Practical Approaches for the Yeast Saccharomyces cerevisiae Genome Modification
Abstract
:1. Introduction
2. Homologous Recombination and Introduction of Directed Double-Stranded Breaks
3. Yeast Transformation
4. Selectable Markers Used for S. cerevisiae Transformation and Transformant Selection
5. Classical Approaches to Genome Modifications in S. cerevisiae
5.1. One-Step Gene Disruption and Deletion Methods

5.2. Two-Step Gene Deletion Methods
5.3. Gene Modifications Methods
5.4. Yeast Libraries
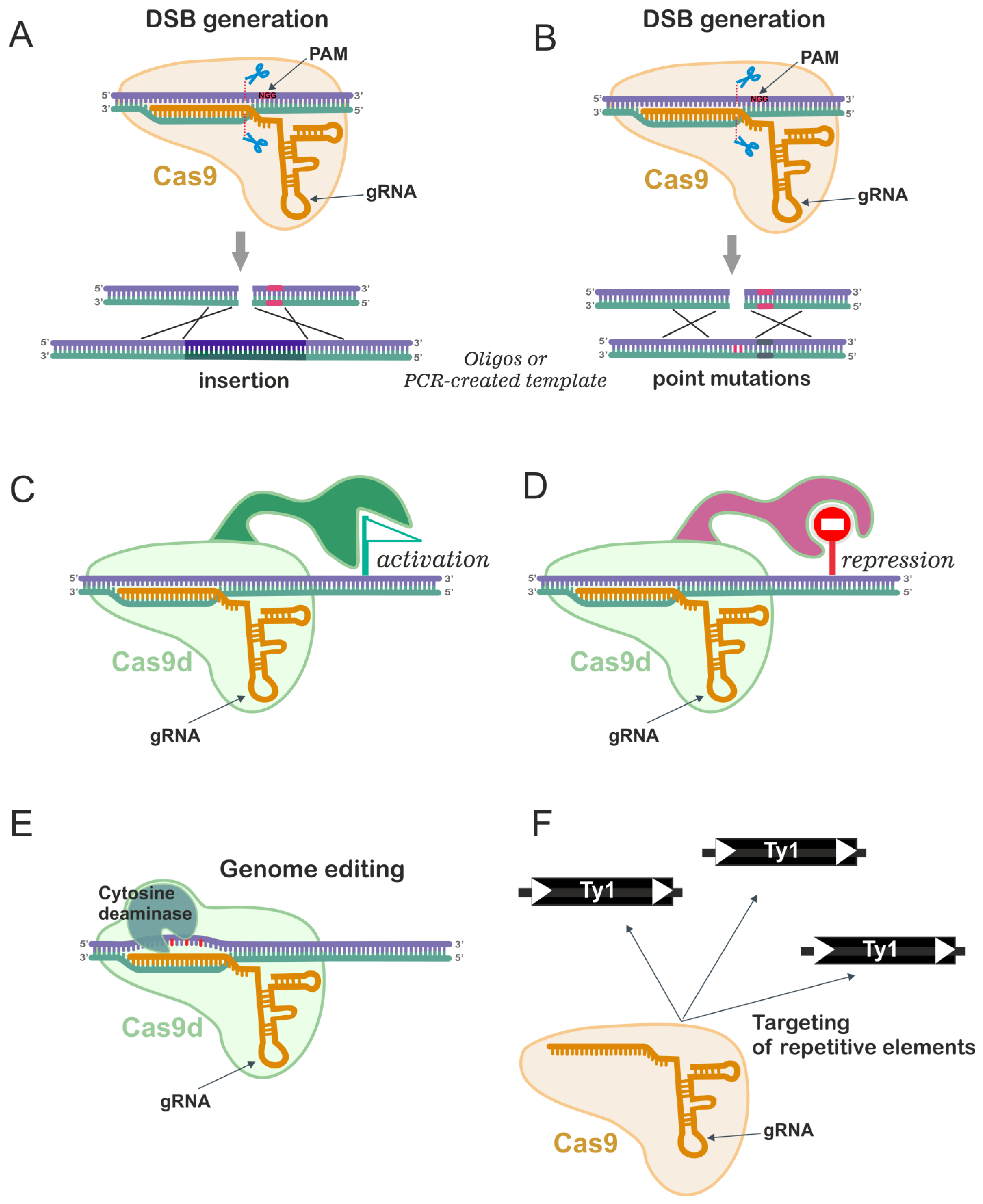
6. CRISPR/Cas9 Genome Editing in Yeast
6.1. The Advantage of CRISPR/Cas9 and Its Usage for Genome Editing
6.2. CRISPR/Cas9 for Multiple Gene Disruptions and Deletions
6.3. CRISPR/Cas9 for Introduction of Point Mutations
6.4. CRISPR/Cas9 in Transcriptional Engineering
6.5. CRISPR/Cas9 in Construction of Yeast Chromosomes
6.6. CRISPR/Cas9 in Metabolic Engineering
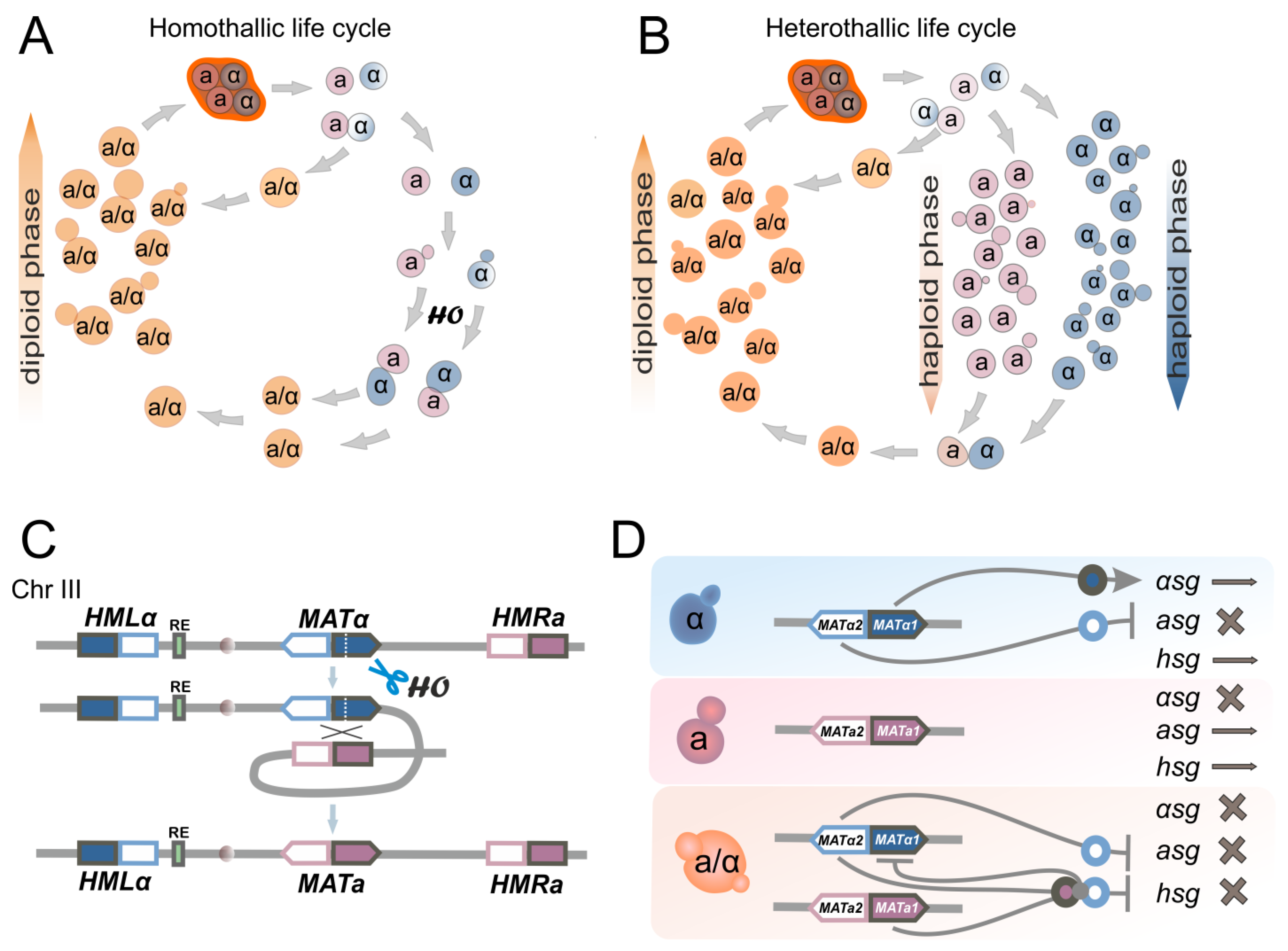
7. Mating-Type Switching in S. cerevisiae
8. Generation of Karyotype Changes in S. cerevisiae
8.1. Ploidy Changes
8.2. Generation of Aneuploid Strains
8.3. Generation of Genome Structural Rearrangements and Karyotype Evolution in Yeast
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fisk, D.G.; Ball, C.A.; Dolinski, K.; Engel, S.R.; Hong, E.L.; Issel-Tarver, L.; Schwartz, K.; Sethuraman, A.; Botstein, D.; Cherry, J.M. Saccharomyces cerevisiae S288C Genome Annotation: A Working Hypothesis. Yeast 2006, 23, 857–865. [Google Scholar] [CrossRef] [Green Version]
- Goffeau, A.; Barrell, G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 Genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef] [Green Version]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The Genomics Resource of Budding Yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef] [Green Version]
- Hirschman, J.E.; Balakrishnan, R.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; Fisk, D.G.; Hong, E.L.; Livstone, M.S.; Nash, R.; et al. Genome Snapshot: A New Resource at the Saccharomyces Genome Database (SGD) Presenting an Overview of the Saccharomyces cerevisiae Genome. Nucleic Acids Res. 2006, 34, D442–D445. [Google Scholar] [CrossRef] [Green Version]
- Haber, J.E. Mating-Type Genes and MAT Switching in Saccharomyces cerevisiae. Genetics 2012, 191, 33–64. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-S.; Haber, J.E. Mating-Type Gene Switching in Saccharomyces cerevisiae. Microbiol. Spectr. 2015, 3, 1–22. [Google Scholar] [CrossRef]
- Harari, Y.; Ram, Y.; Rappoport, N.; Hadany, L.; Kupiec, M. Spontaneous Changes in Ploidy Are Common in Yeast. Curr. Biol. 2018, 28, 825–835.e4. [Google Scholar] [CrossRef] [Green Version]
- Andreychuk, Y.; Zhuk, A.; Tarakhovskaya, E.; Inge-Vechtomov, S.; Stepchenkova, E. Rate of Spontaneous Polyploidization in Haploid Yeast Saccharomyces cerevisiae. Biol. Commun. 2022, 67, 88–96. [Google Scholar] [CrossRef]
- Egorov, A.A.; Alexandrov, A.I.; Urakov, V.N.; Makeeva, D.S.; Edakin, R.O.; Kushchenko, A.S.; Gladyshev, V.N.; Kulakovskiy, I.V.; Dmitriev, S.E. A Standard Knockout Procedure Alters Expression of Adjacent Loci at the Translational Level. Nucleic Acids Res. 2021, 49, 11134–11144. [Google Scholar] [CrossRef]
- Waterman, D.P.; Haber, J.E.; Smolka, M.B. Checkpoint Responses to DNA Double-Strand Breaks. Annu. Rev. Biochem. 2020, 89, 103–133. [Google Scholar] [CrossRef]
- Arbel, M.; Liefshitz, B.; Kupiec, M. How Yeast Cells Deal with Stalled Replication Forks. Curr. Genet. 2020, 66, 911–915. [Google Scholar] [CrossRef]
- Arbel, M.; Liefshitz, B.; Kupiec, M. DNA Damage Bypass Pathways and Their Effect on Mutagenesis in Yeast. FEMS Microbiol. Rev. 2021, 45, fuaa038. [Google Scholar] [CrossRef]
- Jinks-Robertson, S.; Petes, T.D. Mitotic Recombination in Yeast: What We Know and What We Don’t Know. Curr. Opin. Genet. Dev. 2021, 71, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Rawal, C.C.; Liberi, G.; Pellicioli, A. Regulation of DNA Double Strand Breaks Processing: Focus on Barriers. Front. Mol. Biosci. 2019, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Cannavo, E.; Cejka, P. Sae2 Promotes DsDNA Endonuclease Activity within Mre11–Rad50–Xrs2 to Resect DNA Breaks. Nature 2014, 514, 122–125. [Google Scholar] [CrossRef]
- Piazza, A.; Heyer, W.D. Moving Forward One Step Back at a Time: Reversibility during Homologous Recombination. Curr. Genet. 2019, 65, 1333–1340. [Google Scholar] [CrossRef]
- Li, J.; Sun, H.; Huang, Y.; Wang, Y.; Liu, Y.; Chen, X. Pathways and Assays for DNA Double-Strand Break Repair by Homologous Recombination. Acta Biochim. Biophys. Sin. 2019, 51, 879–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramara, J.; Osia, B.; Malkova, A. Break-Induced Replication: The Where, The Why, and The How. Trends Genet. 2018, 34, 518–531. [Google Scholar] [CrossRef]
- Kockler, Z.W.; Osia, B.; Lee, R.; Musmaker, K.; Malkova, A. Repair of DNA Breaks by Break-Induced Replication. Annu. Rev. Biochem. 2021, 90, 165–191. [Google Scholar] [CrossRef]
- Rodgers, K.; McVey, M. Error-Prone Repair of DNA Double-Strand Breaks. J. Cell. Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Schiestl, R.H.; Petes, T.D. Integration of DNA Fragments by Illegitimate Recombination in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1991, 88, 7585–7589. [Google Scholar] [CrossRef] [PubMed]
- Schiestl, R.H.; Dominska, M.; Petes, T.D. Transformation of Saccharomyces cerevisiae with Nonhomologous DNA: Illegitimate Integration of Transforming DNA into Yeast Chromosomes and in Vivo Ligation of Transforming DNA to Mitochondrial DNA Sequences. Mol. Cell. Biol. 1993, 13, 2697–2705. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.; Beach, A.; Li, K.; Haber, J. Rad51-Mediated Double-Strand Break Repair and Mismatch Correction of Divergent Substrates. Nature 2017, 544, 377–380. [Google Scholar] [CrossRef] [Green Version]
- Sajwan, S.; Takasu, Y.; Tamura, T.; Uchino, K.; Sezutsu, H.; Zurovec, M. Efficient Disruption of Endogenous Bombyx Gene by TAL Effector Nucleases. Insect Biochem. Mol. Biol. 2013, 43, 17–23. [Google Scholar] [CrossRef]
- Aouida, M.; Piatek, M.J.; Bangarusamy, D.K.; Mahfouz, M.M. Activities and Specificities of Homodimeric TALENs in Saccharomyces cerevisiae. Curr. Genet. 2014, 60, 61–74. [Google Scholar] [CrossRef]
- Ye, W.; Zhang, W.; Liu, T.; Tan, G.; Li, H.; Huang, Z. Improvement of Ethanol Production in Saccharomyces cerevisiae by High-Efficient Disruption of the ADH2 Gene Using a Novel Recombinant TALEN Vector. Front. Microbiol. 2016, 7, 1067. [Google Scholar] [CrossRef] [Green Version]
- Guha, T.K.; Edgell, D.R. Applications of Alternative Nucleases in the Age of CRISPR/Cas9. Int. J. Mol. Sci. 2017, 18, 2565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinnen, A.; Hicks, J.B.; Fink, G.R. Transformation of Yeast. Proc. Natl. Acad. Sci. USA 1978, 75, 1929–1933. [Google Scholar] [CrossRef]
- Kawai, S.; Hashimoto, W.; Murata, K. Transformation of Saccharomyces cerevisiae and Other Fungi: Methods and Possible Underlying Mechanism. Bioeng. Bugs 2010, 1, 395–403. [Google Scholar] [CrossRef] [Green Version]
- Gietz, R.D.; Woods, R.A. Genetic Transformation of Yeast. Biotechniques 2001, 30, 816–831. [Google Scholar] [CrossRef] [Green Version]
- Gietz, R.D.; Woods, R.A. Transformation of Yeast by Lithium Acetate/Single-Stranded Carrier DNA/Polyethylene Glycol Method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. High-Efficiency Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. Large-Scale High-Efficiency Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2007, 2, 38–41. [Google Scholar] [CrossRef]
- Rivera, A.L.; Magaña-Ortíz, D.; Gómez-Lim, M.; Fernández, F.; Loske, A.M. Physical Methods for Genetic Transformation of Fungi and Yeast. Phys. Life Rev. 2014, 11, 184–203. [Google Scholar] [CrossRef] [PubMed]
- Delorme, E. Transformation of Saccharomyces cerevisiae by Electroporation. Appl. Environ. Microbiol. 1989, 55, 2242. [Google Scholar] [CrossRef]
- Simon, J.R.; McEntee, K. A Rapid and Efficient Procedure for Transformation of Intact Saccharomyces cerevisiae by Electroporation. Biochem. Biophys. Res. Commun. 1989, 164, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Rech, E.L.; Dobson, M.J.; Davey, M.R.; Mulligan, B.J. Introduction of a Yeast Artificial Chromosome Vector into Saccharomyces cerevisiae Cells by Electroporation. Nucleic Acids Res. 1990, 18, 1313. [Google Scholar] [CrossRef]
- Meilhoc, E.; Teissie, J. Electrotransformation of Saccharomyces cerevisiae. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2020; Volume 2050, pp. 187–193. [Google Scholar]
- Grey, M.; Brendel, M. A Ten-Minute Protocol for Transforming Saccharomyces cerevisiae by Electroporation. Curr. Genet. 1992, 22, 335–336. [Google Scholar] [CrossRef]
- Poderyte, M.; Valiūnienė, A.; Ramanavicius, A. Scanning Electrochemical Microscope as a Tool for the Electroporation of Living Yeast Cells. Biosens. Bioelectron. 2022, 205, 114096. [Google Scholar] [CrossRef]
- Bundock, P.; Den Dulk-Ras, A.; Beijersbergen, A.; Hooykaas, P.J.J. Trans-Kingdom T-DNA Transfer from Agrobacterium Tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995, 14, 3206–3214. [Google Scholar] [CrossRef]
- Bundock, P.; Hooykaas, P.J.J. Integration of Agrobacterium Tumefaciens T-DNA in the Saccharomyces cerevisiae Genome by Illegitimate Recombination. Proc. Natl. Acad. Sci. USA 1996, 93, 15272–15275. [Google Scholar] [CrossRef]
- Piers, K.L.; Heath, J.D.; Liang, X.; Stephens, K.M.; Nester, E.W. Agrobacterium tumefaciens-Mediated Transformation of Yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 1613. [Google Scholar] [CrossRef]
- Roushan, M.R.; Shao, S.; Poledri, I.; Hooykaas, P.J.J.; van Heusden, G.P.H. Increased Agrobacterium-mediated Transformation of Saccharomyces cerevisiae after Deletion of the Yeast ADA2 Gene. Lett. Appl. Microbiol. 2022, 74, 228–237. [Google Scholar] [CrossRef]
- Siewers, V. An Overview on Selection Marker Genes for Transformation of Saccharomyces cerevisiae. Methods Mol. Biol. 2022, 2513, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Solis-Escalante, D.; Kuijpers, N.G.A.; Bongaerts, N.; Bolat, I.; Bosman, L.; Pronk, J.T.; Daran, J.M.; Daran-Lapujade, P. AmdSYM, a New Dominant Recyclable Marker Cassette for Saccharomyces cerevisiae. FEMS Yeast Res. 2013, 13, 126–139. [Google Scholar] [CrossRef] [Green Version]
- Kanda, K.; Ishida, T.; Hirota, R.; Ono, S.; Motomura, K.; Ikeda, T.; Kitamura, K.; Kuroda, A. Application of a Phosphite Dehydrogenase Gene as a Novel Dominant Selection Marker for Yeasts. J. Biotechnol. 2014, 182–183, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Leite, F.C.B.; dos Anjos, R.S.G.; Basilio, A.C.M.; Leal, G.F.C.; Simões, D.A.; de Morais, M.A. Construction of Integrative Plasmids Suitable for Genetic Modification of Industrial Strains of Saccharomyces cerevisiae. Plasmid 2013, 69, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Z.Y.; He, X.P.; Liu, N.; Zhang, B.R. New Industrial Brewing Yeast Strains with ILV2 Disruption and LSD1 Expression. Int. J. Food Microbiol. 2008, 123, 18–24. [Google Scholar] [CrossRef]
- Boeke, J.D.; LaCroute, F.; Fink, G.R. A Positive Selection for Mutants Lacking Orotidine-5′-Phosphate Decarboxylase Activity in Yeast: 5-Fluoro-Orotic Acid Resistance. MGG Mol. Gen. Genet. 1984, 197, 345–346. [Google Scholar] [CrossRef]
- Boeke, J.D.; Trueheart, J.; Natsoulis, G.; Fink, G.R. 5-Fluoroorotic Acid as a Selective Agent in Yeast Molecular Genetics. Methods Enzymol. 1987, 154, 164–175. [Google Scholar] [CrossRef]
- Wellington, M.; Rustchenko, E. 5-Fluoro-Orotic Acid Induces Chromosome Alterations in Candida Albicans. Yeast 2005, 22, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Toyn, J.H.; Gunyuzlu, P.; White, W.H.; Thompson, L.A.; Hollis, G.F. A Counterselection for the Tryptophan Pathway in Yeast: 5-Fluoroanthranilic Acid Resistance. Yeast 2000, 16, 553–560. [Google Scholar] [CrossRef]
- Chattoo, B.B.; Sherman, F.; Azubalis, D.A. Selection of Lys2 Mutants of the Yeast Saccharomyces cerevisiae by the Utilization of α-Aminoadipate. Genetics 1979, 93, 51–65. [Google Scholar] [CrossRef]
- Suizu, T.; Iimura, Y.; Gomi, K.; Takahashi, K.; Hara, S.; Yoshizawa, K. L-Canavanine Resistance as a Positive Selectable Marker in Diploid Yeast Transformation through Integral Disruption of the Can1 Gene. Agric. Biol. Chem. 1989, 53, 431–436. [Google Scholar] [CrossRef]
- Nora, L.C.; Westmann, C.A.; Martins-Santana, L.; Alves, L.D.F.; Monteiro, L.M.O.; Guazzaroni, M.E.; Silva-Rocha, R. The Art of Vector Engineering: Towards the Construction of next-Generation Genetic Tools. Microb. Biotechnol. 2019, 12, 125–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shortle, D.; Haber, J.E.; Botstein, D. Lethal Disruption of the Yeast Actin Gene by Integrative DNA Transformation. Science 1982, 217, 371–373. [Google Scholar] [CrossRef]
- Rothstein, R.J. One-Step Gene Disruption in Yeast. Methods Enzymol. 1983, 101, 202–211. [Google Scholar] [CrossRef]
- Rothstein, R. Targeting, Disruption, Replacement, and Allele Rescue: Integrative DNA Transformation in Yeast. Methods Enzymol. 1991, 194, 281–301. [Google Scholar] [CrossRef]
- Johnston, M.; Riles, L.; Hegemann, J.H. Gene Disruption. Methods Enzymol. 2002, 350, 290–315. [Google Scholar] [CrossRef]
- Manivasakam, P.; Weber, S.C.; Mcelver, J.; Schiestl, R.H. Micro-Homology Mediated PCR Targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 1995, 23, 2799. [Google Scholar] [CrossRef] [Green Version]
- Storici, F.; Resnick, M.A. The Delitto Perfetto Approach to in Vivo Site-Directed Mutagenesis and Chromosome Rearrangements with Synthetic Oligonucleotides in Yeast. Methods Enzymol. 2006, 409, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Khmelinskii, A.; Meurer, M.; Duishoev, N.; Delhomme, N.; Knop, M. Seamless Gene Tagging by Endonuclease-Driven Homologous Recombination. PLoS ONE 2011, 6, e23794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yofe, I.; Weill, U.; Meurer, M.; Chuartzman, S.; Zalckvar, E.; Goldman, O.; Ben-Dor, S.; Schütze, C.; Wiedemann, N.; Knop, M.; et al. One Library to Make Them All: Streamlining the Creation of Yeast Libraries via a SWAp-Tag Strategy. Nat. Methods 2016, 13, 371–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baudin, A.; Ozier-Kalogeropoulos, O.; Denouel, A.; Lacroute, F.; Cullin, C. A Simple and Efficient Method for Direct Gene Deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993, 21, 3329–3330. [Google Scholar] [CrossRef] [Green Version]
- Lorenz, M.C.; Muir, R.S.; Lim, E.; McElver, J.; Weber, S.C.; Heitman, J. Gene Disruption with PCR Products in Saccharomyces cerevisiae. Gene 1995, 158, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Hegemann, J.H.; Heick, S.B.; Pöhlmann, J.; Langen, M.M.; Fleig, U. Targeted Gene Deletion in Saccharomyces cerevisiae and Schizosaccharomyces Pombe. Methods Mol. Biol. 2014, 1163, 45–73. [Google Scholar] [CrossRef]
- Wach, A.; Brachat, A.; Pöhlmann, R.; Philippsen, P. New Heterologous Modules for Classical or PCR-Based Gene Disruptions in Saccharomyces cerevisiae. Yeast 1994, 10, 1793–1808. [Google Scholar] [CrossRef]
- Wach, A.; Brachat, A.; Alberti-Segui, C.; Rebischung, C.; Philippsen, P. Heterologous HIS3 Marker and GFP Reporter Modules for PCR-Targeting in Saccharomyces cerevisiae. Yeast 1997, 13, 1065–1075. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A.; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional Modules for Versatile and Economical PCR-Based Gene Deletion and Modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Goldstein, A.L.; McCusker, J.H. Three New Dominant Drug Resistance Cassettes for Gene Disruption in Saccharomyces cerevisiae. Yeast 1999, 15, 1541–1553. [Google Scholar] [CrossRef]
- Hadfield, C.; Jordan, B.E.; Mount, R.C.; Pretorius, G.H.J.; Burak, E. G418-Resistance as a Dominant Marker and Reporter for Gene Expression in Saccharomyces cerevisiae. Curr. Genet. 1990, 18, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional Characterization of the S. cerevisiae Genome by Gene Deletion and Parallel Analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Véronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; André, B.; et al. Functional Profiling of the Saccharomyces cerevisiae Genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Nislow, C. The Yeast Deletion Collection: A Decade of Functional Genomics. Genetics 2014, 197, 451. [Google Scholar] [CrossRef] [Green Version]
- Scherer, S.; Davis, R.W. Replacement of Chromosome Segments with Altered DNA Sequences Constructed in Vitro. Proc. Natl. Acad. Sci. USA 1979, 76, 4951–4955. [Google Scholar] [CrossRef] [PubMed]
- Alani, E.; Cao, L.; Kleckner, N. A Method for Gene Disruption That Allows Repeated Use of URA3 Selection in the Construction of Multiply Disrupted Yeast Strains. Genetics 1987, 116, 541–545. [Google Scholar] [CrossRef]
- Längle-Rouault, F.; Jacobs, E. A Method for Performing Precise Alterations in the Yeast Genome Using a Recycable Selectable Marker. Nucleic Acids Res. 1995, 23, 3079. [Google Scholar] [CrossRef] [Green Version]
- Güldener, U.; Heck, S.; Fiedler, T.; Beinhauer, J.; Hegemann, J.H.; Gießen, J.; Straße, F. A New Efficient Gene Disruption Cassette for Repeated Use in Budding Yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef] [Green Version]
- Gueldener, U.; Heinisch, J.; Koehler, G.J.; Voss, D.; Hegemann, J.H. A Second Set of LoxP Marker Cassettes for Cre-Mediated Multiple Gene Knockouts in Budding Yeast. Nucleic Acids Res. 2002, 30, 23. [Google Scholar] [CrossRef] [Green Version]
- Delneri, D.; Tomlin, G.C.; Wixon, J.L.; Hutter, A.; Sefton, M.; Louis, E.J.; Oliver, S.G. Exploring Redundancy in the Yeast Genome: An Improved Strategy for Use of the Cre–LoxP System. Gene 2000, 252, 127–135. [Google Scholar] [CrossRef]
- Carter, Z.; Delneri, D. New Generation of LoxP-Mutated Deletion Cassettes for the Genetic Manipulation of Yeast Natural Isolates. Yeast 2010, 27, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Salmon, K.; Shen, M.W.Y.; Aeling, K.A.; Ito, E.; Irwin, B.; Tran, U.P.C.; Hatfield, G.W.; Da Silva, N.A.; Sandmeyer, S. A Vector Set for Systematic Metabolic Engineering in Saccharomyces cerevisiae. Yeast 2011, 28, 123–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.N.; Masison, D.; Eisenberg, E.; Greene, L.E. Application of the FLP/FRT System for Conditional Gene Deletion in Yeast Saccharomyces cerevisiae. Yeast 2011, 28, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Davidson, J.F.; Schiestl, R.H. Mis-Targeting of Multiple Gene Disruption Constructs Containing HisG. Curr. Genet. 2000, 38, 188–190. [Google Scholar] [CrossRef] [PubMed]
- Delneri, D.; Colson, I.; Grammenoudi, S.; Roberts, I.N.; Louis, E.J.; Oliver, S.G. Engineering Evolution to Study Speciation in Yeasts. Nature 2003, 422, 68–72. [Google Scholar] [CrossRef]
- Storici, F.; Lewis, L.K.; Resnick, M.A. In Vivo Site-Directed Mutagenesis Using Oligonucleotides. Nat. Biotechnol. 2001, 19, 773–776. [Google Scholar] [CrossRef]
- Storici, F.; Durham, C.L.; Gordenin, D.A.; Resnick, M.A. Chromosomal Site-Specific Double-Strand Breaks Are Efficiently Targeted for Repair by Oligonucleotides in Yeast. Proc. Natl. Acad. Sci. USA 2003, 100, 14994–14999. [Google Scholar] [CrossRef]
- Carvalho, Â.; Pereira, F.; Johansson, B. The MX4blaster Cassette: Repeated and Clean Saccharomyces cerevisiae Genome Modification Using the Genome-Wide Deletion Collection. FEMS Yeast Res. 2013, 13, 711–719. [Google Scholar] [CrossRef] [Green Version]
- Mnaimneh, S.; Davierwala, A.P.; Haynes, J.; Moffat, J.; Peng, W.T.; Zhang, W.; Yang, X.; Pootoolal, J.; Chua, G.; Lopez, A.; et al. Exploration of Essential Gene Functions via Titratable Promoter Alleles. Cell 2004, 118, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Janke, C.; Magiera, M.M.; Rathfelder, N.; Taxis, C.; Reber, S.; Maekawa, H.; Moreno-Borchart, A.; Doenges, G.; Schwob, E.; Schiebel, E.; et al. A Versatile Toolbox for PCR-Based Tagging of Yeast Genes: New Fluorescent Proteins, More Markers and Promoter Substitution Cassettes. Yeast 2004, 21, 947–962. [Google Scholar] [CrossRef]
- Van Driessche, B.; Tafforeau, L.; Hentges, P.; Carr, A.M.; Vandenhaute, J. Additional Vectors for PCR-Based Gene Tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe Using Nourseothricin Resistance. Yeast 2005, 22, 1061–1068. [Google Scholar] [CrossRef]
- Kaufmann, A.; Knop, M. Genomic Promoter Replacement Cassettes to Alter Gene Expression in the Yeast Saccharomyces cerevisiae. Methods Mol. Biol. 2011, 765, 275–294. [Google Scholar] [CrossRef]
- Knop, M.; Siegers, K.; Pereira, G.; Zachariae, W.; Winsor, B.; Nasmyth, K.; Schiebel, E. Epitope Tagging of Yeast Genes Using a PCR-Based Strategy: More Tags and Improved Practical Routines. Yeast 1999, 15, 963–972. [Google Scholar] [CrossRef]
- Gauss, R.; Trautwein, M.; Sommer, T.; Spang, A. New Modules for the Repeated Internal and N-Terminal Epitope Tagging of Genes in Saccharomyces cerevisiae. Yeast 2005, 22, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Khmelinskii, A.; Keller, P.J.; Bartosik, A.; Meurer, M.; Barry, J.D.; Mardin, B.R.; Kaufmann, A.; Trautmann, S.; Wachsmuth, M.; Pereira, G.; et al. Tandem Fluorescent Protein Timers for in Vivo Analysis of Protein Dynamics. Nat. Biotechnol. 2012, 30, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.H.Y.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Pagé, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.V.; Bussey, H.; et al. Systematic Genetic Analysis with Ordered Arrays of Yeast Deletion Mutants. Science 2001, 294, 2364–2368. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.H.Y.; Boone, C. 16 High-Throughput Strain Construction and Systematic Synthetic Lethal Screening in Saccharomyces cerevisiae. Methods Microbiol. 2007, 36, 369–707. [Google Scholar] [CrossRef]
- Baryshnikova, A.; Costanzo, M.; Dixon, S.; Vizeacoumar, F.J.; Myers, C.L.; Andrews, B.; Boone, C. Synthetic Genetic Array (SGA) Analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe, 2nd ed.; Academic Press: San Diego, CA, USA; Burlington, MA, USA; London, UK, 2010; Volume 470. [Google Scholar]
- Norman, K.L.; Kumar, A. Mutant Power: Using Mutant Allele Collections for Yeast Functional Genomics. Brief. Funct. Genom. 2016, 15, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Tarassov, K.; Messier, V.; Landry, C.R.; Radinovic, S.; Molina, M.M.S.; Shames, I.; Malitskaya, Y.; Vogel, J.; Bussey, H.; Michnick, S.W. An in Vivo Map of the Yeast Protein Interactome. Science 2008, 320, 1465–1470. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Snyder, M. Genome-Wide Transposon Mutagenesis in Yeast. Curr. Protoc. Mol. Biol. 2000, 51, 13.3. 1–13.3. 15. [Google Scholar] [CrossRef]
- Bidlingmaier, S.; Snyder, M. Large-Scale Identification of Genes Important for Apical Growth in Saccharomyces cerevisiae by Directed Allele Replacement Technology (DART) Screening. Funct. Integr. Genom. 2002, 1, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Ross-Macdonald, P.; Sheehan, A.; Roeder, G.S.; Snyder, M. A Multipurpose Transposon System for Analyzing Protein Production, Localization, and Function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1997, 94, 190. [Google Scholar] [CrossRef]
- Ross-Macdonald, P.; Coelho, P.S.R.; Roemer, T.; Agarwal, S.; Kumar, A.; Jansen, R.; Cheung, K.H.; Sheehan, A.; Symonlatis, D.; Umansky, L.; et al. Large-Scale Analysis of the Yeast Genome by Transposon Tagging and Gene Disruption. Nature 1999, 402, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Burns, N.; Grimwade, B.; Ross-Macdonald, P.B.; Choi, E.Y.; Finberg, K.; Roeder, G.S.; Snyder, M. Large-Scale Analysis of Gene Expression, Protein Localization, and Gene Disruption in Saccharomyces cerevisiae. Genes Dev. 1994, 8, 1087–1105. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Seringhaus, M.; Biery, M.C.; Sarnovsky, R.J.; Umansky, L.; Piccirillo, S.; Heidtman, M.; Cheung, K.H.; Dobry, C.J.; Gerstein, M.B.; et al. Large-Scale Mutagenesis of the Yeast Genome Using a Tn7-Derived Multipurpose Transposon. Genome Res. 2004, 14, 1975–1986. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Agarwal, S.; Heyman, J.A.; Matson, S.; Heidtman, M.; Piccirillo, S.; Umansky, L.; Drawid, A.; Jansen, R.; Liu, Y.; et al. Subcellular Localization of the Yeast Proteome. Genes Dev. 2002, 16, 707. [Google Scholar] [CrossRef] [Green Version]
- Meurer, M.; Duan, Y.; Sass, E.; Kats, I.; Herbst, K.; Buchmuller, B.C.; Dederer, V.; Huber, F.; Kirrmaier, D.; Štefl, M.; et al. Genome-Wide C-SWAT Library for High-Throughput Yeast Genome Tagging. Nat. Methods 2018, 15, 598–600. [Google Scholar] [CrossRef]
- Weill, U.; Yofe, I.; Sass, E.; Stynen, B.; Davidi, D.; Natarajan, J.; Ben-Menachem, R.; Avihou, Z.; Goldman, O.; Harpaz, N.; et al. Genome-Wide SWAp-Tag Yeast Libraries for Proteome Exploration. Nat. Methods 2018, 15, 617–622. [Google Scholar] [CrossRef]
- Antony, J.S.; Hinz, J.M.; Wyrick, J.J. Tips, Tricks, and Potential Pitfalls of CRISPR Genome Editing in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 2022, 10, 950. [Google Scholar] [CrossRef]
- Da Silva, N.A.; Srikrishnan, S. Introduction and Expression of Genes for Metabolic Engineering Applications in Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 197–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solis-Escalante, D.; Kuijpers, N.G.A.; van der Linden, F.H.; Pronk, J.T.; Daran, J.M.; Daran-Lapujade, P. Efficient Simultaneous Excision of Multiple Selectable Marker Cassettes Using I-SceI-Induced Double-Strand DNA Breaks in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 741–754. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA Maturation by Trans-Encoded Small RNA and Host Factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-CrRNA Ribonucleoprotein Complex Mediates Specific DNA Cleavage for Adaptive Immunity in Bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef] [PubMed]
- Dicarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome Engineering in Saccharomyces cerevisiae Using CRISPR-Cas Systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef] [Green Version]
- Jessop-Fabre, M.M.; Jakočiūnas, T.; Stovicek, V.; Dai, Z.; Jensen, M.K.; Keasling, J.D.; Borodina, I. EasyClone-MarkerFree: A Vector Toolkit for Marker-Less Integration of Genes into Saccharomyces cerevisiae via CRISPR-Cas9. Biotechnol. J. 2016, 11, 1110–1117. [Google Scholar] [CrossRef] [Green Version]
- Stovicek, V.; Holkenbrink, C.; Borodina, I. CRISPR/Cas System for Yeast Genome Engineering: Advances and Applications. FEMS Yeast Res. 2017, 17, fox030. [Google Scholar] [CrossRef] [Green Version]
- Mans, R.; van Rossum, H.M.; Wijsman, M.; Backx, A.; Kuijpers, N.G.A.; van den Broek, M.; Daran-Lapujade, P.; Pronk, J.T.; van Maris, A.J.A.; Daran, J.M.G. CRISPR/Cas9: A Molecular Swiss Army Knife for Simultaneous Introduction of Multiple Genetic Modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov004. [Google Scholar] [CrossRef] [Green Version]
- Utomo, J.C.; Hodgins, C.L.; Ro, D.K. Multiplex Genome Editing in Yeast by CRISPR/Cas9—A Potent and Agile Tool to Reconstruct Complex Metabolic Pathways. Front. Plant Sci. 2021, 12, 719148. [Google Scholar] [CrossRef]
- Laughery, M.F.; Wyrick, J.J. Simple CRISPR-Cas9 Genome Editing in Saccharomyces cerevisiae. Curr. Protoc. Mol. Biol. 2019, 129, e110. [Google Scholar] [CrossRef]
- Bao, Z.; Xiao, H.; Liang, J.; Zhang, L.; Xiong, X.; Sun, N.; Si, T.; Zhao, H. Homology-Integrated CRISPR-Cas (HI-CRISPR) System for One-Step Multigene Disruption in Saccharomyces cerevisiae. ACS Synth. Biol. 2015, 4, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Ronda, C.; Maury, J.; Jakočiunas, T.; Baallal Jacobsen, S.A.; Germann, S.M.; Harrison, S.J.; Borodina, I.; Keasling, J.D.; Jensen, M.K.; Nielsen, A.T. CrEdit: CRISPR Mediated Multi-Loci Gene Integration in Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maestroni, L.; Butti, P.; Senatore, V.G.; Branduardi, P. PCEC-Red: A New Vector for Easier and Faster CRISPR-Cas9 Genome Editing in Saccharomyces cerevisiae. FEMS Yeast Res. 2023, 23, foad002. [Google Scholar] [CrossRef] [PubMed]
- Verwaal, R.; Buiting-Wiessenhaan, N.; Dalhuijsen, S.; Roubos, J.A. CRISPR/Cpf1 Enables Fast and Simple Genome Editing of Saccharomyces cerevisiae. Yeast 2018, 35, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.H.; Wang, F.Q.; Wei, D.Z. Self-Cloning CRISPR/Cpf1 Facilitated Genome Editing in Saccharomyces cerevisiae. Bioresour. Bioprocess. 2018, 5, 36. [Google Scholar] [CrossRef] [Green Version]
- Zetsche, B.; Heidenreich, M.; Mohanraju, P.; Fedorova, I.; Kneppers, J.; Degennaro, E.M.; Winblad, N.; Choudhury, S.R.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Multiplex Gene Editing by CRISPR-Cpf1 Using a Single CrRNA Array. Nat. Biotechnol. 2017, 35, 31–34. [Google Scholar] [CrossRef] [Green Version]
- Ryan, O.W.; Skerker, J.M.; Maurer, M.J.; Li, X.; Tsai, J.C.; Poddar, S.; Lee, M.E.; DeLoache, W.; Dueber, J.E.; Arkin, A.P.; et al. Selection of Chromosomal DNA Libraries Using a Multiplex CRISPR System. Elife 2014, 3, e03703. [Google Scholar] [CrossRef]
- Generoso, W.C.; Gottardi, M.; Oreb, M.; Boles, E. Simplified CRISPR-Cas Genome Editing for Saccharomyces cerevisiae. J. Microbiol. Methods 2016, 127, 203–205. [Google Scholar] [CrossRef]
- Jakočinas, T.; Bonde, I.; Herrgård, M.; Harrison, S.J.; Kristensen, M.; Pedersen, L.E.; Jensen, M.K.; Keasling, J.D. Multiplex Metabolic Pathway Engineering Using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab. Eng. 2015, 28, 213–222. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Wang, Z.; Zhang, Y.; Shi, S.; Nielsen, J.; Liu, Z. A GRNA-TRNA Array for CRISPR-Cas9 Based Rapid Multiplexed Genome Editing in Saccharomyces cerevisiae. Nat. Commun. 2019, 10, 1053. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.C.; Kong, I.I.; Kim, H.; Liu, J.J.; Cate, J.H.D.; Jin, Y.S. Construction of a Quadruple Auxotrophic Mutant of an Industrial Polyploid Saccharomyces cerevisiae Strain by Using RNA-Guided Cas9 Nuclease. Appl. Environ. Microbiol. 2014, 80, 7694–7701. [Google Scholar] [CrossRef] [Green Version]
- Lian, J.; Bao, Z.; Hu, S.; Zhao, H. Engineered CRISPR/Cas9 System for Multiplex Genome Engineering of Polyploid Industrial Yeast Strains. Biotechnol. Bioeng. 2018, 115, 1630–1635. [Google Scholar] [CrossRef]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted Nucleotide Editing Using Hybrid Prokaryotic and Vertebrate Adaptive Immune Systems. Science 2016, 353, aaf8729. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Schubert, O.T.; Bloom, J.S.; Sadhu, M.J.; Kruglyak, L. Genome-Wide Base Editor Screen Identifies Regulators of Protein Abundance in Yeast. Elife 2022, 11, e79525. [Google Scholar] [CrossRef]
- Bao, Z.; HamediRad, M.; Xue, P.; Xiao, H.; Tasan, I.; Chao, R.; Liang, J.; Zhao, H. Genome-Scale Engineering of Saccharomyces cerevisiae with Single-Nucleotide Precision. Nat. Biotechnol. 2018, 36, 505–508. [Google Scholar] [CrossRef]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR Interference (CRISPRi) for Sequence-Specific Control of Gene Expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Farzadfard, F.; Perli, S.D.; Lu, T.K. Tunable and Multifunctional Eukaryotic Transcription Based on CRISPR/Cas. ACS Synth. Biol. 2013, 2, 604. [Google Scholar] [CrossRef]
- Howe, F.S.; Russell, A.; Lamstaes, A.R.; El-Sagheer, A.; Nair, A.; Brown, T.; Mellor, J. CRISPRi Is Not Strand-Specific at All Loci and Redefines the Transcriptional Landscape. Elife 2017, 6, e29878. [Google Scholar] [CrossRef]
- Li, P.; Fu, X.; Zhang, L.; Li, S. CRISPR/Cas-Based Screening of a Gene Activation Library in Saccharomyces cerevisiae Identifies a Crucial Role of OLE1 in Thermotolerance. Microb. Biotechnol. 2019, 12, 1154–1163. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.; Skrekas, C.; Hedin, A.; Sánchez, B.J.; Siewers, V.; Nielsen, J.; David, F. Model-Assisted Fine-Tuning of Central Carbon Metabolism in Yeast through DCas9-Based Regulation. ACS Synth. Biol. 2019, 8, 2457–2463. [Google Scholar] [CrossRef] [Green Version]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.R.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [Green Version]
- Vanegas, K.G.; Lehka, B.J.; Mortensen, U.H. SWITCH: A Dynamic CRISPR Tool for Genome Engineering and Metabolic Pathway Control for Cell Factory Construction in Saccharomyces cerevisiae. Microb. Cell Fact. 2017, 16, 25. [Google Scholar] [CrossRef] [Green Version]
- Lian, J.; Hamedirad, M.; Hu, S.; Zhao, H. Combinatorial Metabolic Engineering Using an Orthogonal Tri-Functional CRISPR System. Nat. Commun. 2017, 8, 1688. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.X.; Li, B.Z.; Mitchell, L.A.; Wu, Y.; Qi, X.; Jin, Z.; Jia, B.; Wang, X.; Zeng, B.X.; Liu, H.M.; et al. “Perfect” Designer Chromosome v and Behavior of a Ring Derivative. Science 2017, 355, eaaf4704. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Sun, X.; Cormack, B.P.; Boeke, J.D. Karyotype Engineering by Chromosome Fusion Leads to Reproductive Isolation in Yeast. Nature 2018, 560, 392–396. [Google Scholar] [CrossRef]
- Shao, Y.; Lu, N.; Wu, Z.; Cai, C.; Wang, S.; Zhang, L.L.; Zhou, F.; Xiao, S.; Liu, L.; Zeng, X.; et al. Creating a Functional Single-Chromosome Yeast. Nature 2018, 560, 331–335. [Google Scholar] [CrossRef]
- Shao, Y.; Lu, N.; Cai, C.; Zhou, F.; Wang, S.; Zhao, Z.; Zhao, G.; Zhou, J.Q.; Xue, X.; Qin, Z. A Single Circular Chromosome Yeast. Cell Res. 2019, 29, 87–89. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Lu, N.; Xue, X.; Qin, Z. Creating Functional Chromosome Fusions in Yeast with CRISPR–Cas9. Nat. Protoc. 2019, 14, 2521–2545. [Google Scholar] [CrossRef]
- Otto, M.; Liu, D.; Siewers, V. Saccharomyces cerevisiae as a Heterologous Host for Natural Products. Methods Mol. Biol. 2022, 2489, 333–367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.X.; Wang, L.R.; Xu, Y.S.; Jiang, W.T.; Shi, T.Q.; Sun, X.M.; Huang, H. Recent Advances in the Application of Multiplex Genome Editing in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2021, 105, 3873–3882. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.W.; Kang, W.K.; Jang, H.W.; Turner, T.L.; Kim, H.J. CAR1 Deletion by CRISPR/Cas9 Reduces Formation of Ethyl Carbamate from Ethanol Fermentation by Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2016, 43, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Yuan, X.; Liang, L.; Fang, J.; Chen, Y.; He, W.; Xue, T. Using CRISPR/Cas9 for Multiplex Genome Engineering to Optimize the Ethanol Metabolic Pathway in Saccharomyces cerevisiae. Biochem. Eng. J. 2019, 145, 120–126. [Google Scholar] [CrossRef]
- Horwitz, A.A.; Walter, J.M.; Schubert, M.G.; Kung, S.H.; Hawkins, K.; Platt, D.M.; Hernday, A.D.; Mahatdejkul-Meadows, T.; Szeto, W.; Chandran, S.S.; et al. Efficient Multiplexed Integration of Synergistic Alleles and Metabolic Pathways in Yeasts via CRISPR-Cas. Cell Syst. 2015, 1, 88–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, N.B.; Strucko, T.; Kildegaard, K.R.; David, F.; Maury, J.; Mortensen, U.H.; Forster, J.; Nielsen, J.; Borodina, I. EasyClone: Method for Iterative Chromosomal Integration of Multiple Genes in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 238–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stovicek, V.; Borja, G.M.; Forster, J.; Borodina, I. EasyClone 2.0: Expanded Toolkit of Integrative Vectors for Stable Gene Expression in Industrial Saccharomyces cerevisiae Strains. J. Ind. Microbiol. Biotechnol. 2015, 42, 1519. [Google Scholar] [CrossRef] [Green Version]
- Qi, M.; Zhang, B.; Jiang, L.; Xu, S.; Dong, C.; Du, Y.L.; Zhou, Z.; Huang, L.; Xu, Z.; Lian, J. PCR & Go: A Pre-Installed Expression Chassis for Facile Integration of Multi-Gene Biosynthetic Pathways. Front. Bioeng. Biotechnol. 2021, 8, 1490. [Google Scholar] [CrossRef]
- Shi, S.; Liang, Y.; Zhang, M.M.; Ang, E.L.; Zhao, H. A Highly Efficient Single-Step, Markerless Strategy for Multi-Copy Chromosomal Integration of Large Biochemical Pathways in Saccharomyces cerevisiae. Metab. Eng. 2016, 33, 19–27. [Google Scholar] [CrossRef]
- Shi, S.; Liang, Y.; Ang, E.L.; Zhao, H. Delta Integration CRISPR-Cas (Di-CRISPR) in Saccharomyces cerevisiae. Methods Mol. Biol. 2019, 1927, 73–91. [Google Scholar] [CrossRef]
- Huang, S.; Geng, A. High-Copy Genome Integration of 2,3-Butanediol Biosynthesis Pathway in Saccharomyces cerevisiae via in Vivo DNA Assembly and Replicative CRISPR-Cas9 Mediated Delta Integration. J. Biotechnol. 2020, 310, 13–20. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Wang, Y.Z.; Xu, Y.S.; Sun, X.M.; Huang, H. Developing GDi-CRISPR System for Multi-Copy Integration in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 2021, 193, 2379–2388. [Google Scholar] [CrossRef]
- Hanasaki, M.; Masumoto, H. CRISPR/Transposon Gene Integration (CRITGI) Can Manage Gene Expression in a Retrotransposon-Dependent Manner. Sci. Rep. 2019, 9, 15300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, L.; Sui, Y.; Tang, X.X.; McGinty, R.J.; Liang, X.Z.; Dominska, M.; Zhang, K.; Mirkin, S.M.; Zheng, D.Q.; Petes, T.D. Shuffling the Yeast Genome Using CRISPR/Cas9-Generated DSBs That Target the Transposable Ty1 Elements. PLoS Genet. 2023, 19, e1010590. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Deng, A.; Zhang, Y.; Liu, S.; Liang, Y.; Bai, H.; Cui, D.; Qiu, Q.; Shang, X.; Yang, Z.; et al. Efficient CRISPR-Cas9 Mediated Multiplex Genome Editing in Yeasts. Biotechnol. Biofuels 2018, 11, 277. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; Charlop-Powers, Z.; Brady, S.F. Multiplexed CRISPR/Cas9- and TAR-Mediated Promoter Engineering of Natural Product Biosynthetic Gene Clusters in Yeast. ACS Synth. Biol. 2016, 5, 1002–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct Cloning and Refactoring of a Silent Lipopeptide Biosynthetic Gene Cluster Yields the Antibiotic Taromycin A. Proc. Natl. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef]
- Sasaki, Y.; Mitsui, R.; Yamada, R.; Ogino, H. Secretory Overexpression of the Endoglucanase by Saccharomyces cerevisiae via CRISPR-δ-Integration and Multiple Promoter Shuffling. Enzyme Microb. Technol. 2019, 121, 17–22. [Google Scholar] [CrossRef]
- Nickoloff, J.A.; Chen, E.Y.; Heffron, F. A 24-Base-Pair DNA Sequence from the MAT Locus Stimulates Intergenic Recombination in Yeast. Proc. Natl. Acad. Sci. USA 1986, 83, 7831–7835. [Google Scholar] [CrossRef]
- Jensen, R.; Sprague, G.F.; Herskowitz, I. Regulation of Yeast Mating-Type Interconversion: Feedback Control of HO Gene Expression by the Mating-Type Locus. Proc. Natl. Acad. Sci. USA 1983, 80, 3035–3039. [Google Scholar] [CrossRef]
- Klar, A.J.S.; Hicks, J.B.; Strathern, J.N. Directionality of Yeast Mating-Type Interconversion. Cell 1982, 28, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Bobola, N.; Jansen, R.P.; Shin, T.H.; Nasmyth, K. Asymmetric Accumulation of Ash1p in Postanaphase Nuclei Depends on a Myosin and Restricts Yeast Mating-Type Switching to Mother Cells. Cell 1996, 84, 699–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herskowitz, I.; Jensen, R.E. Putting the HO Gene to Work: Practical Uses for Mating-Type Switching. Methods Enzymol. 1991, 194, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Sandell, L.L.; Zakian, V.A. Loss of a Yeast Telomere: Arrest, Recovery, and Chromosome Loss. Cell 1993, 75, 729–739. [Google Scholar] [CrossRef]
- Russell, D.W.; Jensen, R.; Zoller, M.J.; Burke, J.; Errede, B.; Smith, M.; Herskowitz, I. Structure of the Saccharomyces cerevisiae HO Gene and Analysis of Its Upstream Regulatory Region. Mol. Cell. Biol. 1986, 6, 4281–4294. [Google Scholar] [CrossRef]
- Nickoloff, J.A.; Singer, J.D.; Hoekstra, M.F.; Heffron, F. Double-Strand Breaks Stimulate Alternative Mechanisms of Recombination Repair. J. Mol. Biol. 1989, 207, 527–541. [Google Scholar] [CrossRef]
- Hou, L.; Li, X.; Wang, C.; Cao, X.; Wang, H. Construction of Ploidy Series of Saccharomyces cerevisiae by the Plasmid YCplac33-GHK. J. Ind. Microbiol. Biotechnol. 2013, 40, 393–397. [Google Scholar] [CrossRef]
- Gnügge, R.; Symington, L.S. Efficient DNA Double-Strand Break Formation at Single or Multiple Defined Sites in the Saccharomyces cerevisiae Genome. Nucleic Acids Res. 2020, 48, e115. [Google Scholar] [CrossRef]
- Rattray, A.; Santoyo, G.; Shafer, B.; Strathern, J.N. Elevated Mutation Rate during Meiosis in Saccharomyces cerevisiae. PLoS Genet. 2015, 11, e1004910. [Google Scholar] [CrossRef] [Green Version]
- Mansour, O.; Morciano, L.; Zion, K.; Elgrabli, R.; Zenvirth, D.; Simchen, G.; Arbel-Eden, A. Timing of Appearance of New Mutations during Yeast Meiosis and Their Association with Recombination. Curr. Genet. 2020, 66, 577–592. [Google Scholar] [CrossRef]
- Hiraoka, M.; Watanabe, K.I.; Umezu, K.; Maki, H. Spontaneous Loss of Heterozygosity in Diploid Saccharomyces cerevisiae Cells. Genetics 2000, 156, 1531–1548. [Google Scholar] [CrossRef]
- Xie, Z.X.; Mitchell, L.A.; Liu, H.M.; Li, B.Z.; Liu, D.; Agmon, N.; Wu, Y.; Li, X.; Zhou, X.; Li, B.; et al. Rapid and Efficient CRISPR/Cas9-Based Mating-Type Switching of Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2018, 8, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Krogerus, K.; Fletcher, E.; Rettberg, N.; Gibson, B.; Preiss, R. Efficient Breeding of Industrial Brewing Yeast Strains Using CRISPR/Cas9-Aided Mating-Type Switching. Appl. Microbiol. Biotechnol. 2021, 105, 8359–8376. [Google Scholar] [CrossRef] [PubMed]
- Haber, J.E.; Savage, W.T.; Raposa, S.M.; Weiffenbach, B.; Rowe, L.B. Mutations Preventing Transpositions of Yeast Mating Type Alleles. Proc. Natl. Acad. Sci. USA 1980, 77, 2824–2828. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N. Apparent Diameter and Cell Density of Yeast Strains with Different Ploidy. Sci. Rep. 2023, 13, 1513. [Google Scholar] [CrossRef] [PubMed]
- Sharp, N.P.; Sandell, L.; James, C.G.; Otto, S.P. The Genome-Wide Rate and Spectrum of Spontaneous Mutations Differ between Haploid and Diploid Yeast. Proc. Natl. Acad. Sci. USA 2018, 115, E5046–E5055. [Google Scholar] [CrossRef] [Green Version]
- Lada, A.G.; Stepchenkova, E.I.; Waisertreiger, I.S.R.; Noskov, V.N.; Dhar, A.; Eudy, J.D.; Boissy, R.J.; Hirano, M.; Rogozin, I.B.; Pavlov, Y.I. Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase. PLoS Genet. 2013, 9, e1003736. [Google Scholar] [CrossRef] [Green Version]
- Andalis, A.A.; Storchova, Z.; Styles, C.; Galitski, T.; Pellman, D.; Fink, G.R. Defects Arising from Whole-Genome Duplications in Saccharomyces cerevisiae. Genetics 2004, 167, 1109–1121. [Google Scholar] [CrossRef] [Green Version]
- Mayer, V.W.; Aguilera, A. High Levels of Chromosome Instability in Polyploids of Saccharomyces cerevisiae. Mutat. Res. 1990, 231, 177–186. [Google Scholar] [CrossRef]
- Storchová, Z.; Breneman, A.; Cande, J.; Dunn, J.; Burbank, K.; O’Toole, E.; Pellman, D. Genome-Wide Genetic Analysis of Polyploidy in Yeast. Nature 2006, 443, 541–547. [Google Scholar] [CrossRef]
- Fukuda, N.; Honda, S. Artificial Mating-Type Conversion and Repetitive Mating for Polyploid Generation. ACS Synth. Biol. 2018, 7, 1413–1423. [Google Scholar] [CrossRef]
- Oya, K.; Matsuura, A. Artificial Control of Mating Type and Repeated Mating to Produce Polyploid Cells in Saccharomyces cerevisiae. STAR Protoc. 2023, 4, 102085. [Google Scholar] [CrossRef] [PubMed]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome Evolution across 1,011 Saccharomyces cerevisiae Isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [Green Version]
- Wilkening, S.; Tekkedil, M.M.; Lin, G.; Fritsch, E.S.; Wei, W.; Gagneur, J.; Lazinski, D.W.; Camilli, A.; Steinmetz, L.M. Genotyping 1000 Yeast Strains by Next-Generation Sequencing. BMC Genom. 2013, 14, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasch, A.P.; Hose, J.; Newton, M.A.; Sardi, M.; Yong, M.; Wang, Z. Further Support for Aneuploidy Tolerance in Wild Yeast and Effects of Dosage Compensation on Gene Copy-Number Evolution. Elife 2016, 5, e14409. [Google Scholar] [CrossRef] [PubMed]
- Scopel, E.F.C.; Hose, J.; Bensasson, D.; Gasch, A.P. Genetic Variation in Aneuploidy Prevalence and Tolerance across Saccharomyces cerevisiae Lineages. Genetics 2021, 217, iyab015. [Google Scholar] [CrossRef]
- Hose, J.; Escalante, L.E.; Clowers, K.J.; Dutcher, H.A.; Robinson, D.; Bouriakov, V.; Coon, J.J.; Shishkova, E.; Gasch, A.P. The Genetic Basis of Aneuploidy Tolerance in Wild Yeast. Elife 2020, 9, e52063. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.; Stelkens, R. Aneuploidy in Yeast: Segregation Error or Adaptation Mechanism? Yeast 2019, 36, 525–539. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.J.; Nelliat, A. A Double-Edged Sword: Aneuploidy Is a Prevalent Strategy in Fungal Adaptation. Genes 2019, 10, 787. [Google Scholar] [CrossRef] [Green Version]
- Barney, J.B.; Chandrashekarappa, D.G.; Soncini, S.R.; Schmidt, M.C. Drug Resistance in Diploid Yeast Is Acquired through Dominant Alleles, Haploinsufficiency, Gene Duplication and Aneuploidy. PLoS Genet. 2021, 17, e1009800. [Google Scholar] [CrossRef]
- Fukuda, N.; Honda, S.; Fujiwara, M.; Yoshimura, Y.; Nakamura, T. Polyploid Engineering by Increasing Mutant Gene Dosage in Yeasts. Microb. Biotechnol. 2021, 14, 979–992. [Google Scholar] [CrossRef]
- Torres, E.M.; Sokolsky, T.; Tucker, C.M.; Chan, L.Y.; Boselli, M.; Dunham, M.J.; Amon, A. Effects of Aneuploidy on Cellular Physiology and Cell Division in Haploid Yeast. Science 2007, 317, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Oromendia, A.B.; Dodgson, S.E.; Amon, A. Aneuploidy Causes Proteotoxic Stress in Yeast. Genes Dev. 2012, 26, 2696–2708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrimore, K.E.; Barattin-Voynova, N.S.; Reid, D.W.; Ng, D.T.W. Aneuploidy-Induced Proteotoxic Stress Can Be Effectively Tolerated without Dosage Compensation, Genetic Mutations, or Stress Responses. BMC Biol. 2020, 18, 117. [Google Scholar] [CrossRef]
- Heasley, L.R.; Watson, R.A.; Argueso, J.L. Punctuated Aneuploidization of the Budding Yeast Genome. Genetics 2020, 216, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.C.; Amon, A. Gene Copy-Number Alterations: A Cost-Benefit Analysis. Cell 2013, 152, 394–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.J.; Regan, S.; Liu, G.; Alemara, S.; Heng, H.H. Understanding Aneuploidy in Cancer through the Lens of System Inheritance, Fuzzy Inheritance and Emergence of New Genome Systems. Mol. Cytogenet. 2018, 11, 31. [Google Scholar] [CrossRef] [Green Version]
- Clarke, M.N.; Marsoner, T.; Adell, M.A.Y.; Ravichandran, M.C.; Campbell, C.S. Adaptation to High Rates of Chromosomal Instability and Aneuploidy through Multiple Pathways in Budding Yeast. EMBO J. 2023, 42, e111500. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, A.C.; McBride, R.M.; Otto, S.P. Ploidy Reduction in Saccharomyces cerevisiae. Biol. Lett. 2008, 4, 91–94. [Google Scholar] [CrossRef] [Green Version]
- Selmecki, A.M.; Maruvka, Y.E.; Richmond, P.A.; Guillet, M.; Shoresh, N.; Sorenson, A.L.; De, S.; Kishony, R.; Michor, F.; Dowell, R.; et al. Polyploidy Can Drive Rapid Adaptation in Yeast. Nature 2015, 519, 349–351. [Google Scholar] [CrossRef] [Green Version]
- Dutta, A.; Dutreux, F.; Schacherer, J. Loss of Heterozygosity Spectrum Depends on Ploidy Level in Natural Yeast Populations. Mol. Biol. Evol. 2022, 39, msac214. [Google Scholar] [CrossRef]
- Wickner, R.B. Mapping Chromosomal Genes of Saccharomyces cerevisiae Using an Improved Genetic Mapping Method. Genetics 1979, 92, 803–821. [Google Scholar] [CrossRef] [PubMed]
- Dutcher, S.K. Internuclear Transfer of Genetic Information in Kar1-1/KAR1 Heterokaryons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1981, 1, 245–253. [Google Scholar] [CrossRef]
- Dorweiler, J.E.; Manogaran, A.L. Cytoduction and Plasmiduction in Yeast. Bio-Protocol 2021, 11, e4146. [Google Scholar] [CrossRef]
- Zang, Y.; Garr, M.; Gjuracic, K.; Bruschi, C.V. Chromosome V Loss Due to Centromere Knockout or MAD2-Deletion Is Immediately Followed by Restitution of Homozygous Diploidy in Saccharomyces cerevisiae. Yeast 2002, 19, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Reid, R.J.D.; Sunjevaric, I.; Voth, W.P.; Ciccone, S.; Du, W.; Olsen, A.E.; Stillman, D.J.; Rothstein, R. Chromosome-Scale Genetic Mapping Using a Set of 16 Conditionally Stable Saccharomyces cerevisiae Chromosomes. Genetics 2008, 180, 1799–1808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schild, D.; Mortimer, R.K. A Mapping Method for Saccharomyces cerevisiae Using Rad52-Induced Chromosome Loss. Genetics 1985, 110, 569–589. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Han, M.; Zhou, S.; Li, B.Z.; Wu, Y.; Yuan, Y.J. Chromosome Drives via CRISPR-Cas9 in Yeast. Nat. Commun. 2020, 11, 4344. [Google Scholar] [CrossRef]
- Anders, K.R.; Kudrna, J.R.; Keller, K.E.; Kinghorn, B.A.A.; Miller, E.M.; Pauw, D.; Peck, A.T.; Shellooe, C.E.; Strong, I.J.T. A Strategy for Constructing Aneuploid Yeast Strains by Transient Nondisjunction of a Target Chromosome. BMC Genet. 2009, 10, 36. [Google Scholar] [CrossRef] [Green Version]
- Fleiss, A.; O’Donnell, S.; Fournier, T.; Lu, W.; Agier, N.; Delmas, S.; Schachere, J.; Fischer, G. Reshuffling Yeast Chromosomes with CRISPR/Cas9. PLoS Genet. 2019, 15, e1008332. [Google Scholar] [CrossRef] [Green Version]
- Agier, N.; Fleiss, A.; Delmas, S.; Fischer, G. A Versatile Protocol to Generate Translocations in Yeast Genomes Using CRISPR/Cas9. Methods Mol. Biol. 2021, 2196, 181–198. [Google Scholar] [CrossRef] [PubMed]
- Dymond, J.S.; Richardson, S.M.; Coombes, C.E.; Babatz, T.; Muller, H.; Annaluru, N.; Blake, W.J.; Schwerzmann, J.W.; Dai, J.; Lindstrom, D.L.; et al. Synthetic Chromosome Arms Function in Yeast and Generate Phenotypic Diversity by Design. Nature 2011, 477, 471–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dymond, J.; Boeke, J. The Saccharomyces cerevisiae SCRaMbLE System and Genome Minimization. Bioeng. Bugs 2012, 3, 168. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Wang, L.; Wang, Y.; Zhang, W.; Guo, Y.; Shen, Y.; Jiang, L.; Wu, Q.; Zhang, C.; Cai, Y.; et al. Identifying and Characterizing SCRaMbLEd Synthetic Yeast Using ReSCuES. Nat. Commun. 2018, 9, 1930. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Wu, Y.; Li, B.Z.; Mitchell, L.A.; Liu, H.; Pan, S.; Wang, J.; Zhang, H.R.; Jia, N.; Li, B.; et al. Precise Control of SCRaMbLE in Synthetic Haploid and Diploid Yeast. Nat. Commun. 2018, 9, 1933. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.J.; Wu, Y.; Yang, K.; Li, Y.; Xu, H.; Zhang, H.; Li, B.Z.; Li, X.; Xiao, W.H.; Zhou, X.; et al. Heterozygous Diploid and Interspecies SCRaMbLEing. Nat. Commun. 2018, 9, 1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Li, Y.; Chen, X.; Ding, M.; Wu, Y.; Yuan, Y.J. SCRaMbLE Generates Evolved Yeasts with Increased Alkali Tolerance. Microb. Cell Fact. 2019, 18, 52. [Google Scholar] [CrossRef] [Green Version]
- Blount, B.A.; Gowers, G.O.F.; Ho, J.C.H.; Ledesma-Amaro, R.; Jovicevic, D.; McKiernan, R.M.; Xie, Z.X.; Li, B.Z.; Yuan, Y.J.; Ellis, T. Rapid Host Strain Improvement by in Vivo Rearrangement of a Synthetic Yeast Chromosome. Nat. Commun. 2018, 9, 1932. [Google Scholar] [CrossRef]
- Kutyna, D.R.; Onetto, C.A.; Williams, T.C.; Goold, H.D.; Paulsen, I.T.; Pretorius, I.S.; Johnson, D.L.; Borneman, A.R. Construction of a Synthetic Saccharomyces cerevisiae Pan-Genome Neo-Chromosome. Nat. Commun. 2022, 13, 3628. [Google Scholar] [CrossRef]
- Hochrein, L.; Mitchell, L.A.; Schulz, K.; Messerschmidt, K.; Mueller-Roeber, B. L-SCRaMbLE as a Tool for Light-Controlled Cre-Mediated Recombination in Yeast. Nat. Commun. 2018, 9, 1931. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Fu, X.; Gong, X.; Wang, Y.; Zhang, H.; Zhao, Y.; Shen, Y. Systematic Dissection of Key Factors Governing Recombination Outcomes by GCE-SCRaMbLE. Nat. Commun. 2022, 13, 5836. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.M.; Mitchell, L.A.; Stracquadanio, G.; Yang, K.; Dymond, J.S.; DiCarlo, J.E.; Lee, D.; Huang, C.L.V.; Chandrasegaran, S.; Cai, Y.; et al. Design of a Synthetic Yeast Genome. Science 2017, 355, 1040–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Mitchell, L.A.; Bader, J.S.; Boeke, J.D. Synthetic Genomes. Annu. Rev. Biochem. 2020, 89, 77–101. [Google Scholar] [CrossRef]
- Hoess, R.H.; Wierzbicki, A.; Abremski, K. The Role of the LoxP Spacer Region in P1 Site-Specific Recombination. Nucleic Acids Res. 1986, 14, 2287–2300. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, Z.X.; Ma, Y.; Chen, X.R.; Huang, Y.Q.; He, B.; Jia, B.; Li, B.Z.; Yuan, Y.J. Ring Synthetic Chromosome V SCRaMbLE. Nat. Commun. 2018, 9, 3783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muramoto, N.; Oda, A.; Tanaka, H.; Nakamura, T.; Kugou, K.; Suda, K.; Kobayashi, A.; Yoneda, S.; Ikeuchi, A.; Sugimoto, H.; et al. Phenotypic Diversification by Enhanced Genome Restructuring after Induction of Multiple DNA Double-Strand Breaks. Nat. Commun. 2018, 9, 1995. [Google Scholar] [CrossRef] [Green Version]
- Laureau, R.; Loeillet, S.; Salinas, F.; Bergström, A.; Legoix-Né, P.; Liti, G.; Nicolas, A. Extensive Recombination of a Yeast Diploid Hybrid through Meiotic Reversion. PLoS Genet. 2016, 12, e1005781. [Google Scholar] [CrossRef] [Green Version]
- Mozzachiodi, S.; Tattini, L.; Llored, A.; Irizar, A.; Škofljanc, N.; D’Angiolo, M.; De Chiara, M.; Barré, B.P.; Yue, J.X.; Lutazi, A.; et al. Aborting Meiosis Allows Recombination in Sterile Diploid Yeast Hybrids. Nat. Commun. 2021, 12, 6564. [Google Scholar] [CrossRef]
- Mozzachiodi, S.; Krogerus, K.; Gibson, B.; Nicolas, A.; Liti, G. Unlocking the Functional Potential of Polyploid Yeasts. Nat. Commun. 2022, 13, 2580. [Google Scholar] [CrossRef]
- Natesuntorn, W.; Iwami, K.; Matsubara, Y.; Sasano, Y.; Sugiyama, M.; Kaneko, Y.; Harashima, S. Genome-Wide Construction of a Series of Designed Segmental Aneuploids in Saccharomyces cerevisiae. Sci. Rep. 2015, 5, 12510. [Google Scholar] [CrossRef]
- Aksenova, A.Y.; Greenwell, P.W.; Dominska, M.; Shishkin, A.A.; Kim, J.C.; Petes, T.D.; Mirkin, S.M. Genome Rearrangements Caused by Interstitial Telomeric Sequences in Yeast. Proc. Natl. Acad. Sci. USA 2013, 110, 19866–19871. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.; Dominska, M.; Greenwell, P.; Aksenova, A.Y.; Mirkin, S.; Petes, T. Genetic Control of Genomic Alterations Induced in Yeast by Interstitial Telomeric Sequences. Genetics 2018, 209, 425–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stepchenkova, E.I.; Zadorsky, S.P.; Shumega, A.R.; Aksenova, A.Y. Practical Approaches for the Yeast Saccharomyces cerevisiae Genome Modification. Int. J. Mol. Sci. 2023, 24, 11960. https://doi.org/10.3390/ijms241511960
Stepchenkova EI, Zadorsky SP, Shumega AR, Aksenova AY. Practical Approaches for the Yeast Saccharomyces cerevisiae Genome Modification. International Journal of Molecular Sciences. 2023; 24(15):11960. https://doi.org/10.3390/ijms241511960
Chicago/Turabian StyleStepchenkova, Elena I., Sergey P. Zadorsky, Andrey R. Shumega, and Anna Y. Aksenova. 2023. "Practical Approaches for the Yeast Saccharomyces cerevisiae Genome Modification" International Journal of Molecular Sciences 24, no. 15: 11960. https://doi.org/10.3390/ijms241511960
APA StyleStepchenkova, E. I., Zadorsky, S. P., Shumega, A. R., & Aksenova, A. Y. (2023). Practical Approaches for the Yeast Saccharomyces cerevisiae Genome Modification. International Journal of Molecular Sciences, 24(15), 11960. https://doi.org/10.3390/ijms241511960





