Standardization of a Sex-Sorting Protocol for Stallion Spermatozoa by Means of Absolute RT-qPCR
Abstract
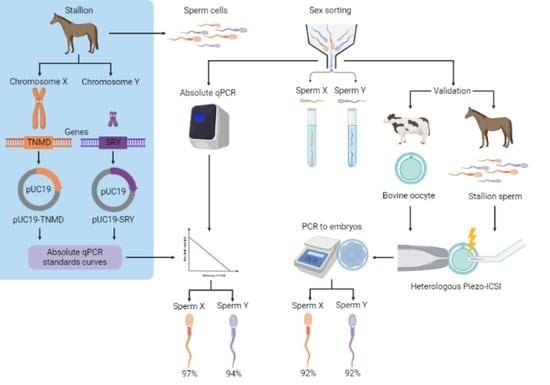
1. Introduction
2. Results
2.1. Design of Standards Curves
2.2. Determination of the Sex-Sorting Accuracy
2.3. Sexing of Equine/Bovine Hybrid Embryos Produced by PIEZO-ICSI
3. Discussion
4. Materials and Methods
4.1. Primers Design
4.2. Design of Standards for Absolute qPCR
4.3. Collection of Semen Samples and Cryopreservation
4.4. Sex-Sorting of Equine Sperm
4.5. DNA Extraction from Equine Spermatozoa
4.6. Determination of Sex-Sorting Error Rate
4.7. Absolute RT-qPCR
4.8. Oocyte Collection and In Vitro Maturation (IVM)
4.9. Heterologous ICSI, Bovine Parthenotes Production, and Embryo Culture
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naniwa, Y.; Sakamoto, Y.; Toda, S.; Uchiyama, K. Bovine sperm sex-selection technology in Japan. Reprod. Med. Biol. 2019, 18, 17–26. [Google Scholar] [CrossRef]
- Hirst, C.E.; Major, A.T.; Smith, C.A. Sex determination and gonadal sex differentiation in the chicken model. Int. J. Dev. Biol. 2018, 62, 153–166. [Google Scholar] [CrossRef] [PubMed]
- González-Marín, C.; Góngora, C.E.; Moreno, J.F.; Vishwanath, R. Small ruminant SexedULTRA™ sperm sex-sorting: Status report and recent developments. Theriogenology 2021, 162, 67–73. [Google Scholar] [CrossRef]
- Quelhas, J.; Santiago, J.; Matos, B.; Rocha, A.; Lopes, G.; Fardilha, M. Bovine semen sexing: Sperm membrane proteomics as candidates for immunological selection of X- and Y-chromosome-bearing sperm. Vet. Med. Sci. 2021, 7, 1633–1641. [Google Scholar] [CrossRef]
- Pozdyshev, D.V.; Kombarova, N.A.; Muronetz, V.I. Biochemical Features of X or Y Chromosome-Bearing Spermatozoa for Sperm Sexing. Biochemistry 2023, 88, 655–666. [Google Scholar] [CrossRef]
- Bhat, Y.; Sharma, M. X-sperm enrichment of bovine semen by Percoll density gradient method and its effect on semen quality, sex ratio and conception rate. Indian J. Anim. Res. 2020, 54, 1181–1187. [Google Scholar] [CrossRef]
- Ohlweiler, L.U.; Mezzalira, J.C.; Mezzalira, A. Porcine IVF embryo development and estrogen receptors are influenced by the concentration of percoll gradients during sperm selection. Mol. Reprod. Dev. 2020, 87, 135–141. [Google Scholar] [CrossRef]
- Thomas, J.M.; Locke, J.W.C.; Bonacker, R.C.; Knickmeyer, E.R.; Wilson, D.J.; Vishwanath, R.; Arnett, A.M.; Smith, M.F.; Patterson, D.J. Evaluation of SexedULTRA 4M™ sex-sorted semen in timed artificial insemination programs for mature beef cows. Theriogenology 2019, 123, 100–107. [Google Scholar] [CrossRef]
- Kalbfleisch, T.S.; Rice, E.; DePriest, M.S.; Walenz, B.P.; Hestand, M.S.; Vermeesch, J.R.; O’Connell, B.L.; Fiddes, I.T.; Vershinina, A.O.; Petersen, J.L.; et al. EquCab3, an updated reference genome for the domestic horse. BioRxiv 2018. [Google Scholar] [CrossRef]
- Raudsepp, T.; Santani, A.; Wallner, B.; Kata, S.R.; Ren, C.; Zhang, H.B.; Womack, J.E.; Skow, L.C.; Chowdhary, B.P. A detailed physical map of the horse Y chromosome. Proc. Natl. Acad. Sci. USA 2004, 101, 9321–9326. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, A. The hunt for dark DNA. New Sci. 2018, 237, 29–31. [Google Scholar] [CrossRef]
- Khamlor, T.; Pongpiachan, P.; Sangsritavong, S.; Chokesajjawatee, N. Determination of sperm sex ratio in bovine semen using multiplex real-time polymerase chain reaction. Asian-Australas. J. Anim. Sci. 2014, 27, 1411. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, J.H.; Lee, J.H.; Choi, K.M.; Joung, S.Y.; Kim, J.Y.; Chung, G.M.; Jin, D.I.; Im, K.S. Rapid sexing of preimplantation bovine embryo using consecutive and multiplex polymerase chain reaction (PCR) with biopsied single blastomere. Theriogenology 2001, 55, 1843–1853. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Xi, H.; Ren, Y.; Li, Y.; Wen, F.; Xian, M.; Zhao, M.; Zhu, D.; Wang, L.; Lei, A.; et al. TLR7/8 signalling affects X-sperm motility via the GSK3 α/β-hexokinase pathway for the efficient production of sexed dairy goat embryos. J. Anim. Sci. Biotechnol. 2021, 12, 89. [Google Scholar] [CrossRef]
- Yadav, S.K.; Gangwar, D.K.; Singh, J.; Tikadar, C.K.; Khanna, V.V.; Saini, S.; Dholpuria, S.; Palta, P.; Manik, R.S.; Singh, M.K.; et al. An immunological approach of sperm sexing and different methods for identification of X- and Y-chromosome bearing sperm. Vet. World 2017, 10, 498. [Google Scholar] [CrossRef]
- Kurtz, S.; Petersen, B. Pre-determination of sex in pigs by application of CRISPR/Cas system for genome editing. Theriogenology 2019, 137, 67–74. [Google Scholar] [CrossRef]
- Azam, A.; Awan, M.A.; Khalid, M.; Sami, A.; Shahzad, Q.; Ansari, M.S.; Rakha, B.A.; Naqvi, S.M.S.; Akhter, S. Efficiency of sucrose density gradient method for sex separation of buffalo spermatozoa as validated by SYBR Green Real Time PCR. Anim. Sci. Pap. Rep. 2020, 38, 22–23. [Google Scholar]
- Gaddam, N.; ST, V.R.; Putty, K.; Sakaram, D.; Ganji, V.K. Quantitative and qualitative assessment of sex sorted semen of Bos indicus and Bos taurus breeds. Res. Sq. 2022, 1–13. [Google Scholar] [CrossRef]
- Saffari, M.; Heidari, F.; Shamsara, M.; Hashemi, E. The Effect of Diffrent Factors on X-and Y Bearing Sperm in Bull Semen. Iran. J. Vet. Med. 2018, 12, 219–227. [Google Scholar] [CrossRef]
- Laghi, L.; Randolph, A.E.; Malesci, A.; Boland, C.R. Constraints imposed by supercoiling on in vitro amplification of polyomavirus DNA. J. Gen. Virol. 2004, 85, 3383–3388. [Google Scholar] [CrossRef]
- Hou, Y.; Zhang, H.; Miranda, L.; Lin, S. Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: Microalgal pcna as the model gene. PLoS ONE 2010, 5, e9545. [Google Scholar] [CrossRef] [PubMed]
- Barrachina, F.; Soler-Ventura, A.; Oliva, R.; Jodar, M. Sperm nucleoproteins (histones and protamines). In A Clinician’s Guide to Sperm DNA and Chromatin Damage; Springer: Cham, Switzerland, 2018; pp. 31–51. [Google Scholar] [CrossRef]
- Wang, D.; Zhu, H.; Guo, J.; Lin, B.; Zhang, L.; Hao, H.; Du, W.; Zhao, X. A study of a method to assess the purity of sorted bovine semen using rapid single-sperm sexing PCR. J. Anim. Vet. Adv. 2011, 10, 750–756. [Google Scholar] [CrossRef]
- Vázquez, M. Optimización de una Técnica de Extracción de ADN Genómico de Espermatozoides de Toro. Doctoral Dissertation, Instituto de Fisiología Celular, Mexico City, Mexico, 2014. [Google Scholar]
- Arias, M.E.; Sánchez, R.; Felmer, R. Effect of anisomycin, a protein synthesis inhibitor, on the in vitro developmental potential, ploidy and embryo quality of bovine ICSI embryos. Zygote 2016, 24, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, M.; Fissore, R.A. ICSI-generated mouse zygotes exhibit altered calcium oscillations, inositol 1,4,5-trisphosphate receptor-1 down-regulation, and embryo development. Mol. Hum. Reprod. 2003, 9, 523–533. [Google Scholar] [CrossRef]







| Sexing | Primers Used | % of Correct Sex |
|---|---|---|
| Absolute RT-qPCR of sexed spermatozoa X | qPCR-TNMD | 96.63% |
| Absolute RT-qPCR of unsexed spermatozoa | qPCR-TNMD | 46.57% |
| Absolute RT-qPCR of sexed spermatozoa Y | qPCR-SRY | 93.57% |
| Absolute RT-qPCR of unsexed spermatozoa | qPCR-SRY | 47.57% |
| PCR of equine/bovine embryos generated with a sperm X | qPCR-TNMD | 92.1% |
| PCR of equine/bovine embryos generated with a sperm Y | qPCR-SRY | 92.0% |
| Name | Sequence 5′ to 3′ | Use | Amplicon |
|---|---|---|---|
| qPCR-SRY-F | AGGACAGCAACATACCGTTCTCG | absolute qPCR of sperm embryo sexing. | 141 bp |
| qPCR-SRY-R | GCATTCATGGGTCGTTTGACA | absolute qPCR of sperm embryo sexing. | 141 bp |
| qPCR-TNMD-F | GCGGGTTATCTGTCGTGTCATC | absolute qPCR of sperm, embryo sexing. | 89 bp |
| qPCR-TNMD-R | GCTCTTGTGCTCGAACTTGC | absolute qPCR of sperm, embryo sexing. | 89 bp |
| SRY-F | GCAATGGCGCCCGGGAAGCGGTTTGTCACTTTTCT | Cloning of the SRY gene from Equus caballus. | 695 bp |
| SRY-R | AACTAGGATCCTGGGGATTAGAAGTAGGGCACAGA | Cloning of the SRY gene from Equus caballus. | 695 bp |
| TNMD-F | GCAATGGCGCCTCACAGCCCCTCAGATCAAAGTCA | Cloning of the TNMD gene from Equus caballus | 564 bp |
| TNMD-R | AACTAGGATCCGCTACCAGGAGCCAAATGCCTTAT | Cloning of the TNMD gene from Equus caballus. | 564 bp |
| Quantity of Standard pUC19-SRY | Number of Molecules of pUC19-SRY Vector | Quantity of Standard pUC19-TNMD | Number of Molecules of pUC19-TNMD Vector |
|---|---|---|---|
| 10 pg | 2,890,000 | 10 pg | 3,010,000 |
| 8 pg | 2,310,000 | 8 pg | 2,410,000 |
| 6 pg | 1,730,000 | 6 pg | 1,810,000 |
| 4 pg | 1,160,000 | 4 pg | 1,210,000 |
| 2 pg | 578,000 | 2 pg | 603,000 |
| 0 pg | 0 | 0 pg | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz, E.; Castro, M.; Aguila, L.; Contreras, M.J.; Fuentes, F.; Arias, M.E.; Felmer, R. Standardization of a Sex-Sorting Protocol for Stallion Spermatozoa by Means of Absolute RT-qPCR. Int. J. Mol. Sci. 2023, 24, 11947. https://doi.org/10.3390/ijms241511947
Muñoz E, Castro M, Aguila L, Contreras MJ, Fuentes F, Arias ME, Felmer R. Standardization of a Sex-Sorting Protocol for Stallion Spermatozoa by Means of Absolute RT-qPCR. International Journal of Molecular Sciences. 2023; 24(15):11947. https://doi.org/10.3390/ijms241511947
Chicago/Turabian StyleMuñoz, Erwin, Macarena Castro, Luis Aguila, María José Contreras, Fernanda Fuentes, María Elena Arias, and Ricardo Felmer. 2023. "Standardization of a Sex-Sorting Protocol for Stallion Spermatozoa by Means of Absolute RT-qPCR" International Journal of Molecular Sciences 24, no. 15: 11947. https://doi.org/10.3390/ijms241511947
APA StyleMuñoz, E., Castro, M., Aguila, L., Contreras, M. J., Fuentes, F., Arias, M. E., & Felmer, R. (2023). Standardization of a Sex-Sorting Protocol for Stallion Spermatozoa by Means of Absolute RT-qPCR. International Journal of Molecular Sciences, 24(15), 11947. https://doi.org/10.3390/ijms241511947