cortazar, A.R.; Schaub-Clerigué, A.; Pujana-Vaquerizo, M.; Fernández-Ruiz, S.; Lacasa-Viscasillas, I.; Santos-Martin, A.; Loizaga-Iriarte, A.; et al. Integrative analysis of transcriptomics and clinical data uncovers the tumor-suppressive activity of MITF in prostate cancer. Cell Death Dis. 2018, 9, 1041. [Google Scholar] [CrossRef]
- Alkhateeb, A.; Rezaeian, I.; Singireddy, S.; Cavallo-Medved, D.; Porter, L.A.; Rueda, L. Transcriptomics Signature from Next-Generation Sequencing Data Reveals New Transcriptomic Biomarkers Related to Prostate Cancer. Cancer Inf. 2019, 18, 1176935119835522. [Google Scholar] [CrossRef]
- Hamzeh, O.; Alkhateeb, A.; Zheng, J.Z.; Kandalam, S.; Leung, C.; Atikukke, G.; Cavallo-Medved, D.; Palanisamy, N.; Rueda, L. A Hierarchical Machine Learning Model to Discover Gleason Grade-Specific Biomarkers in Prostate Cancer. Diagnostics 2019, 9, 219. [Google Scholar] [CrossRef]
- Zhu, D.; Zheng, S.; Fang, C.; Guo, X.; Han, D.; Tang, M.; Fu, H.; Jiang, M.; Xie, N.; Nie, Y.; et al. Dysbindin promotes pancreatic ductal adenocarcinoma metastasis by activating NF-κB/MDM2 via miR-342-3p. Cancer Lett. 2020, 477, 107–121. [Google Scholar] [CrossRef]
- Talbot, K.; Ong, W.Y.; Blake, D.J.; Tang, J.; Louneva, N.; Carlson, G.C.; Arnold, S.E. Dysbindin-1 and Its Protein Family. In Handbook of Neurochemistry and Molecular Neurobiology: Schizophrenia; Lajtha, A., Javitt, D., Kantrowitz, J., Eds.; Springer: Boston, MA, USA, 2009; pp. 107–241. [Google Scholar] [CrossRef]
- Wang, H.; Xu, J.; Lazarovici, P.; Zheng, W. Dysbindin-1 Involvement in the Etiology of Schizophrenia. Int. J. Mol. Sci. 2017, 18, 2044. [Google Scholar] [CrossRef]
- Ito, H.; Morishita, R.; Shinoda, T.; Iwamoto, I.; Sudo, K.; Okamoto, K.; Nagata, K. Dysbindin-1, a schizophrenia-related molecule, is involved in the regulation of neuronal dendritic development. Mol. Psychiatry 2010, 15, 969. [Google Scholar] [CrossRef]
- Mutsuddi, M.; Morris, D.W.; Waggoner, S.G.; Daly, M.J.; Scolnick, E.M.; Sklar, P. Analysis of high-resolution HapMap of DTNBP1 (Dysbindin) suggests no consistency between reported common variant associations and schizophrenia. Am. J. Hum. Genet. 2006, 79, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.W.; McGhee, K.A.; Schwaiger, S.; Scully, P.; Quinn, J.; Meagher, D.; Waddington, J.L.; Gill, M.; Corvin, A.P. No evidence for association of the dysbindin gene [DTNBP1] with schizophrenia in an Irish population-based study. Schizophr. Res. 2003, 60, 167–172. [Google Scholar] [CrossRef]
- Williams, N.M.; O’Donovan, M.C.; Owen, M.J. Is the dysbindin gene (DTNBP1) a susceptibility gene for schizophrenia? Schizophr. Bull. 2005, 31, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Guo, X.; Lv, X.; Yin, R.; Lv, X.; Wang, F.; Zhao, J.; Bai, Q.; Yao, X.; Chen, Y. Dysbindin promotes progression of pancreatic ductal adenocarcinoma via direct activation of PI3K. J. Mol. Cell Biol. 2017, 9, 504–515. [Google Scholar] [CrossRef]
- Cheng, X.; Li, D.; Qi, T.; Sun, J.; Zhou, T.; Zheng, W.V. Objective to identify and verify the regulatory mechanism of DTNBP1 as a prognostic marker for hepatocellular carcinoma. Sci. Rep. 2022, 12, 211. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Guo, X.; Ru, Y.; Zhou, F.; Yang, X.; Ge, J.; Li, J.; Liu, S.; Jiang, K.; Chen, B. Dysbindin facilitates invasion and metastasis by promoting phosphorylation of ERK in epithelial ovarian cancer. J. Cancer 2020, 11, 2821–2829. [Google Scholar] [CrossRef]
- Ta, H.D.K.; Wang, W.J.; Phan, N.N.; An Ton, N.T.; Anuraga, G.; Ku, S.C.; Wu, Y.F.; Wang, C.Y.; Lee, K.H. Potential Therapeutic and Prognostic Values of LSM Family Genes in Breast Cancer. Cancers 2021, 13, 4902. [Google Scholar] [CrossRef]
- Chipperfield, K.; Fletcher, J.; Millar, J.; Brooker, J.; Smith, R.; Frydenberg, M.; Burney, S. Predictors of depression, anxiety and quality of life in patients with prostate cancer receiving androgen deprivation therapy. Psychooncology 2013, 22, 2169–2176. [Google Scholar] [CrossRef]
- Galvão, D.A.; Taaffe, D.R.; Spry, N.; Joseph, D.; Newton, R.U. Cardiovascular and metabolic complications during androgen deprivation: Exercise as a potential countermeasure. Prostate Cancer Prostatic Dis. 2009, 12, 233–240. [Google Scholar] [CrossRef]
- Terrisse, S.; Goubet, A.G.; Ueda, K.; Thomas, A.M.; Quiniou, V.; Thelemaque, C.; Dunsmore, G.; Clave, E.; Gamat-Huber, M.; Yonekura, S.; et al. Immune system and intestinal microbiota determine efficacy of androgen deprivation therapy against prostate cancer. J. Immunother. Cancer 2022, 10, e004191. [Google Scholar] [CrossRef]
- Wang, I.; Song, L.; Wang, B.Y.; Rezazadeh Kalebasty, A.; Uchio, E.; Zi, X. Prostate cancer immunotherapy: A review of recent advancements with novel treatment methods and efficacy. Am. J. Clin. Exp. Urol. 2022, 10, 210–233. [Google Scholar] [PubMed]
- Wang, C.; Zhang, Y.; Gao, W.-Q. The evolving role of immune cells in prostate cancer. Cancer Lett. 2022, 525, 9–21. [Google Scholar] [CrossRef]
- Srigley, J.R.; Delahunt, B.; Samaratunga, H.; Billis, A.; Cheng, L.; Clouston, D.; Evans, A.; Furusato, B.; Kench, J.; Leite, K.; et al. Controversial issues in Gleason and International Society of Urological Pathology (ISUP) prostate cancer grading: Proposed recommendations for international implementation. Pathology 2019, 51, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Moarii, M.; Boeva, V.; Vert, J.-P.; Reyal, F. Changes in correlation between promoter methylation and gene expression in cancer. BMC Genom. 2015, 16, 873. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, W.; Zhang, H.-M.; Xie, G.-Y.; Miao, Y.-R.; Xia, M.; Guo, A.-Y. hTFtarget: A Comprehensive Database for Regulations of Human Transcription Factors and Their Targets. Genom. Proteom. Bioinform. 2020, 18, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Ta, H.D.K.; Minh Xuan, D.T.; Tang, W.C.; Anuraga, G.; Ni, Y.C.; Pan, S.R.; Wu, Y.F.; Fitriani, F.; Putri Hermanto, E.M.; Athoillah, M.; et al. Novel Insights into the Prognosis and Immunological Value of the SLC35A (Solute Carrier 35A) Family Genes in Human Breast Cancer. Biomedicines 2021, 9, 1804. [Google Scholar] [CrossRef]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Nagahashi, M.; Matsuyama, R.; Endo, I.; Takabe, K. The E2F Pathway Score as a Predictive Biomarker of Response to Neoadjuvant Therapy in ER+/HER2- Breast Cancer. Cells 2020, 9, 1643. [Google Scholar] [CrossRef]
- Liang, Y.X.; Lu, J.M.; Mo, R.J.; He, H.C.; Xie, J.; Jiang, F.N.; Lin, Z.Y.; Chen, Y.R.; Wu, Y.D.; Luo, H.W.; et al. E2F1 promotes tumor cell invasion and migration through regulating CD147 in prostate cancer. Int. J. Oncol. 2016, 48, 1650–1658. [Google Scholar] [CrossRef]
- Libertini, S.J.; Chen, H.; al-Bataina, B.; Koilvaram, T.; George, M.; Gao, A.C.; Mudryj, M. The interleukin 6 receptor is a direct transcriptional target of E2F3 in prostate tumor derived cells. Prostate 2012, 72, 649–660. [Google Scholar] [CrossRef]
- Majumder, S.; Bhowal, A.; Basu, S.; Mukherjee, P.; Chatterji, U.; Sengupta, S. Deregulated E2F5/p38/SMAD3 Circuitry Reinforces the Pro-Tumorigenic Switch of TGFβ Signaling in Prostate Cancer. J. Cell. Physiol. 2016, 231, 2482–2492. [Google Scholar] [CrossRef]
- Wang, Y.; Pei, X.; Xu, P.; Tan, Z.; Zhu, Z.; Zhang, G.; Jiang, Z.; Deng, Z. E2F7, regulated by miR-30c, inhibits apoptosis and promotes cell cycle of prostate cancer cells. Oncol. Rep. 2020, 44, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Frame, S. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001, 2, 769–776. [Google Scholar] [CrossRef]
- Tan, R.; Nie, M.; Long, W. The role of B cells in cancer development. Front. Oncol. 2022, 12, 958756. [Google Scholar] [CrossRef] [PubMed]
- De Visser, K.E.; Korets, L.V.; Coussens, L.M. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell 2005, 7, 411–423. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Ravetch, J.V. Fc-receptors as regulators of immunity. Adv. Immunol. 2007, 96, 179–204. [Google Scholar] [CrossRef]
- Bonizzi, G.; Bebien, M.; Otero, D.C.; Johnson-Vroom, K.E.; Cao, Y.; Vu, D.; Jegga, A.G.; Aronow, B.J.; Ghosh, G.; Rickert, R.C.; et al. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 2004, 23, 4202–4210. [Google Scholar] [CrossRef]
- Ammirante, M.; Luo, J.L.; Grivennikov, S.; Nedospasov, S.; Karin, M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature 2010, 464, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, H.; Jamiyan, T.; Yamaguchi, R.; Kakumoto, A.; Abe, A.; Harada, O.; Masunaga, A. Tumor-infiltrating B cells and T cells correlate with postoperative prognosis in triple-negative carcinoma of the breast. BMC Cancer 2021, 21, 286. [Google Scholar] [CrossRef]
- Anuraga, G.; Wang, W.J.; Phan, N.N.; An Ton, N.T.; Ta, H.D.K.; Berenice Prayugo, F.; Minh Xuan, D.T.; Ku, S.C.; Wu, Y.F.; Andriani, V.; et al. Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer. J. Pers. Med. 2021, 11, 1089. [Google Scholar] [CrossRef]
- Bai, B.; Chen, Q.; Jing, R.; He, X.; Wang, H.; Ban, Y.; Ye, Q.; Xu, W.; Zheng, C. Molecular Basis of Prostate Cancer and Natural Products as Potential Chemotherapeutic and Chemopreventive Agents. Front. Pharmacol. 2021, 12, 738235. [Google Scholar] [CrossRef]
- Bouras, E.; Karakioulaki, M.; Bougioukas, K.I.; Aivaliotis, M.; Tzimagiorgis, G.; Chourdakis, M. Gene promoter methylation and cancer: An umbrella review. Gene 2019, 710, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.J. Treatment effects in prostate cancer. Mod. Pathol. 2018, 31, S110–S121. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Weinert, T.A. Checkpoints: Controls that ensure the order of cell cycle events. Science 1989, 246, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. p53, the Cellular Gatekeeper for Growth and Division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Jallepalli, P.V.; Lengauer, C. Chromosome segregation and cancer: Cutting through the mystery. Nat. Rev. Cancer 2001, 1, 109–117. [Google Scholar] [CrossRef]
- Ong, M.S.; Deng, S.; Halim, C.E.; Cai, W.; Tan, T.Z.; Huang, R.Y.; Sethi, G.; Hooi, S.C.; Kumar, A.P.; Yap, C.T. Cytoskeletal Proteins in Cancer and Intracellular Stress: A Therapeutic Perspective. Cancers 2020, 12, 238. [Google Scholar] [CrossRef]
- Xu, J.; Yang, X.; Deshmukh, D.; Chen, H.; Fang, S.; Qiu, Y. The Role of Crosstalk between AR3 and E2F1 in Drug Resistance in Prostate Cancer Cells. Cells 2020, 9, 1094. [Google Scholar] [CrossRef]
- Ramos-Montoya, A.; Lamb, A.D.; Russell, R.; Carroll, T.; Jurmeister, S.; Galeano-Dalmau, N.; Massie, C.E.; Boren, J.; Bon, H.; Theodorou, V.; et al. HES6 drives a critical AR transcriptional programme to induce castration-resistant prostate cancer through activation of an E2F1-mediated cell cycle network. EMBO Mol. Med. 2014, 6, 651–661. [Google Scholar] [CrossRef]
- Mandigo, A.C.; Shafi, A.A.; McCann, J.J.; Yuan, W.; Laufer, T.S.; Bogdan, D.; Gallagher, L.; Dylgjeri, E.; Semenova, G.; Vasilevskaya, I.A.; et al. Novel Oncogenic Transcription Factor Cooperation in RB-Deficient Cancer. Cancer Res. 2022, 82, 221–234. [Google Scholar] [CrossRef]
- Qiu, K.; Zheng, Z.; Huang, Y. Long intergenic noncoding RNA 00844 promotes apoptosis and represses proliferation of prostate cancer cells through upregulating GSTP1 by recruiting EBF1. J. Cell. Physiol. 2020, 235, 8472–8485. [Google Scholar] [CrossRef]
- Kumar, A.; Coleman, I.; Morrissey, C.; Zhang, X.; True, L.D.; Gulati, R.; Etzioni, R.; Bolouri, H.; Montgomery, B.; White, T.; et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med. 2016, 22, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.; Zoubeidi, A.; Selth, L.A. The epigenetic and transcriptional landscape of neuroendocrine prostate cancer. Endocr. Relat. Cancer 2020, 27, R35–R50. [Google Scholar] [CrossRef] [PubMed]
- Barakat, D.J.; Mendonca, J.; Barberi, T.; Zhang, J.; Kachhap, S.K.; Paz-Priel, I.; Friedman, A.D. C/EBPβ regulates sensitivity to bortezomib in prostate cancer cells by inducing REDD1 and autophagosome-lysosome fusion. Cancer Lett. 2016, 375, 152–161. [Google Scholar] [CrossRef]
- Li, B.; Thrasher, J.B.; Terranova, P. Glycogen synthase kinase-3: A potential preventive target for prostate cancer management. Urol. Oncol. 2015, 33, 456–463. [Google Scholar] [CrossRef]
- Li, R.; Erdamar, S.; Dai, H.; Sayeeduddin, M.; Frolov, A.; Wheeler, T.M.; Ayala, G.E. Cytoplasmic accumulation of glycogen synthase kinase-3beta is associated with aggressive clinicopathological features in human prostate cancer. Anticancer Res. 2009, 29, 2077–2081. [Google Scholar] [PubMed]
- Wu, Z.; Chen, H.; Luo, W.; Zhang, H.; Li, G.; Zeng, F.; Deng, F. The Landscape of Immune Cells Infiltrating in Prostate Cancer. Front. Oncol. 2020, 10, 517637. [Google Scholar] [CrossRef] [PubMed]
- King, A. Could immunotherapy finally break through in prostate cancer? Nature 2022, 609, S42–S44. [Google Scholar] [CrossRef]
- Hagerling, C.; Gonzalez, H.; Salari, K.; Wang, C.Y.; Lin, C.; Robles, I.; van Gogh, M.; Dejmek, A.; Jirström, K.; Werb, Z. Immune effector monocyte-neutrophil cooperation induced by the primary tumor prevents metastatic progression of breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 21704–21714. [Google Scholar] [CrossRef]
- Wang, C.Y.; Chang, Y.C.; Kuo, Y.L.; Lee, K.T.; Chen, P.S.; Cheung, C.H.A.; Chang, C.P.; Phan, N.N.; Shen, M.R.; Hsu, H.P. Mutation of the PTCH1 gene predicts recurrence of breast cancer. Sci. Rep. 2019, 9, 16359. [Google Scholar] [CrossRef]
- Cendrowicz, E.; Sas, Z.; Bremer, E.; Rygiel, T.P. The Role of Macrophages in Cancer Development and Therapy. Cancers 2021, 13, 1946. [Google Scholar] [CrossRef]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef]
- Sargos, P.; Ferretti, L.; Gross-Goupil, M.; Orre, M.; Cornelis, F.; Henriques de Figueiredo, B.; Houédé, N.; Merino, C.; Roubaud, G.; Dallaudiére, B.; et al. Characterization of prostate neuroendocrine cancers and therapeutic management: A literature review. Prostate Cancer Prostatic Dis. 2014, 17, 220–226. [Google Scholar] [CrossRef]
- Saudi, A.; Banday, V.; Zirakzadeh, A.A.; Selinger, M.; Forsberg, J.; Holmbom, M.; Henriksson, J.; Waldén, M.; Alamdari, F.; Aljabery, F.; et al. Immune-Activated B Cells Are Dominant in Prostate Cancer. Cancers 2023, 15, 920. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Bujanda, Z.A.; Haffner, M.C.; Chaimowitz, M.G.; Chowdhury, N.; Venturini, N.J.; Patel, R.A.; Obradovic, A.; Hansen, C.S.; Jacków, J.; Maynard, J.P.; et al. Castration-mediated IL-8 promotes myeloid infiltration and prostate cancer progression. Nat. Cancer 2021, 2, 803–818. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Spataro, C.; Zagato, E.; Di Mitri, D.; Gil, V.; Crespo, M.; De Bernardis, G.; Losa, M.; Mirenda, M.; Pasquini, E.; et al. IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature 2018, 559, 363–369. [Google Scholar] [CrossRef]
- De Cicco, P.; Ercolano, G.; Ianaro, A. The New Era of Cancer Immunotherapy: Targeting Myeloid-Derived Suppressor Cells to Overcome Immune Evasion. Front. Immunol. 2020, 11, 1680. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Wu, Y.H.; Yeh, I.J.; Phan, N.N.; Yen, M.C.; Hung, J.H.; Chiao, C.C.; Chen, C.F.; Sun, Z.; Hsu, H.P.; Wang, C.Y.; et al. Gene signatures and potential therapeutic targets of Middle East respiratory syndrome coronavirus (MERS-CoV)-infected human lung adenocarcinoma epithelial cells. J. Microbiol. Immunol. Infect. 2021, 54, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Bailey, M.H.; Porta-Pardo, E.; Thorsson, V.; Colaprico, A.; Bertrand, D.; Gibbs, D.L.; Weerasinghe, A.; Huang, K.L.; Tokheim, C.; et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 2018, 173, 305–320.e310. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Chiao, C.C.; Phan, N.N.; Li, C.Y.; Sun, Z.D.; Jiang, J.Z.; Hung, J.H.; Chen, Y.L.; Yen, M.C.; Weng, T.Y.; et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am. J. Cancer Res. 2020, 10, 95–113. [Google Scholar] [PubMed]
- Kao, T.J.; Wu, C.C.; Phan, N.N.; Liu, Y.H.; Ta, H.D.K.; Anuraga, G.; Wu, Y.F.; Lee, K.H.; Chuang, J.Y.; Wang, C.Y. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging 2021, 13, 17970. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R.; et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef]
- Kelder, T.; van Iersel, M.P.; Hanspers, K.; Kutmon, M.; Conklin, B.R.; Evelo, C.T.; Pico, A.R. WikiPathways: Building research communities on biological pathways. Nucleic Acids Res. 2011, 40, D1301–D1307. [Google Scholar] [CrossRef]
- Xuan, D.T.M.; Wu, C.C.; Kao, T.J.; Ta, H.D.K.; Anuraga, G.; Andriani, V.; Athoillah, M.; Chiao, C.C.; Wu, Y.F.; Lee, K.H.; et al. Prognostic and immune infiltration signatures of proteasome 26S subunit, non-ATPase (PSMD) family genes in breast cancer patients. Aging 2021, 13, 24882–24913. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Yeh, I.J.; Phan, N.N.; Wu, Y.H.; Yen, M.C.; Hung, J.H.; Chiao, C.C.; Chen, C.F.; Sun, Z.; Jiang, J.Z.; et al. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect. Genet. Evol. 2020, 85, 104438. [Google Scholar] [CrossRef]
- Xuan, D.T.M.; Yeh, I.J.; Wu, C.C.; Su, C.Y.; Liu, H.L.; Chiao, C.C.; Ku, S.C.; Jiang, J.Z.; Sun, Z.; Ta, H.D.K.; et al. Comparison of Transcriptomic Signatures between Monkeypox-Infected Monkey and Human Cell Lines. J. Immunol. Res. 2022, 2022, 3883822. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Ku, S.C.; Liu, H.L.; Su, C.Y.; Yeh, I.J.; Yen, M.C.; Anuraga, G.; Ta, H.D.K.; Chiao, C.C.; Xuan, D.T.M.; Prayugo, F.B.; et al. Comprehensive analysis of prognostic significance of cadherin (CDH) gene family in breast cancer. Aging 2022, 14, 8498–8567. [Google Scholar] [CrossRef]
- Li, C.Y.; Anuraga, G.; Chang, C.P.; Weng, T.Y.; Hsu, H.P.; Ta, H.D.K.; Su, P.F.; Chiu, P.H.; Yang, S.J.; Chen, F.W.; et al. Repurposing nitric oxide donating drugs in cancer therapy through immune modulation. J. Exp. Clin. Cancer Res. 2023, 42, 22. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef] [PubMed]
- Karolchik, D.; Bejerano, G.; Hinrichs, A.S.; Kuhn, R.M.; Miller, W.; Rosenbloom, K.R.; Zweig, A.S.; Haussler, D.; Kent, W.J. Comparative genomic analysis using the UCSC genome browser. Methods Mol. Biol. 2007, 395, 17–34. [Google Scholar] [CrossRef]
- Scott, E.; Hodgson, K.; Calle, B.; Turner, H.; Cheung, K.; Bermudez, A.; Marques, F.J.G.; Pye, H.; Yo, E.C.; Islam, K.; et al. Upregulation of GALNT7 in prostate cancer modifies O-glycosylation and promotes tumour growth. Oncogene 2023, 42, 926–937. [Google Scholar] [CrossRef]
- Wei, Z.; Wang, S.; Xu, Y.; Wang, W.; Soares, F.; Ahmed, M.; Su, P.; Wang, T.; Orouji, E.; Xu, X.; et al. MYC reshapes CTCF-mediated chromatin architecture in prostate cancer. Nat. Commun. 2023, 14, 1787. [Google Scholar] [CrossRef] [PubMed]
- Sang, Q.; Fan, L.; Liu, T.; Qiu, Y.; Du, J.; Mo, B.; Chen, M.; Chen, X. MicroRNA156 conditions auxin sensitivity to enable growth plasticity in response to environmental changes in Arabidopsis. Nat. Commun. 2023, 14, 1449. [Google Scholar] [CrossRef]
- Hunt, G.P.; Grassi, L.; Henkin, R.; Smeraldi, F.; Spargo, T.P.; Kabiljo, R.; Koks, S.; Ibrahim, Z.; Dobson, R.J.B.; Al-Chalabi, A.; et al. GEOexplorer: A webserver for gene expression analysis and visualisation. Nucleic Acids Res. 2022, 50, W367–W374. [Google Scholar] [CrossRef]
- McNair, C.; Xu, K.; Mandigo, A.C.; Benelli, M.; Leiby, B.; Rodrigues, D.; Lindberg, J.; Gronberg, H.; Crespo, M.; De Laere, B.; et al. Differential impact of RB status on E2F1 reprogramming in human cancer. J. Clin. Investig. 2018, 128, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Yu, J.; Laxman, B.; Rhodes, D.R.; Mehra, R.; Tomlins, S.A.; Shah, R.B.; Chandran, U.; Monzon, F.A.; Becich, M.J.; et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005, 8, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Arredouani, M.S.; Lu, B.; Bhasin, M.; Eljanne, M.; Yue, W.; Mosquera, J.M.; Bubley, G.J.; Li, V.; Rubin, M.A.; Libermann, T.A.; et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin. Cancer Res. 2009, 15, 5794–5802. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Xie, R.; Li, B.; Jia, L.; Li, Y. Identification of Core Genes and Pathways in Melanoma Metastasis via Bioinformatics Analysis. Int. J. Mol. Sci. 2022, 23, 794. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tram, V.T.N.; Khoa Ta, H.D.; Anuraga, G.; Dung, P.V.T.; Xuan, D.T.M.; Dey, S.; Wang, C.-Y.; Liu, Y.-N. Dysbindin Domain-Containing 1 in Prostate Cancer: New Insights into Bioinformatic Validation of Molecular and Immunological Features. Int. J. Mol. Sci. 2023, 24, 11930. https://doi.org/10.3390/ijms241511930
Tram VTN, Khoa Ta HD, Anuraga G, Dung PVT, Xuan DTM, Dey S, Wang C-Y, Liu Y-N. Dysbindin Domain-Containing 1 in Prostate Cancer: New Insights into Bioinformatic Validation of Molecular and Immunological Features. International Journal of Molecular Sciences. 2023; 24(15):11930. https://doi.org/10.3390/ijms241511930
Chicago/Turabian StyleTram, Van Thi Ngoc, Hoang Dang Khoa Ta, Gangga Anuraga, Phan Vu Thuy Dung, Do Thi Minh Xuan, Sanskriti Dey, Chih-Yang Wang, and Yen-Nien Liu. 2023. "Dysbindin Domain-Containing 1 in Prostate Cancer: New Insights into Bioinformatic Validation of Molecular and Immunological Features" International Journal of Molecular Sciences 24, no. 15: 11930. https://doi.org/10.3390/ijms241511930
APA StyleTram, V. T. N., Khoa Ta, H. D., Anuraga, G., Dung, P. V. T., Xuan, D. T. M., Dey, S., Wang, C.-Y., & Liu, Y.-N. (2023). Dysbindin Domain-Containing 1 in Prostate Cancer: New Insights into Bioinformatic Validation of Molecular and Immunological Features. International Journal of Molecular Sciences, 24(15), 11930. https://doi.org/10.3390/ijms241511930








