Investigation of Mutated in Colorectal Cancer (MCC) Gene Family Evolution History Indicates a Putative Role in Th17/Treg Differentiation
Abstract
1. Introduction
2. Results
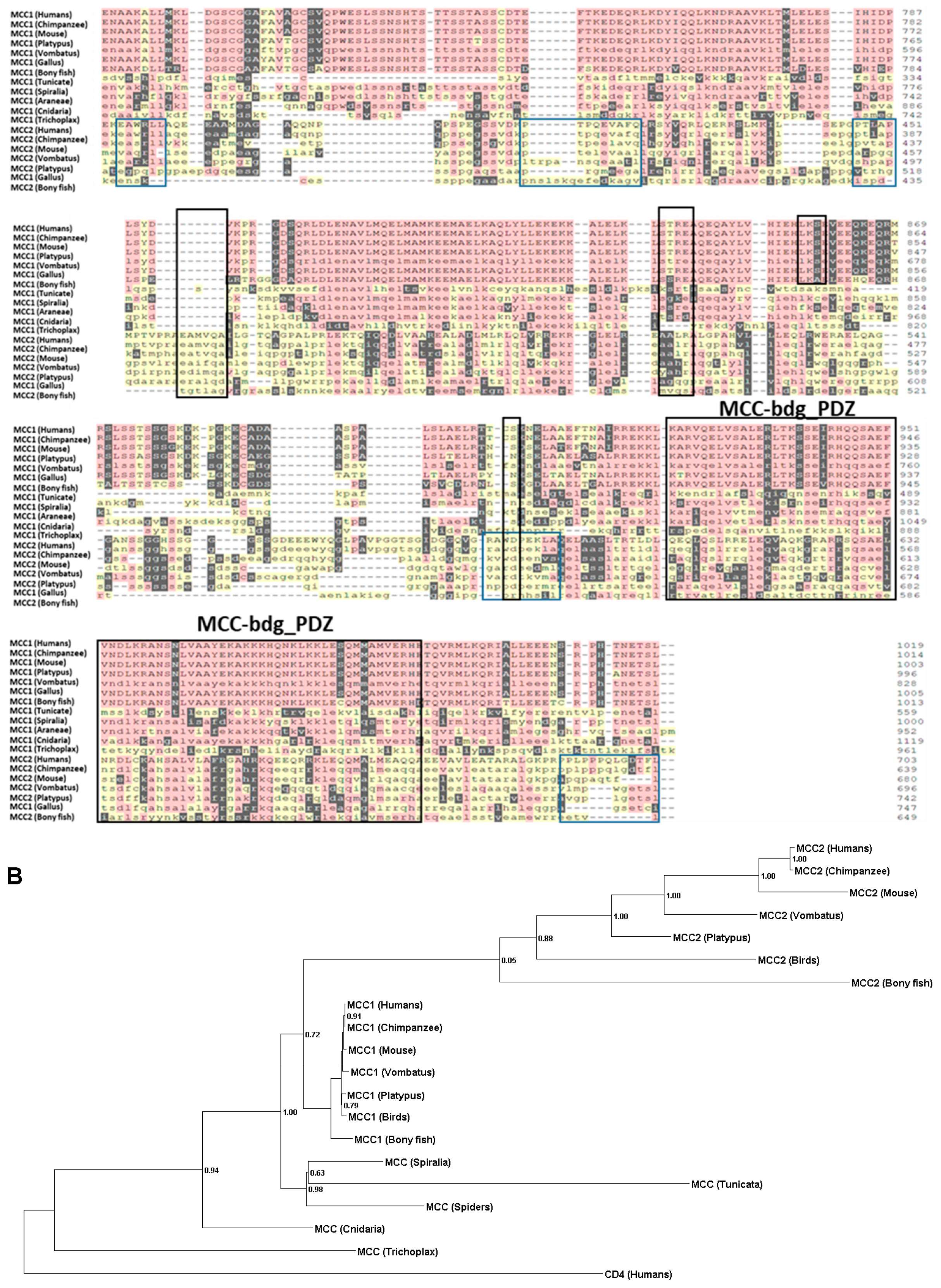
2.1. Phylogenetic Analysis
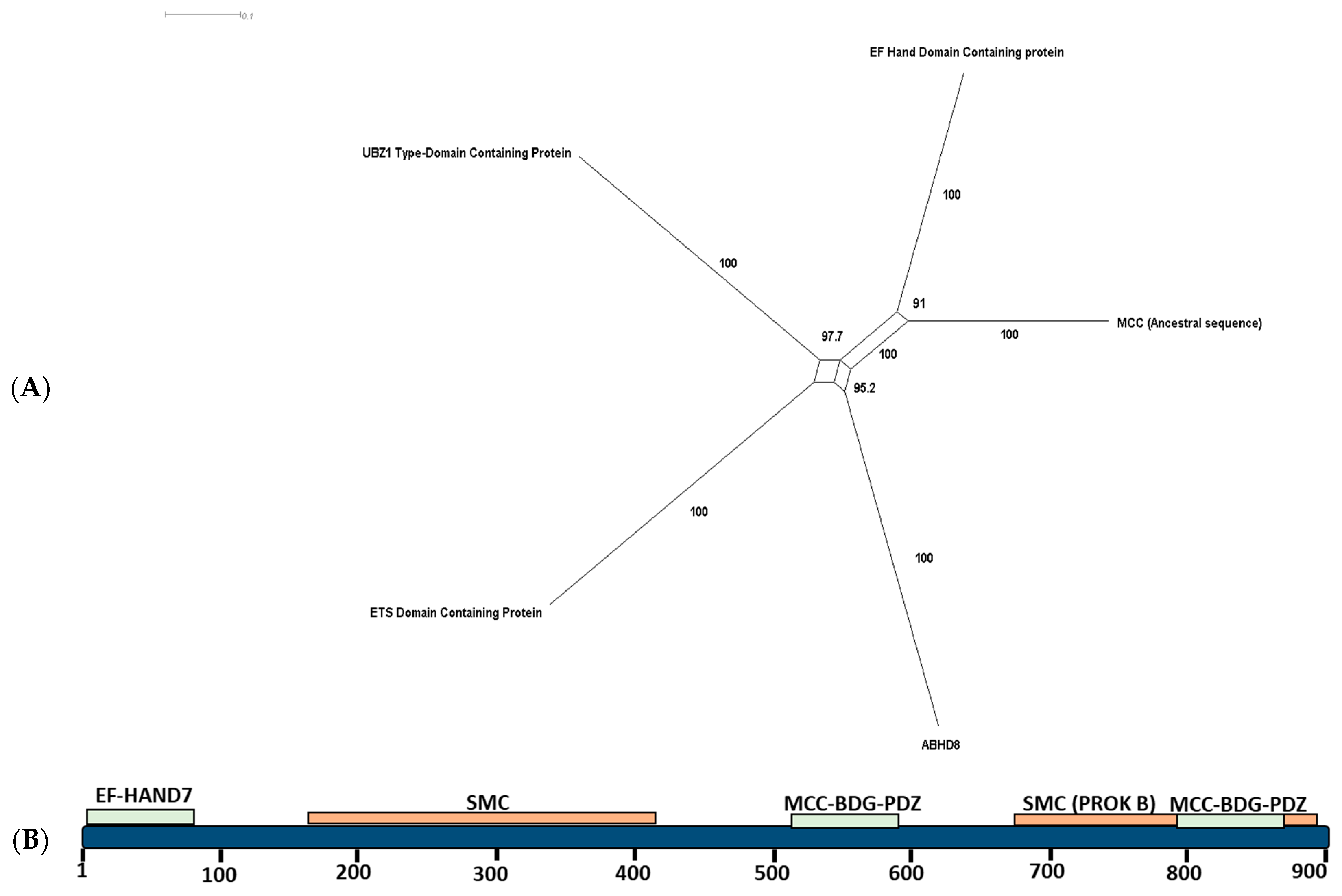
2.2. Ancestral Sequence Reconstruction and Network Split Results
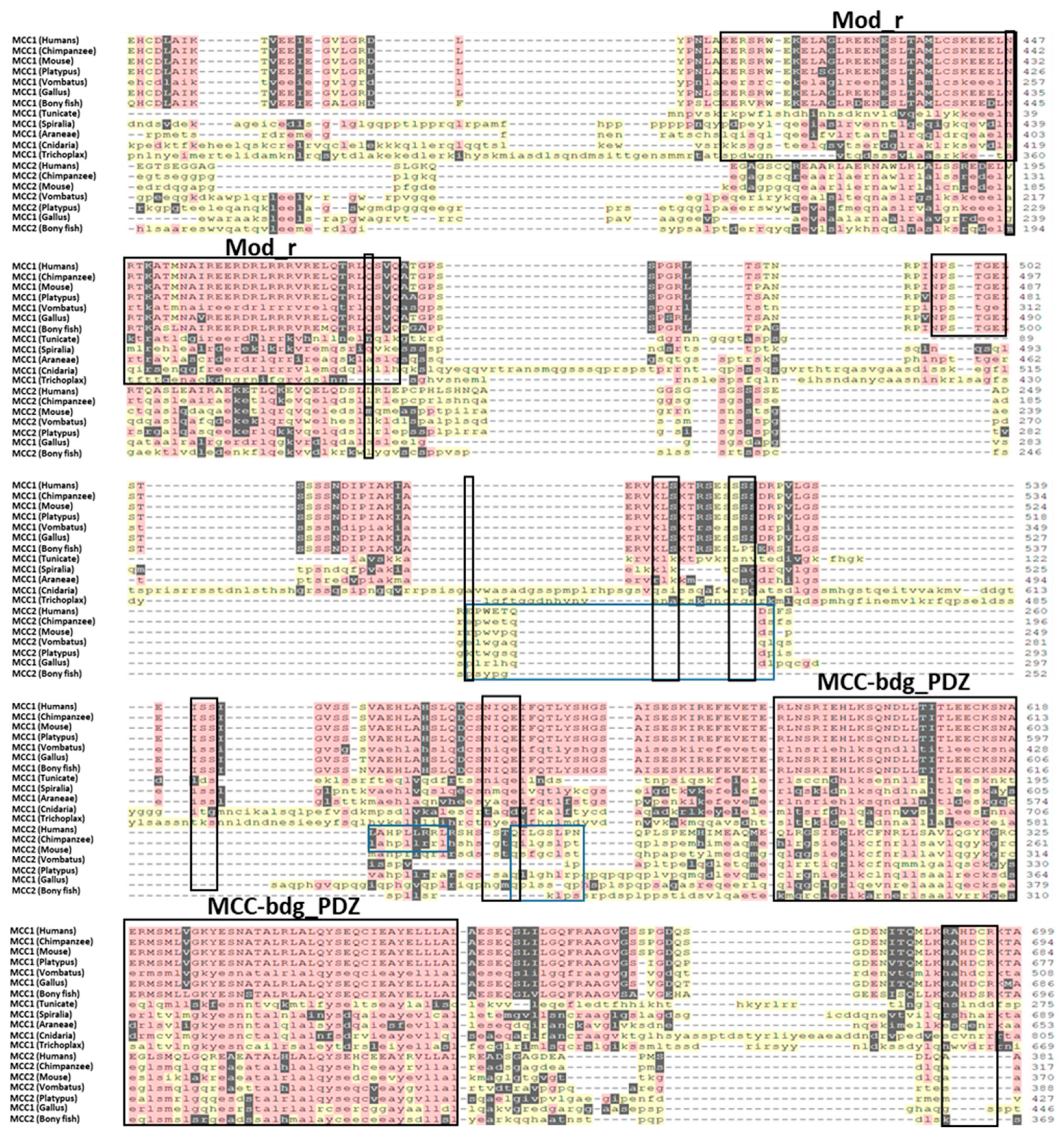
2.2.1. Sites of Functional Divergence
2.2.2. Motifs
2.3. Positive Selection
3. Discussion
4. Methods
4.1. Database Search
4.2. Alignment and Phylogenetic Analysis
4.3. Functional Divergence Estimation and Motif Search
4.4. Positive Selection Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhaumik, S.; Mickael, M.E.; Moran, M.; Spell, M.; Basu, R. RORγt Promotes Foxp3 Expression by Antagonizing the Effector Program in Colonic Regulatory T Cells. J. Immunol. 2021, 207, 2027–2038. [Google Scholar] [CrossRef]
- Mickael, M.-E.; Basu, R.; Bhaumik, S. Retinoid-Related Orphan Receptor RORγt in CD4+ T Cell Mediated Intestinal Homeostasis and Inflammation. Am. J. Pathol. 2020, 190, 1984–1999. [Google Scholar] [CrossRef] [PubMed]
- Mickael, M.E.; Bhaumik, S.; Chakraborti, A.; Umfress, A.A.; van Groen, T.; Macaluso, M.; Totenhagen, J.; Sorace, A.G.; Bibb, J.A.; Standaert, D.G.; et al. RORγt-Expressing Pathogenic CD4+ T Cells Cause Brain Inflammation during Chronic Colitis. J. Immunol. 2022, 208, 2054–2066. [Google Scholar] [CrossRef] [PubMed]
- Amicarella, F.; Muraro, M.G.; Hirt, C.; Cremonesi, E.; Padovan, E.; Mele, V.; Governa, V.; Han, J.; Huber, X.; Droeser, R.A.; et al. Dual Role of Tumour-Infiltrating T Helper 17 Cells in Human Colorectal Cancer. Gut 2017, 66, 692–704. [Google Scholar] [CrossRef]
- Bhaumik, S.; Łazarczyk, M.; Kubick, N.; Klimovich, P.; Gurba, A.; Paszkiewicz, J.; Teodorowicz, P.; Kocki, T.; Horbańczuk, J.O.; Manda, G.; et al. Investigation of the Molecular Evolution of Treg Suppression Mechanisms Indicates a Convergent Origin. Curr. Issues Mol. Biol. 2023, 45, 628–648. [Google Scholar] [CrossRef]
- Olguín, J.E.; Medina-Andrade, I.; Rodríguez, T.; Rodríguez-Sosa, M.; Terrazas, L.I. Relevance of Regulatory T Cells during Colorectal Cancer Development. Cancers 2020, 12, 1888. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, N.; Dasgupta, H.; Bhattacharya, R.; Pal, D.; Roy, R.; Islam, S.; Alam, N.; Biswas, J.; Roy, A.; Roychoudhury, S.; et al. Frequent Inactivation of MCC/CTNNBIP1 and Overexpression of Phospho-Beta-Catenin Y654 Are Associated with Breast Carcinoma: Clinical and Prognostic Significance. Biochim. Biophys. Acta Mol. Basis Dis. 2016, 1862, 1472–1484. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, N.; Panda, C.K. Wnt/β-Catenin Signaling Pathway as Chemotherapeutic Target in Breast Cancer: An Update on Pros and Cons. Clin. Breast Cancer 2020, 20, 361–370. [Google Scholar] [CrossRef]
- Edwards, S.K.E.; Baron, J.; Moore, C.R.; Liu, Y.; Perlman, D.H.; Hart, R.P.; Xie, P. Mutated in Colorectal Cancer (MCC) Is a Novel Oncogene in B Lymphocytes. J. Hematol. Oncol. 2014, 7, 56. [Google Scholar] [CrossRef]
- Young, T.; Poobalan, Y.; Ali, Y.; Siew Tein, W.; Sadasivam, A.; Ee Kim, T.; Erica Tay, P.; Dunn, N.R. Mutated in Colorectal Cancer (Mcc), a Candidate Tumor Suppressor, Is Dynamically Expressed during Mouse Embryogenesis. Dev. Dyn. 2011, 240, 2166–2174. [Google Scholar] [CrossRef] [PubMed]
- Mickael, M.E.; Kubick, N.; Łazarczyk, M.; Sacharczuk, M.; Marchewka, J.; Urbański, P.; Horbańczuk, J.O. Transcriptome Analysis of the Th17/Treg Axis Reveals Multiple Pathways That Ensure Distinct Differentiation Patterns. Anim. Sci. Pap. Rep. 2023, 41, 79–93. [Google Scholar]
- Begley, D.; Murphy, A.M.; Hiu, C.; Tsubota, S.I. Modifier of RudimentaryP1, Mod(r)P1, a Trans-Acting Regulatory Mutation of Rudimentary. Mol. Gen. Genet. MGG 1995, 248, 69–78. [Google Scholar] [CrossRef]
- Bache, K.G.; Slagsvold, T.; Cabezas, A.; Rosendal, K.R.; Raiborg, C.; Stenmark, H. The Growth-Regulatory Protein HCRP1/h Vps37A Is a Subunit of Mammalian ESCRT-I and Mediates Receptor down-Regulation. Mol. Biol. Cell 2004, 15, 4337–4346. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Coelho, M.B.; Kotik-Kogan, O.; Simpson, P.J.; Matthews, S.J.; Smith, C.W.J.; Curry, S. Crystallographic Analysis of Polypyrimidine Tract-Binding Protein-Raver1 Interactions Involved in Regulation of Alternative Splicing. Structure 2011, 19, 1816–1825. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Nelson, E.S.; Maiers, J.L.; DeMali, K.A. New Insights into Vinculin Function and Regulation. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Yang, Y.; Liao, B.; Wang, S.; Yan, B.; Jin, Y.; Shu, H.B.; Wang, Y.Y. E3 Ligase WWP2 Negatively Regulates TLR3-Mediated Innate Immune Response by Targeting TRIF for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. USA 2013, 110, 5115–5120. [Google Scholar] [CrossRef] [PubMed]
- Blair, S.S. Notch Signaling: Fringe Really Is a Glycosyltransferase. Curr. Biol. 2000, 10, R608–R612. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Haltiwanger, R.S. O-Fucose Modifications of Epidermal Growth Factor-like Repeats and Thrombospondin Type 1 Repeats: Unusual Modifications in Unusual Places. Cell. Mol. Life Sci. 2003, 60, 241–250. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Banerjee, S.; Sarkar, F.H. Emerging Role of Notch in Stem Cells and Cancer. Cancer Lett. 2009, 279, 8–12. [Google Scholar] [CrossRef]
- Márton, M.; Tihanyi, N.; Gyulavári, P.; Bánhegyi, G.; Kapuy, O. NRF2-Regulated Cell Cycle Arrest at Early Stage of Oxidative Stress Response Mechanism. PLoS ONE 2018, 13, e0207949. [Google Scholar] [CrossRef]
- Kopacz, A.; Kloska, D.; Forman, H.J.; Jozkowicz, A.; Grochot-Przeczek, A. Beyond Repression of Nrf2: An Update on Keap1. Free Radic. Biol. Med. 2020, 157, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Aristin Revilla, S.; Kranenburg, O.; Coffer, P.J. Colorectal Cancer-Infiltrating Regulatory T Cells: Functional Heterogeneity, Metabolic Adaptation, and Therapeutic Targeting. Front. Immunol. 2022, 13, 903564. [Google Scholar] [CrossRef]
- Lewit-Bentley, A.; Réty, S. EF-Hand Calcium-Binding Proteins. Curr. Opin. Struct. Biol. 2000, 10, 637–643. [Google Scholar] [CrossRef]
- Chazin, W.J. Relating Form and Function of EF-Hand Calcium Binding Proteins. Acc. Chem. Res. 2011, 44, 171–179. [Google Scholar] [CrossRef]
- Yap, K.L.; Ames, J.B.; Swindells, M.B.; Ikura, M. Diversity of Conformational States and Changes within the EF-Hand Protein Superfamily. Proteins Struct. Funct. Genet. 1999, 37, 499–507. [Google Scholar] [CrossRef]
- Miyara, M.; Sakaguchi, S. Natural Regulatory T Cells: Mechanisms of Suppression. Trends Mol. Med. 2007, 13, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, F.; Casadei, F.; Luchinat, C. EF-Hand Protein Dynamics and Evolution of Calcium Signal Transduction: An NMR View. JBIC J. Biol. Inorg. Chem. 2006, 11, 949–962. [Google Scholar] [CrossRef]
- Mickael, M.E.; Rajput, A.; Steyn, J.; Wiemerslage, L.; Bürglin, T. An Optimised Phylogenetic Method Sheds More Light on the Main Branching Events of Rhodopsin-like Superfamily. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 20, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Kubick, N.; Klimovich, P.; Bieńkowska, I.; Poznanski, P.; Łazarczyk, M.; Sacharczuk, M.; Mickael, M.-E. Investigation of Evolutionary History and Origin of the Tre1 Family Suggests a Role in Regulating Hemocytes Cells Infiltration of the Blood–Brain Barrier. Insects 2021, 12, 882. [Google Scholar] [CrossRef] [PubMed]
- Mickael, M.E.; Bieńkowska, I.; Sacharczuk, M. An Update on the Evolutionary History of Bregs. Genes 2022, 13, 890. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG Mapping Tools for Uncovering Hidden Features in Biological Data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Mickael, M.-E.; Kubick, N.; Gurba, A.; Klimovich, P.; Bieńkowska, I.; Kocki, T.; Sacharczuk, M. Fezf2 and Aire1 Evolutionary Trade-off in Negative Selection of T Cells in the Thymus. bioRxiv 2022. [Google Scholar] [CrossRef]


| Pos | Entropy | SH | Rnk | Consensus | ||||
|---|---|---|---|---|---|---|---|---|
| Ali | A | B | AB | Rel. | A | B | ||
| 772 | 1.19 | 1.84 | 2.38 | 1.05 | 0.00 | 11 | Qne | ATlp |
| 770 | 1.21 | 1.84 | 2.39 | 1.05 | 0.00 | 11 | Lefr | DPav |
| 909 | 0.82 | 1.56 | 2.04 | 1.05 | 0.00 | 9 | Tav | PLS |
| 891 | 0.41 | 1.84 | 1.89 | 1.05 | 0.00 | 6 | Rt | QLav |
| 886 | 1.21 | 1.84 | 2.39 | 1.05 | 0.00 | 6 | Kamp | QEgl |
| 949 | 0.00 | 1.15 | 1.37 | 1.05 | 0.00 | 4 | D | Rhk |
| 701 | 0.82 | 0.86 | 1.78 | 1.05 | 0.00 | 4 | Sct | Gr |
| 948 | 0.82 | 1.15 | 1.89 | 1.05 | 0.00 | 4 | Lfv | Pnr |
| 684 | 1.42 | 0.59 | 2.06 | 1.05 | 0.00 | 4 | Ivqt | Ph |
| 698 | 0.82 | 1.66 | 2.08 | 1.05 | 0.00 | 4 | Rms | Qchk |
| 946 | 1.21 | 2.13 | 2.50 | 1.05 | 0.00 | 4 | Qdps | Aegkt |
| 686 | 1.42 | 1.84 | 2.52 | 1.05 | 0.00 | 4 | Edkp | SAtv |
| 1162 | 0.65 | 0.00 | 1.36 | 1.05 | 0.00 | 3 | Kr | Q |
| 900 | 0.41 | 0.99 | 1.57 | 1.05 | 0.00 | 3 | Ki | RQ |
| 1128 | 0.41 | 1.15 | 1.63 | 1.05 | 0.00 | 3 | Ed | Rqt |
| 715 | 0.98 | 0.59 | 1.79 | 1.05 | 0.00 | 3 | IL | Ag |
| 711 | 0.81 | 1.38 | 1.97 | 1.05 | 0.00 | 3 | Dn | Rqe |
| 709 | 0.82 | 1.66 | 2.08 | 1.05 | 0.00 | 3 | Qde | Flrv |
| 898 | 1.21 | 1.15 | 2.14 | 1.05 | 0.00 | 3 | Qeks | Rhn |
| 901 | 1.21 | 1.38 | 2.22 | 1.05 | 0.00 | 3 | Ndks | Eaq |
| 1190 | 1.42 | 1.15 | 2.27 | 1.05 | 0.00 | 3 | Lmft | Asv |
| 1126 | 1.28 | 1.84 | 2.44 | 1.05 | 0.00 | 3 | SNl | KDat |
| 1189 | 1.78 | 1.66 | 2.69 | 1.05 | 0.00 | 3 | Aslnt | Rehq |
| 952 | 0.41 | 0.00 | 1.21 | 1.05 | 0.00 | 2 | Nt | K |
| 954 | 0.00 | 1.15 | 1.37 | 1.05 | 0.00 | 2 | V | Qae |
| 1004 | 0.82 | 0.00 | 1.47 | 1.05 | 0.00 | 2 | Vch | L |
| 1153 | 1.04 | 0.00 | 1.61 | 1.05 | 0.00 | 2 | Edk | R |
| 1154 | 0.41 | 1.15 | 1.63 | 1.05 | 0.00 | 2 | Kr | Gns |
| 1131 | 0.82 | 0.59 | 1.68 | 1.05 | 0.00 | 2 | Hay | Rq |
| 1132 | 0.41 | 1.84 | 1.89 | 1.05 | 0.00 | 2 | Qi | ASrv |
| 410 | 0.82 | 1.38 | 1.97 | 1.05 | 0.00 | 2 | Edn | Rqh |
| 1092 | 0.82 | 1.45 | 2.00 | 1.05 | 0.00 | 2 | Kqr | DEl |
| 876 | 1.58 | 1.84 | 2.63 | 1.05 | 0.00 | 2 | Tlmnv | KApr |
| 737 | 0.00 | 1.38 | 1.46 | 1.05 | 0.00 | 1 | N | Edr |
| 1142 | 0.00 | 1.38 | 1.46 | 1.05 | 0.00 | 1 | K | Cfs |
| 1185 | 0.00 | 1.84 | 1.63 | 1.05 | 0.00 | 1 | K | AEst |
| 734 | 0.41 | 1.38 | 1.72 | 1.05 | 0.00 | 1 | Yf | Rqh |
| 730 | 0.00 | 2.13 | 1.73 | 1.05 | 0.00 | 1 | L | Qekrs |
| Motif | Position | Known Function | Species | Family |
|---|---|---|---|---|
| NPSTGE | 496–501 [A] | Motif binds the Kelch domain of KEAP1 with high affinity. | H, C, M, P, V, G, D | MCC1 |
| AGSSS | 10–14 [A] | SPOP is part of the complex SPOP/Cul3 and it plays an important role in protein degradation ubiquitination. | H and C | MCC1 |
| LQKLLE ALHKLLT | 167–173 [A] 184–190 [A] | LXXLL supports binding to nuclear receptors. RORγt is a well-known nuclear receptor, but it is unknown whether MMC can bind to it. | H, C, M, P, and G | MCC1 |
| CGRK | 364–367 [A] | Peptide C-terminal amidation, possibly to protect against degradation. | H, C, M, P, V, G, D | MCC1 |
| NSC | 117–119 [A] | NSC is a motif for N-glycosylation, a post-translational modification process where glycans are added to specific Asn residues in proteins in the endoplasmic reticulum and Golgi apparatus. | H and C | MCC1 |
| GGSSLH/P | 175–180 [A] | NEK2 is a phosphorylation motif recognized and targeted by protein kinases for phosphorylation. | H, C, M | MCC1 |
| CGRKKSSC | 364–371 [A] | This motif is a site for the attachment of a fucose residue to a serine, indicating the possible involvement of fucosylation in the regulation or function of that protein. | H, C, M, P, V, G, D | MCC1 |
| RKKSSCS KKSSCSL | 366–372 [A] 367–373 [A] | The motif sequence is recognized by PKA as a specific target for phosphorylation. | H, C, M, P, V, G, D | MCC1 |
| LKSE | 857–860 [A] | SUMO-1 may attach to that motif in a Sumoylation process. Sumoylation can have transcriptional regulation, DNA repair, nuclear transport, and protein quality control. | H, C, M, P, V, G | MCC1 |
| GRHP/APPGE | 13–20 [A] | The motif may function as a docking site for Tankyrase-1 and 2, regulating protein interactions, localization, and stability. | H, P, V | MCC2 |
| LPPP | 693–696 [A] | LPPP motif acts as a docking site in calcineurin substrates to allow them to interact with the catalytic and regulatory subunits of calcineurin and facilitate participation in cellular signaling events. | H, C | MCC2 |
| FAPP | 42–45 [A] | This motif is recognized by the EVH1 domains, which are specific protein domains found in the PPP4R3 regulatory subunits of the PP4 holoenzyme. | H | MCC2 |
| MSARA | 1–5 [A] | This motif is found in pro-apoptotic proteins and it counteracts caspase inhibition by the Inhibitor of Apoptosis Proteins in apoptotic cells. | H, M, V, P | MCC2 |
| EPWETQDSF | 251–259 [A] | This motif mediates the coatomer subunit delta construction. Coatomer plays a part in forming vesicle coat. | H, C, M | MCC2 |
| RAWDPEKLA | 583–591 [A] | This short motif is found in cargo proteins and mediates kinesin-1-dependent microtubule transport by binding to the KLC TPR region. | H, C, M | MCC2 |
| PTLAP PPLPP | 447–451 [A] 691–695 [A] | PxLxP motif is recognized by a subset of MYND domain-containing proteins. MYND domain is involved in transcriptional regulation, chromatin remodeling, protein localization, and signal transduction | H, C | MCC2 |
| QILGSLPN | 276–283 [A] | The PTB RRM2 Interacting (PRI) motif is found in some splicing regulators | H, C | MCC2 |
| PPQLGD | 695–700 [A] | TRAF2-binding motif. Members of the tumor necrosis factor receptor (TNFR) superfamily recruit TNFR-associated factors (TRAFs) to initiate cell signaling. | H, C | MCC2 |
| PPLP | 691–694 [A] | PPLP is a motif recognized by WW domains of Group II that play a role in multiple cellular processes. | H, C | MCC2 |
| M/LAHPLL | 261–266 [A] | Dileucine motifs involved in the trafficking and sorting of proteins in the endosomal-basolateral-lysosomal pathway | H, C, M | MCC2 |
| EAW/SRLL | 384–389 [A] | Dileucine motifs involved in the trafficking and sorting of proteins in the endosomal-basolateral-lysosomal pathway | H, C, M | MCC2 |
| Model | ω |
|---|---|
| general | 0.45728, p-value is <0.00001 |
| branch | ω value was 0.89 p-value > 0.005 |
| branch-site | ω value was 0.64046 p-value > 0.005 |
| sites | No sites with ω > 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubick, N.; Paszkiewicz, J.; Bieńkowska, I.; Ławiński, M.; Horbańczuk, J.O.; Sacharczuk, M.; Mickael, M.E. Investigation of Mutated in Colorectal Cancer (MCC) Gene Family Evolution History Indicates a Putative Role in Th17/Treg Differentiation. Int. J. Mol. Sci. 2023, 24, 11940. https://doi.org/10.3390/ijms241511940
Kubick N, Paszkiewicz J, Bieńkowska I, Ławiński M, Horbańczuk JO, Sacharczuk M, Mickael ME. Investigation of Mutated in Colorectal Cancer (MCC) Gene Family Evolution History Indicates a Putative Role in Th17/Treg Differentiation. International Journal of Molecular Sciences. 2023; 24(15):11940. https://doi.org/10.3390/ijms241511940
Chicago/Turabian StyleKubick, Norwin, Justyna Paszkiewicz, Irmina Bieńkowska, Michał Ławiński, Jarosław Olav Horbańczuk, Mariusz Sacharczuk, and Michel Edwar Mickael. 2023. "Investigation of Mutated in Colorectal Cancer (MCC) Gene Family Evolution History Indicates a Putative Role in Th17/Treg Differentiation" International Journal of Molecular Sciences 24, no. 15: 11940. https://doi.org/10.3390/ijms241511940
APA StyleKubick, N., Paszkiewicz, J., Bieńkowska, I., Ławiński, M., Horbańczuk, J. O., Sacharczuk, M., & Mickael, M. E. (2023). Investigation of Mutated in Colorectal Cancer (MCC) Gene Family Evolution History Indicates a Putative Role in Th17/Treg Differentiation. International Journal of Molecular Sciences, 24(15), 11940. https://doi.org/10.3390/ijms241511940






