Molecular Descriptors Property Prediction Using Transformer-Based Approach
Abstract
:1. Introduction
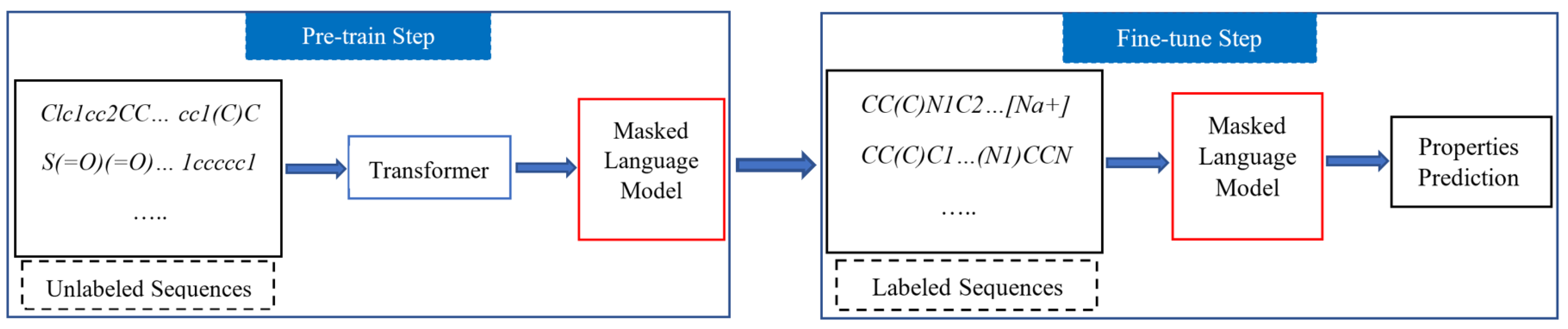
- We introduce a two-stage (pre-training and fine-tuning) model to utilize a large amount of both labeled and unlabeled data for molecular properties prediction.
- We propose a new approach to calculate the attention score for the transformer layer taking advantage of 3D structure information of the compounds.
- We show that without relying on the computational expense of a pre-trained and fine-tuned model using the proposed attention score, our model is able to achieve comparable performance in predicting anti-malaria drug candidates.
Related Work
Machine Learning in Malaria Drug Discovery
2. Results and Discussion
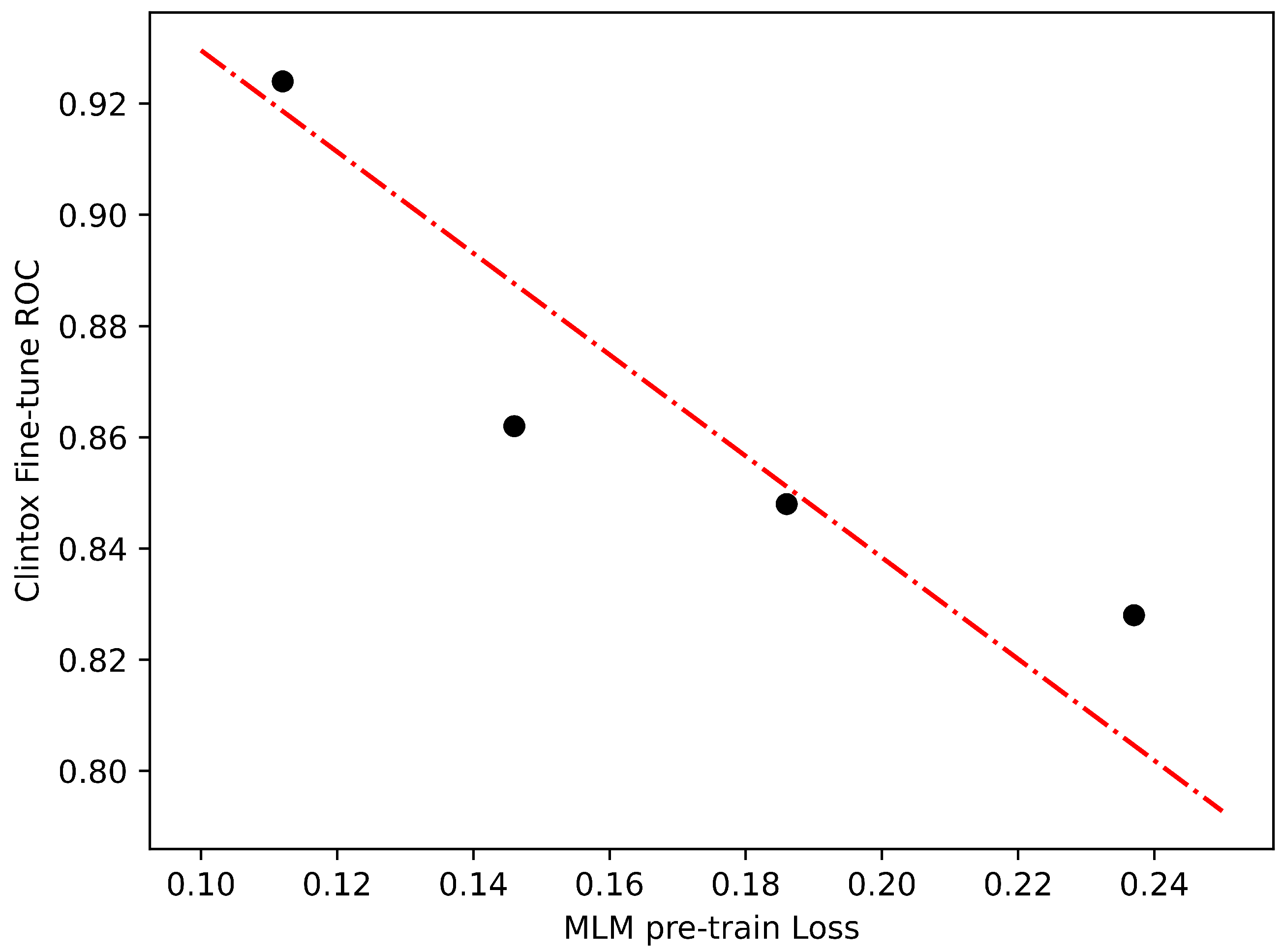
2.1. Classification Problem
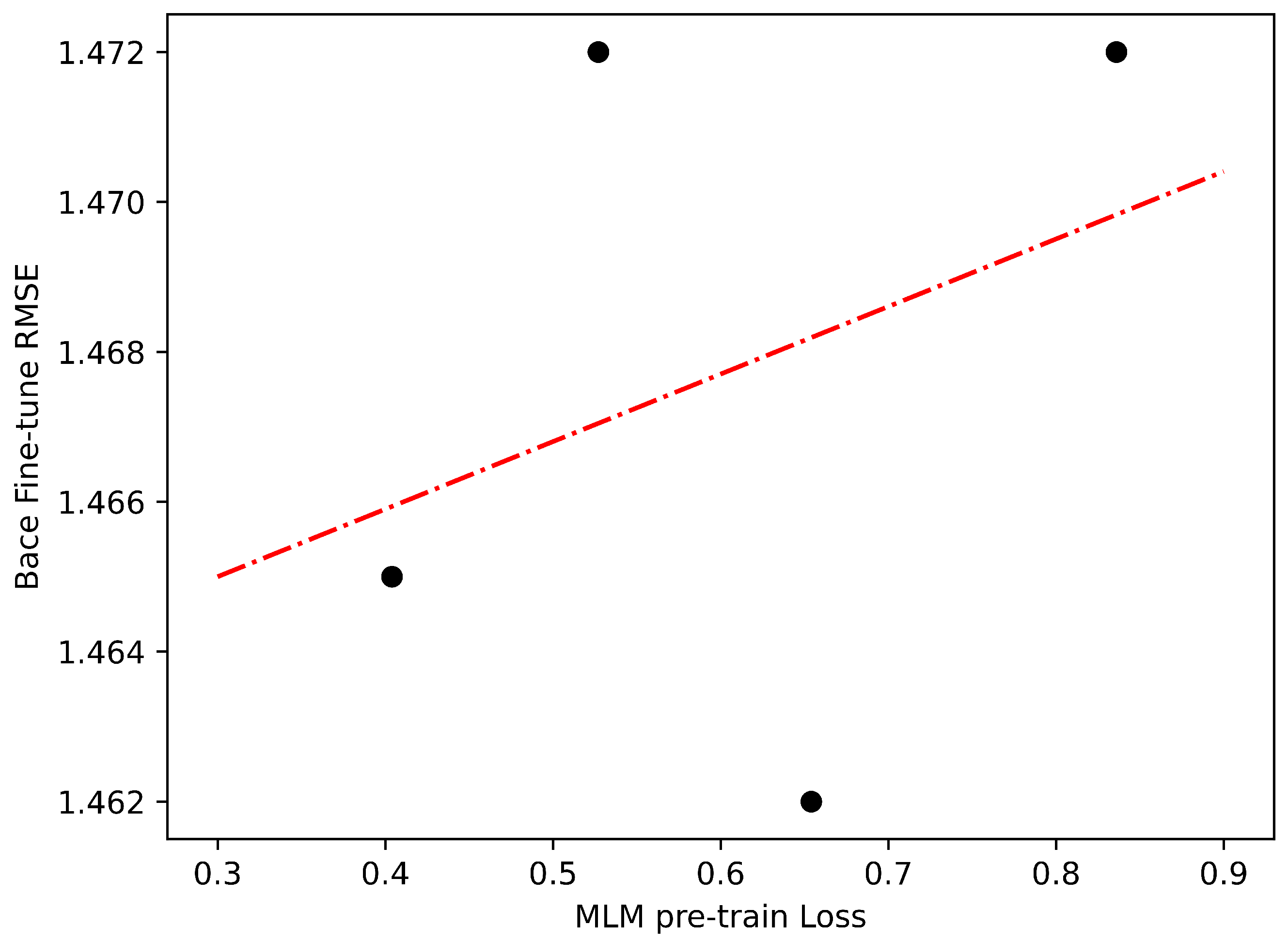
2.2. Regression Problem
Case Study: Anti-Malaria Drug Target Classification
3. Methods and Materials
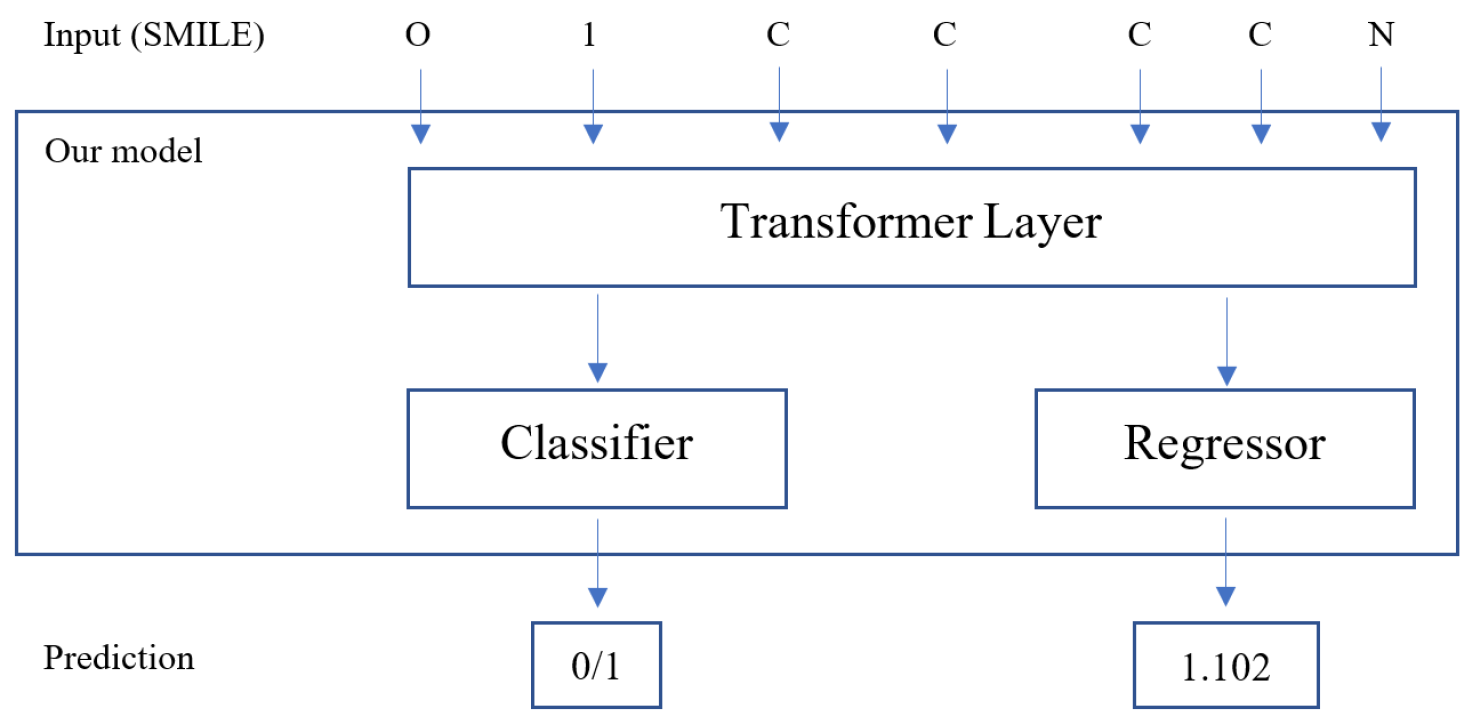
3.1. Methodology
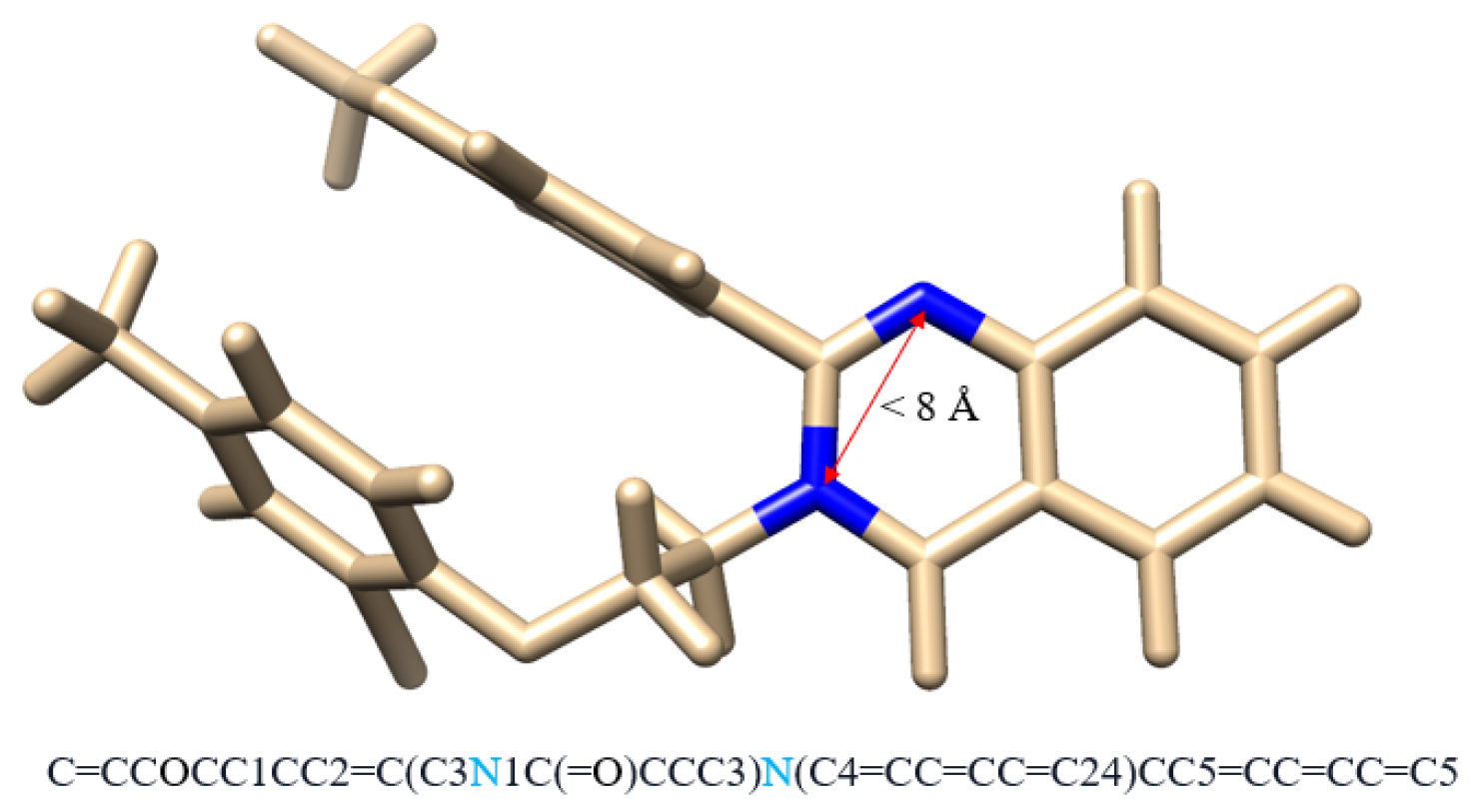
3.1.1. Transformer Layers
3.1.2. Pre-Training Setup
3.1.3. Fine-Tuned Model
3.1.4. Case Study: Anti-Malaria Drug Target Classification
3.2. Evaluation Dataset
3.2.1. Pre-Trained Dataset
3.2.2. Fine-Tuned Dataset
- BACE (Classification and Regression): The BACE dataset provides quantitative IC50 and qualitative binding results for a set of inhibitors of human beta-secretase 1 (BACE-1). It has 1513 compounds.
- Clearance (Regression): The dataset contains human clearance, which is the parameter that determines total systemic exposure to the drug. It has 837 compounds.
- Delaney (Regression): The Delaney dataset contains structures and water solubility data. It has 1128 compounds.
- Lipophilicity (Regression): The lipophilicity dataset provides experimental results of the octanol/water distribution coefficient. It has 4200 compounds.
- BBBP (Classification): The blood–brain barrier penetration (BBBP) dataset consists of binary labels for the prediction of barrier permeability. It has 2039 compounds.
- ClinTox (Classification): The ClinTox dataset compares drugs approved by the FDA and drugs that have failed clinical trials for toxicity reasons. It has 1478 compounds.
- Tox21 (Classification): The “Toxicology in the 21st Century” (Tox21) contains qualitative toxicity measurements on 12 biological targets, including nuclear receptors and stress response pathways. It has 7831 compounds.
- Antimalarial (Classification): The antimalarial dataset is a given experimentally verified antimalarial drug candidate from public chemical databases, ChEMBL and PubChem. It has 4794 compounds.
3.3. Implementation
3.3.1. Model Details
3.3.2. Tokenizer
3.3.3. Case Study: Anti-Malaria Drug Target Classification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| PF | Plasmodium Falciparum |
| CDC | Centers for Disease Control and Prevention |
| SMILES | Simplified Molecular-Input Line-Entry |
| GNN | Graph Neural Network |
| MLM | Masked Language Modeling |
| ROC-AUC | Area Under the Receiver Operating Characteristic Curve |
| MTR | Multi-Task Regression |
| XGB | Extreme Gradient Boosting |
| ANN | Artificial Neural Network |
References
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2019, 20, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Danishuddin; Khan, A.U. Descriptors and their selection methods in QSAR analysis: Paradigm for drug design. Drug Discov. Today 2016, 21, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Mswahili, M.E.; Lee, M.J.; Martin, G.L.; Kim, J.; Kim, P.; Choi, G.J.; Jeong, Y.S. Cocrystal prediction using machine learning models and descriptors. Appl. Sci. 2021, 11, 1323. [Google Scholar] [CrossRef]
- Liu, Q.; Deng, J.; Liu, M. Classification models for predicting the antimalarial activity against Plasmodium falciparum. SAR QSAR Environ. Res. 2020, 31, 313–324. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, S.; Zhu, F.; Huang, J. Seq2seq fingerprint: An unsupervised deep molecular embedding for drug discovery. In Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Boston, MA, USA, 20–23 August 2017; pp. 285–294. [Google Scholar]
- Zhang, X.; Wang, S.; Zhu, F.; Xu, Z.; Wang, Y.; Huang, J. Seq3seq fingerprint: Towards end-to-end semi-supervised deep drug discovery. In Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Washington, DC, USA, 29 August–1 September 2018; pp. 404–413. [Google Scholar]
- Tran, T.; Ekenna, C. Protein binding pose prediction via conditional variational autoencoding for plasmodium falciparum. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 2448–2455. [Google Scholar]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Wang, S.; Guo, Y.; Wang, Y.; Sun, H.; Huang, J. SMILES-BERT: Large scale unsupervised pre-training for molecular property prediction. In Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Niagara Falls, NY, USA, 7–10 September 2019; pp. 429–436. [Google Scholar]
- Gupta, S.; Hill, A.; Kwiatkowski, D.; Greenwood, A.M.; Greenwood, B.M.; Day, K.P. Parasite virulence and disease patterns in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. USA 1994, 91, 3715–3719. [Google Scholar] [CrossRef] [PubMed]
- Blasco, B.; Leroy, D.; Fidock, D.A. Antimalarial drug resistance: Linking Plasmodium falciparum parasite biology to the clinic. Nat. Med. 2017, 23, 917–928. [Google Scholar] [CrossRef]
- Pallarès, I.; De Groot, N.S.; Iglesias, V.; Sant’Anna, R.; Biosca, A.; Fernàndez-Busquets, X.; Ventura, S. Discovering putative prion-like proteins in Plasmodium falciparum: A computational and experimental analysis. Front. Microbiol. 2018, 9, 1737. [Google Scholar] [CrossRef] [Green Version]
- Halfmann, R.; Alberti, S.; Krishnan, R.; Lyle, N.; O’Donnell, C.W.; King, O.D.; Berger, B.; Pappu, R.V.; Lindquist, S. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol. Cell 2011, 43, 72–84. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural message passing for quantum chemistry. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 1263–1272. [Google Scholar]
- Mansimov, E.; Mahmood, O.; Kang, S.; Cho, K. Molecular geometry prediction using a deep generative graph neural network. Sci. Rep. 2019, 9, 20381. [Google Scholar] [CrossRef] [Green Version]
- Velickovic, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph attention networks. Stat 2017, 1050, 10-48550. [Google Scholar]
- Shang, C.; Liu, Q.; Chen, K.S.; Sun, J.; Lu, J.; Yi, J.; Bi, J. Edge attention-based multi-relational graph convolutional networks. arXiv 2018, arXiv:1802.04944. [Google Scholar]
- Hu, W.; Liu, B.; Gomes, J.; Zitnik, M.; Liang, P.; Pande, V.; Leskovec, J. Strategies for pre-training graph neural networks. arXiv 2019, arXiv:1905.12265. [Google Scholar]
- Heller, S.; McNaught, A.; Stein, S.; Tchekhovskoi, D.; Pletnev, I. InChI-the worldwide chemical structure identifier standard. J. Cheminform. 2013, 5, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krenn, M.; Ai, Q.; Barthel, S.; Carson, N.; Frei, A.; Frey, N.C.; Friederich, P.; Gaudin, T.; Gayle, A.A.; Jablonka, K.M.; et al. SELFIES and the future of molecular string representations. Patterns 2022, 3, 100588. [Google Scholar] [CrossRef]
- Li, X.; Fourches, D. Inductive transfer learning for molecular activity prediction: Next-Gen QSAR Models with MolPMoFiT. J. Cheminform. 2020, 12, 1–15. [Google Scholar] [CrossRef]
- Morris, P.; St. Clair, R.; Hahn, W.E.; Barenholtz, E. Predicting binding from screening assays with transformer network embeddings. J. Chem. Inf. Model. 2020, 60, 4191–4199. [Google Scholar] [CrossRef]
- Blanchard, A.E.; Shekar, M.C.; Gao, S.; Gounley, J.; Lyngaas, I.; Glaser, J.; Bhowmik, D. Automating Genetic Algorithm Mutations for Molecules Using a Masked Language Model. IEEE Trans. Evol. Comput. 2022, 26, 793–799. [Google Scholar] [CrossRef]
- Schneider, N.; Sayle, R.A.; Landrum, G.A. Get Your Atoms in Order An Open-Source Implementation of a Novel and Robust Molecular Canonicalization Algorithm. J. Chem. Inf. Model. 2015, 55, 2111–2120. [Google Scholar] [CrossRef]
- Neglur, G.; Grossman, R.L.; Liu, B. Assigning unique keys to chemical compounds for data integration: Some interesting counter examples. In Proceedings of the International Workshop on Data Integration in the Life Sciences, San Diego, CA, USA, 20–22 July 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 145–157. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Fabian, B.; Edlich, T.; Gaspar, H.; Segler, M.; Meyers, J.; Fiscato, M.; Ahmed, M. Molecular representation learning with language models and domain-relevant auxiliary tasks. arXiv 2020, arXiv:2011.13230. [Google Scholar]
- Ahmad, W.; Simon, E.; Chithrananda, S.; Grand, G.; Ramsundar, B. ChemBERTa-2: Towards Chemical Foundation Models. arXiv 2022, arXiv:2209.01712. [Google Scholar]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef] [PubMed]
- Burden, F.R.; Winkler, D.A. Robust QSAR models using Bayesian regularized neural networks. J. Med. Chem. 1999, 42, 3183–3187. [Google Scholar] [PubMed]
- Alves, V.M.; Muratov, E.; Fourches, D.; Strickland, J.; Kleinstreuer, N.; Andrade, C.H.; Tropsha, A. Predicting chemically-induced skin reactions. Part I: QSAR models of skin sensitization and their application to identify potentially hazardous compounds. Toxicol. Appl. Pharmacol. 2015, 284, 262–272. [Google Scholar] [CrossRef] [Green Version]
- Hartung, T. Making big sense from big data in toxicology by read-across. ALTEX-Altern. Anim. Exp. 2016, 33, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Goh, G.B.; Hodas, N.O.; Vishnu, A. Deep learning for computational chemistry. J. Comput. Chem. 2017, 38, 1291–1307. [Google Scholar] [CrossRef] [Green Version]
- Neves, B.J.; Braga, R.C.; Alves, V.M.; Lima, M.N.; Cassiano, G.C.; Muratov, E.N.; Costa, F.T.; Andrade, C.H. Deep Learning-driven research for drug discovery: Tackling Malaria. Plos Comput. Biol. 2020, 16, e1007025. [Google Scholar]
- Mason, D.J.; Eastman, R.T.; Lewis, R.P.; Stott, I.P.; Guha, R.; Bender, A. Using machine learning to predict synergistic antimalarial compound combinations with novel structures. Front. Pharmacol. 2018, 9, 1096. [Google Scholar] [CrossRef] [PubMed]
- Segler, M.H.; Kogej, T.; Tyrchan, C.; Waller, M.P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 2018, 4, 120–131. [Google Scholar] [CrossRef] [Green Version]
- Keshavarzi Arshadi, A.; Salem, M.; Collins, J.; Yuan, J.S.; Chakrabarti, D. DeepMalaria: Artificial intelligence driven discovery of potent antiplasmodials. Front. Pharmacol. 2020, 10, 1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mswahili, M.E.; Martin, G.L.; Woo, J.; Choi, G.J.; Jeong, Y.S. Antimalarial Drug Predictions Using Molecular Descriptors and Machine Learning against Plasmodium Falciparum. Biomolecules 2021, 11, 1750. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.N.; Cassiano, G.C.; Tomaz, K.C.; Silva, A.C.; Sousa, B.K.; Ferreira, L.T.; Tavella, T.A.; Calit, J.; Bargieri, D.Y.; Neves, B.J.; et al. Integrative multi-kinase approach for the identification of potent antiplasmodial hits. Front. Chem. 2019, 7, 773. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Swanson, K.; Jin, W.; Coley, C.; Eiden, P.; Gao, H.; Guzman-Perez, A.; Hopper, T.; Kelley, B.; Mathea, M.; et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 2019, 59, 3370–3388. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.K. Random decision forests. In Proceedings of the 3rd International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; Volume 1, pp. 278–282. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Graves, A. Generating sequences with recurrent neural networks. arXiv 2013, arXiv:1308.0850. [Google Scholar]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. Roberta: A robustly optimized bert pretraining approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Chithrananda, S.; Grand, G.; Ramsundar, B. ChemBERTa: Large-scale self-supervised pretraining for molecular property prediction. arXiv 2020, arXiv:2010.09885. [Google Scholar]
- Rao, R.; Bhattacharya, N.; Thomas, N.; Duan, Y.; Chen, P.; Canny, J.; Abbeel, P.; Song, Y. Evaluating protein transfer learning with TAPE. Adv. Neural Inf. Process. Syst. 2019, 32, 9689–9701. [Google Scholar]
- Fuchs, F.; Worrall, D.; Fischer, V.; Welling, M. Se (3)-transformers: 3d roto-translation equivariant attention networks. Adv. Neural Inf. Process. Syst. 2020, 33, 1970–1981. [Google Scholar]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A benchmark for molecular machine learning. Chem. Sci. 2018, 9, 513–530. [Google Scholar] [CrossRef] [Green Version]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020; pp. 38–45. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Tran, T.; Ekenna, C. Molecular Descriptors Property Prediction via a Natural Language Processing Approach. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; pp. 3492–3497. [Google Scholar]


| BACE | BBBP | ClinTox | Tox21 | |
|---|---|---|---|---|
| D-MPNN | 0.812 | 0.697 | 0.906 | 0.719 |
| RF | 0.851 | 0.719 | 0.783 | 0.724 |
| GCN | 0.818 | 0.676 | 0.907 | 0.688 |
| MLM-5M | 0.793 | 0.701 | 0.341 | 0.762 |
| MLM-10M | 0.729 | 0.696 | 0.349 | 0.748 |
| MLM-77M | 0.735 | 0.698 | 0.239 | 0.749 |
| MTR-5M | 0.734 | 0.742 | 0.552 | 0.834 |
| MTR-10M | 0.783 | 0.733 | 0.601 | 0.827 |
| MTR-77M | 0.799 | 0.728 | 0.563 | 0.817 |
| Our model | 0.808 | 0.683 | 0.914 | 0.781 |
| BACE | Clearance | Delaney | Lipophilicity | |
|---|---|---|---|---|
| D-MPNN | 2.253 | 49.754 | 1.105 | 1.212 |
| RF | 1.318 | 52.077 | 1.741 | 0.962 |
| GCN | 1.645 | 51.227 | 0.885 | 0.781 |
| MLM-5M | 1.451 | 54.601 | 0.946 | 0.986 |
| MLM-10M | 1.611 | 53.859 | 0.961 | 1.009 |
| MLM-77M | 1.509 | 52.754 | 1.025 | 0.987 |
| MTR-5M | 1.477 | 50.154 | 0.874 | 0.758 |
| MTR-10M | 1.417 | 48.934 | 0.858 | 0.744 |
| MTR-77M | 1.363 | 48.515 | 0.889 | 0.798 |
| Our model | 1.481 | 56.063 | 1.066 | 0.908 |
| BACE | BBBP | ClinTox | Tox21 | Clearance | Delaney | Lipophilicity | |
|---|---|---|---|---|---|---|---|
| Classification | 0.033 | 0.025 | 0.013 | 0.026 | N/A | N/A | N/A |
| Regression | 0.173 | N/A | N/A | N/A | 1.341 | 0.125 | 0.037 |
| Acc | F1 | AUC | |
|---|---|---|---|
| XGB | 0.8318 | 0.8412 | N/A |
| ANN | 0.8223 | 0.8445 | N/A |
| With_pre-training | 0.8601 | 0.8721 | 0.9012 |
| Without_pre-training | 0.8553 | 0.8471 | 0.8937 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, T.; Ekenna, C. Molecular Descriptors Property Prediction Using Transformer-Based Approach. Int. J. Mol. Sci. 2023, 24, 11948. https://doi.org/10.3390/ijms241511948
Tran T, Ekenna C. Molecular Descriptors Property Prediction Using Transformer-Based Approach. International Journal of Molecular Sciences. 2023; 24(15):11948. https://doi.org/10.3390/ijms241511948
Chicago/Turabian StyleTran, Tuan, and Chinwe Ekenna. 2023. "Molecular Descriptors Property Prediction Using Transformer-Based Approach" International Journal of Molecular Sciences 24, no. 15: 11948. https://doi.org/10.3390/ijms241511948
APA StyleTran, T., & Ekenna, C. (2023). Molecular Descriptors Property Prediction Using Transformer-Based Approach. International Journal of Molecular Sciences, 24(15), 11948. https://doi.org/10.3390/ijms241511948






