Comparative Biochemical, Structural, and Functional Analysis of Recombinant Phospholipases D from Three Loxosceles Spider Venoms
Abstract
1. Introduction
2. Results
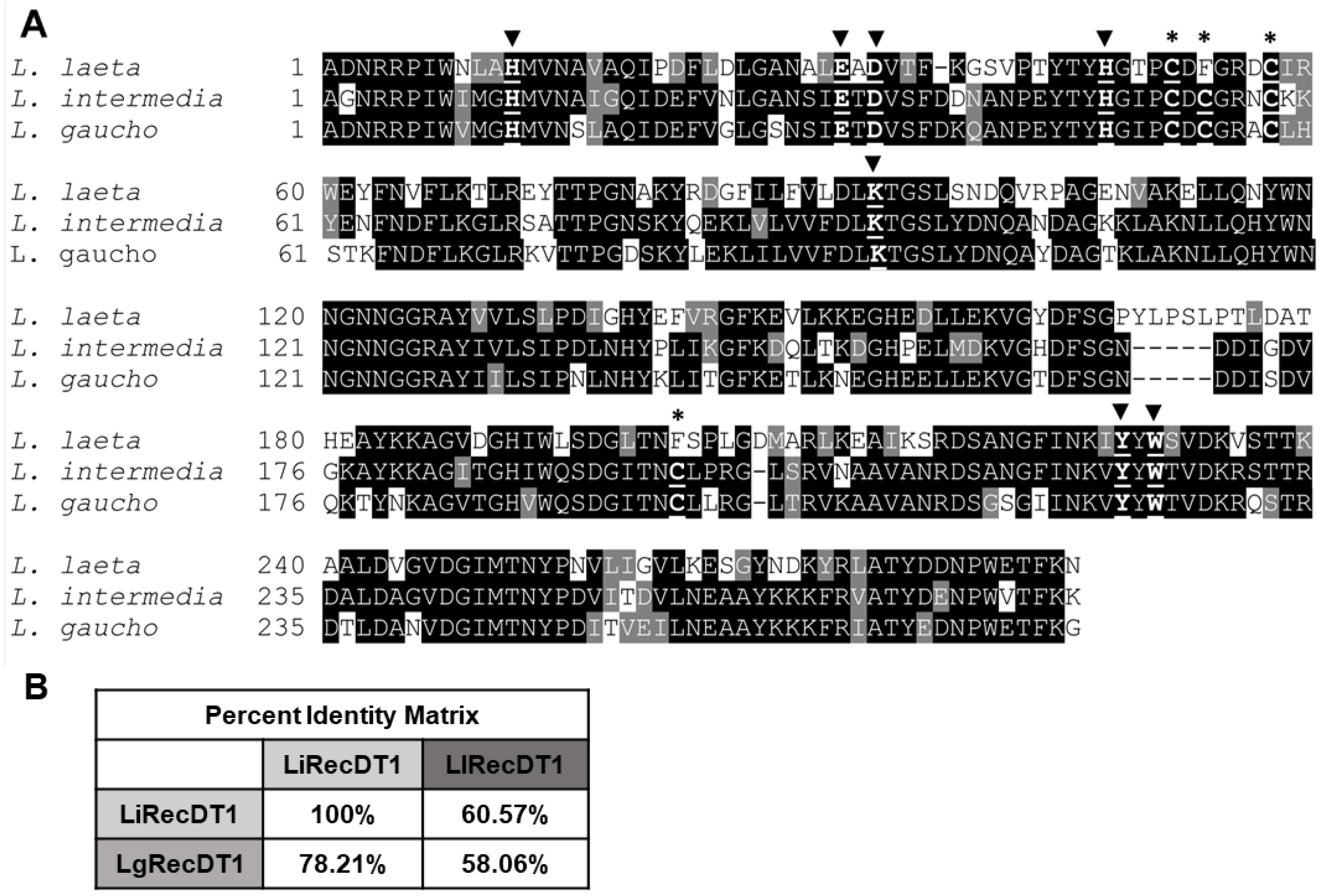
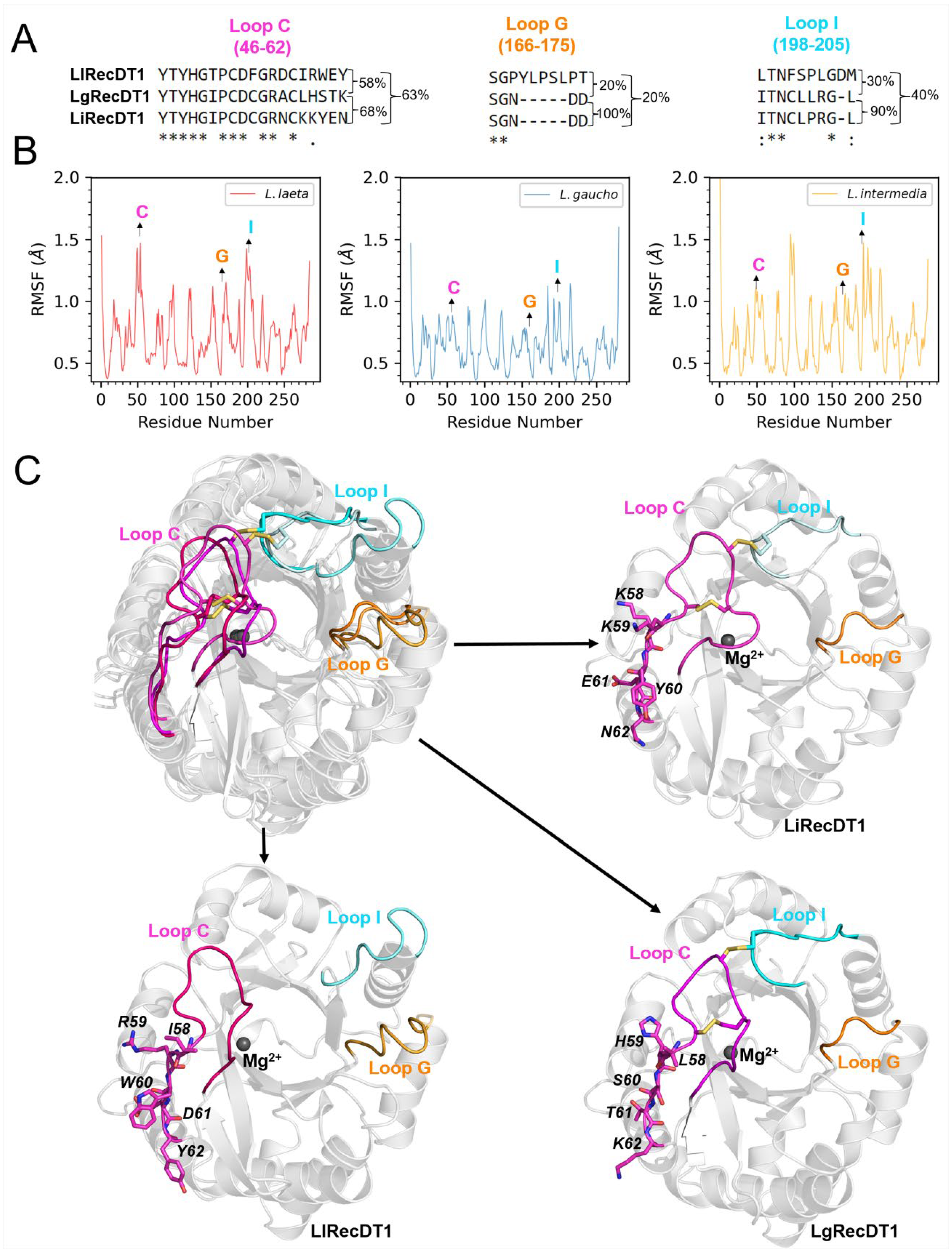
2.1. Amino Acid Sequence Comparison of PLDs
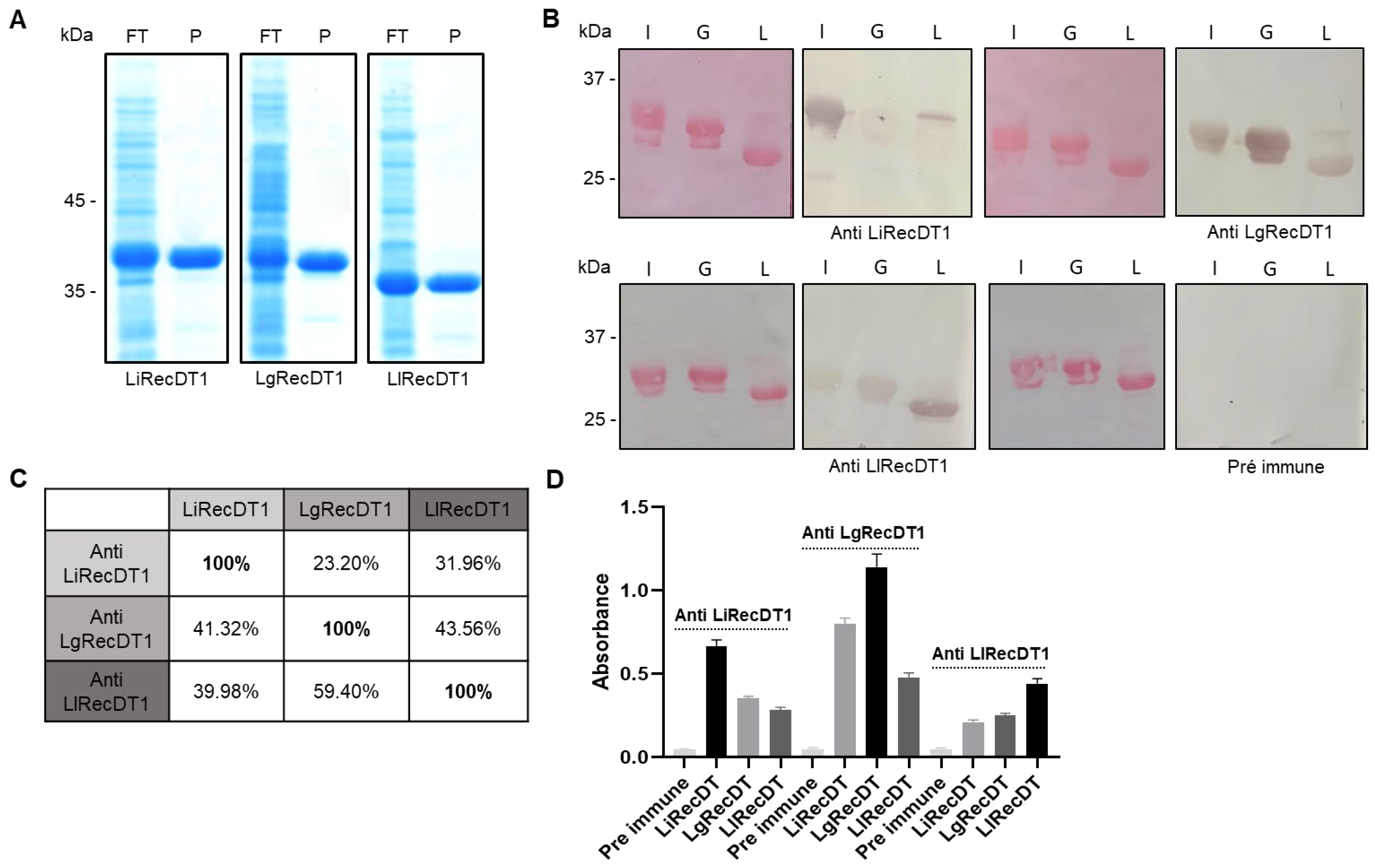
2.2. Immunological Relationship of Phospholipases D
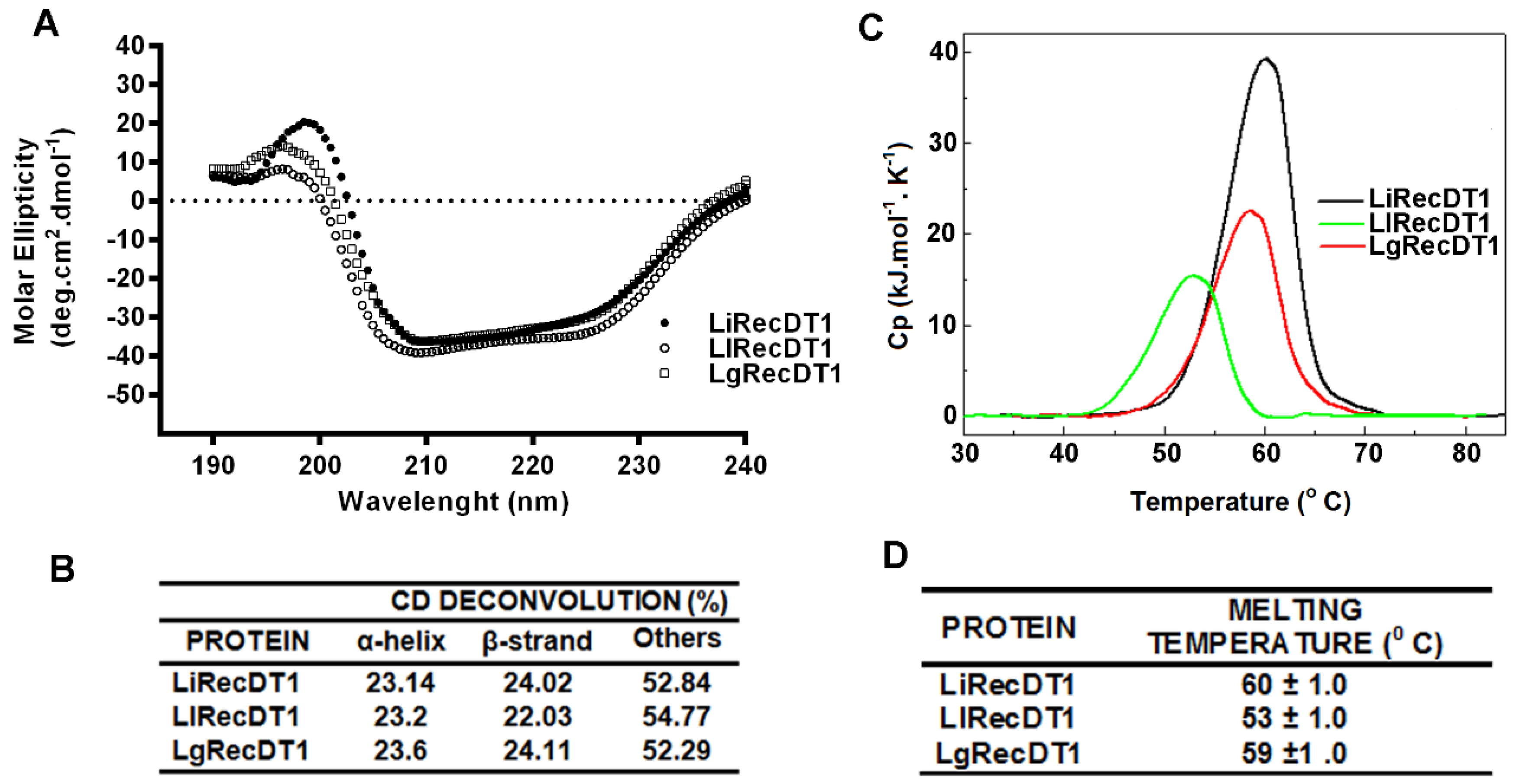
2.3. Biophysical Properties of PLDs
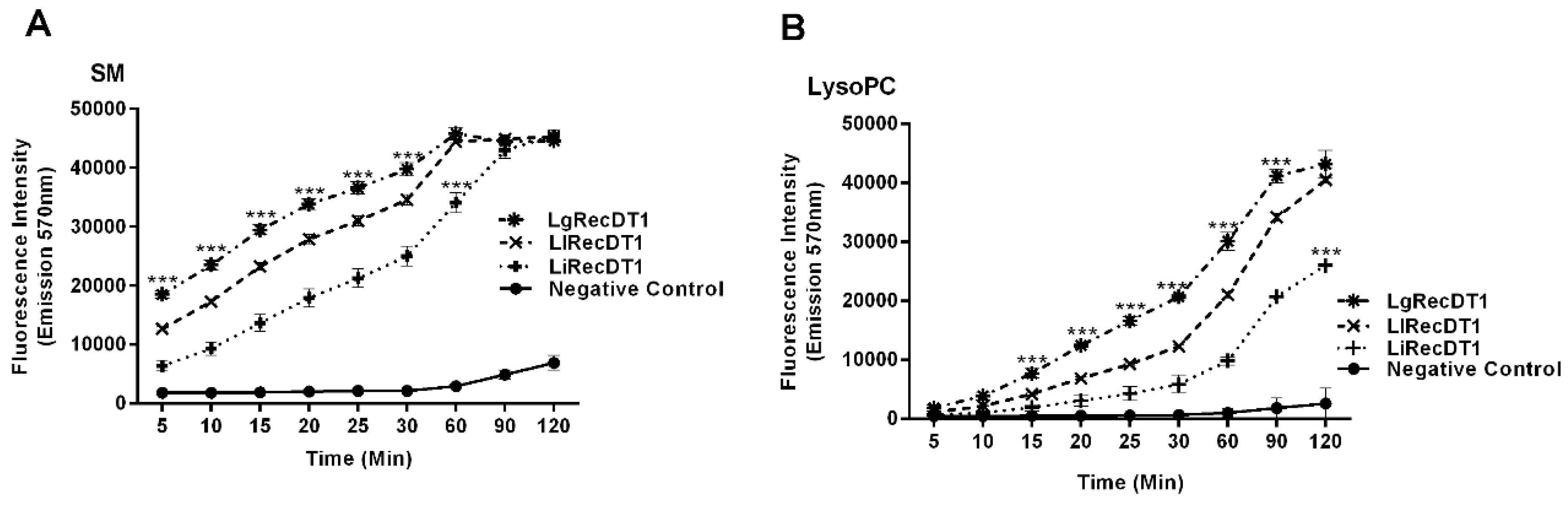
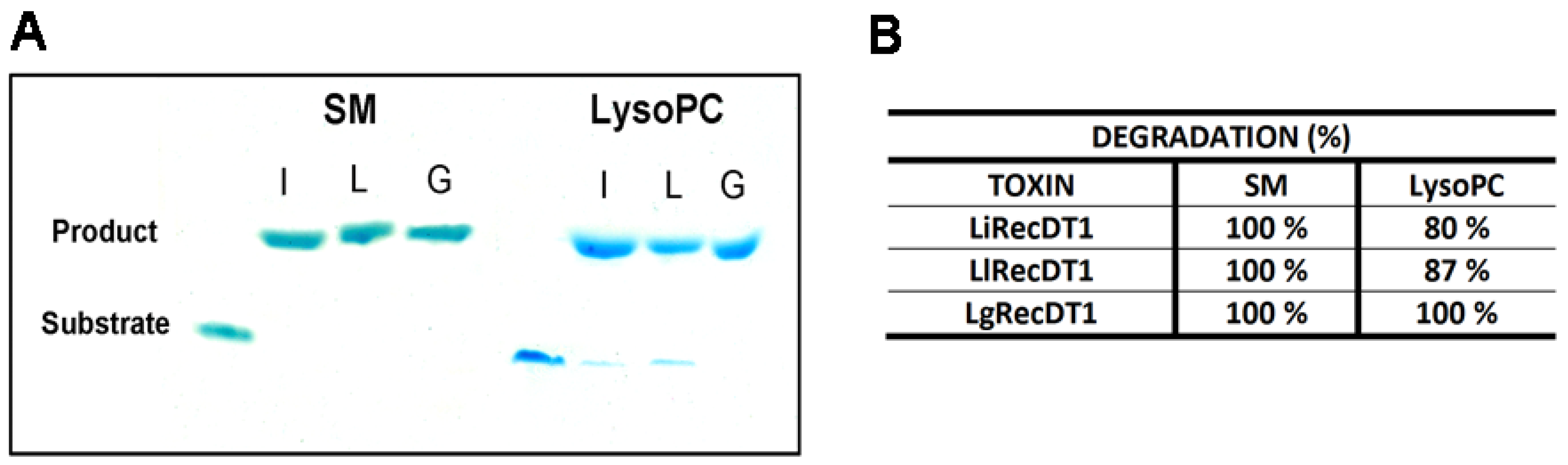
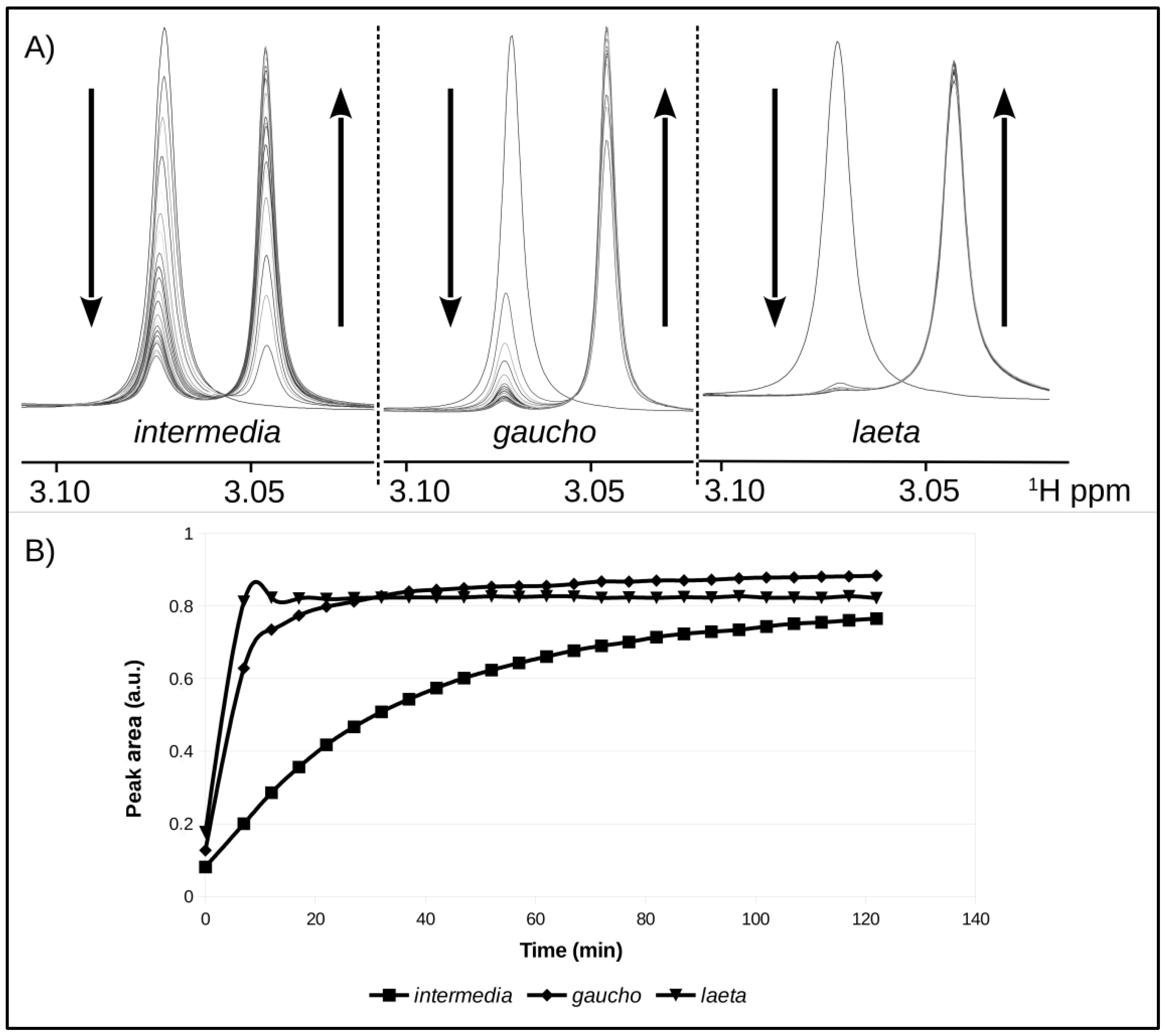
2.4. Enzymatic Activity of Recombinant PLDs
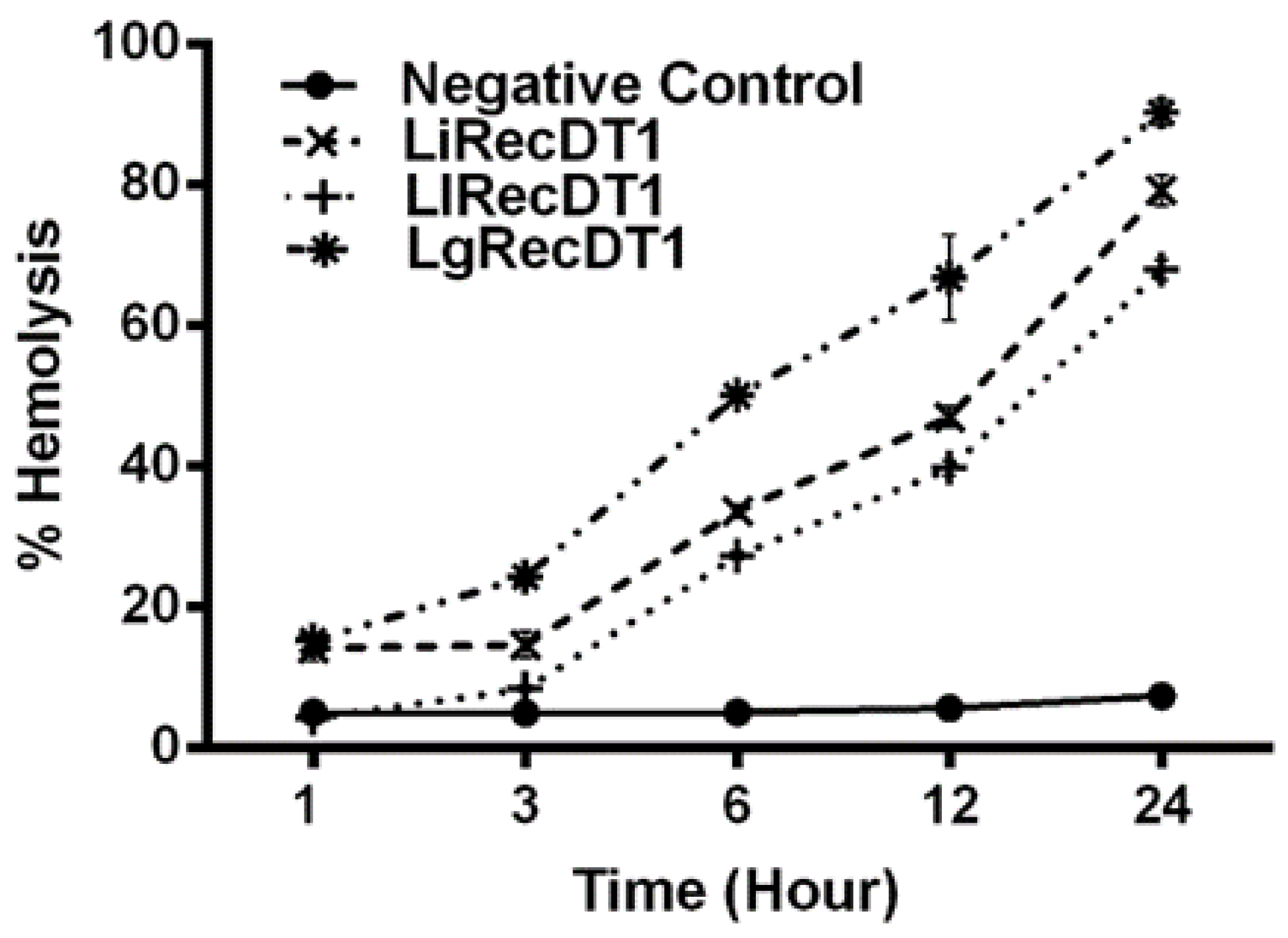
2.5. Hemolytic Activity
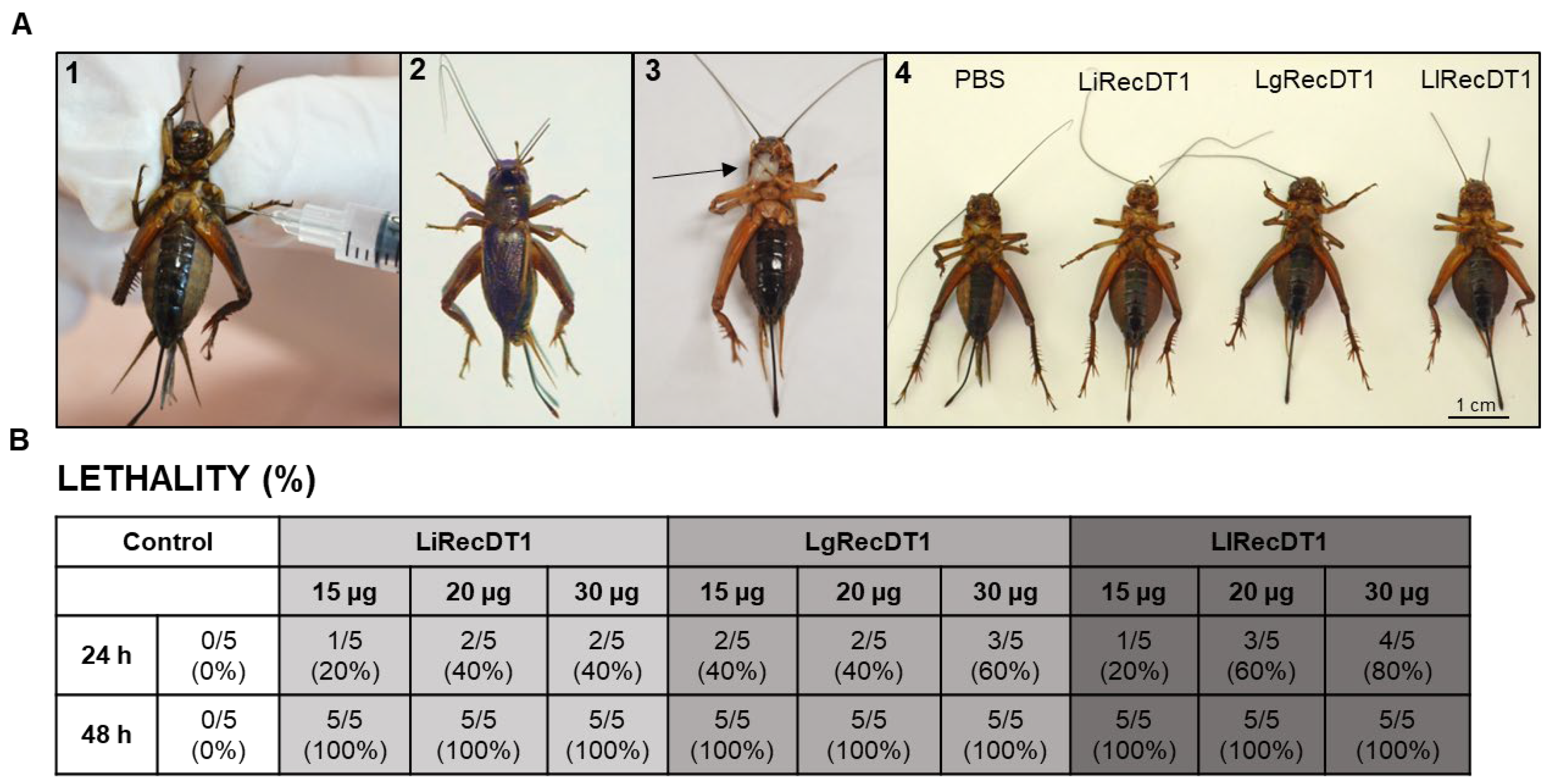
2.6. Animal Lethality
2.7. Insecticidal Activity of Recombinant PLDs
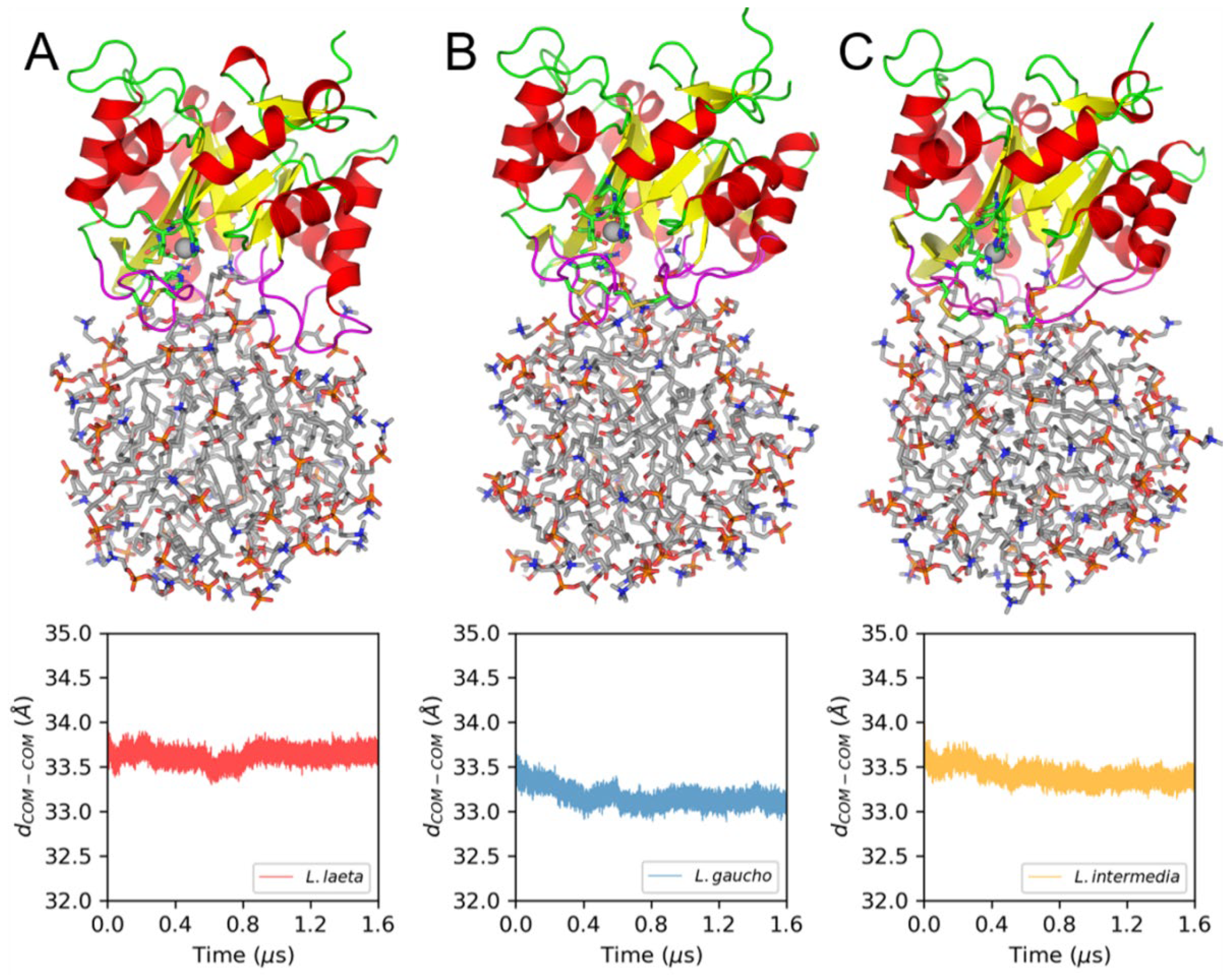
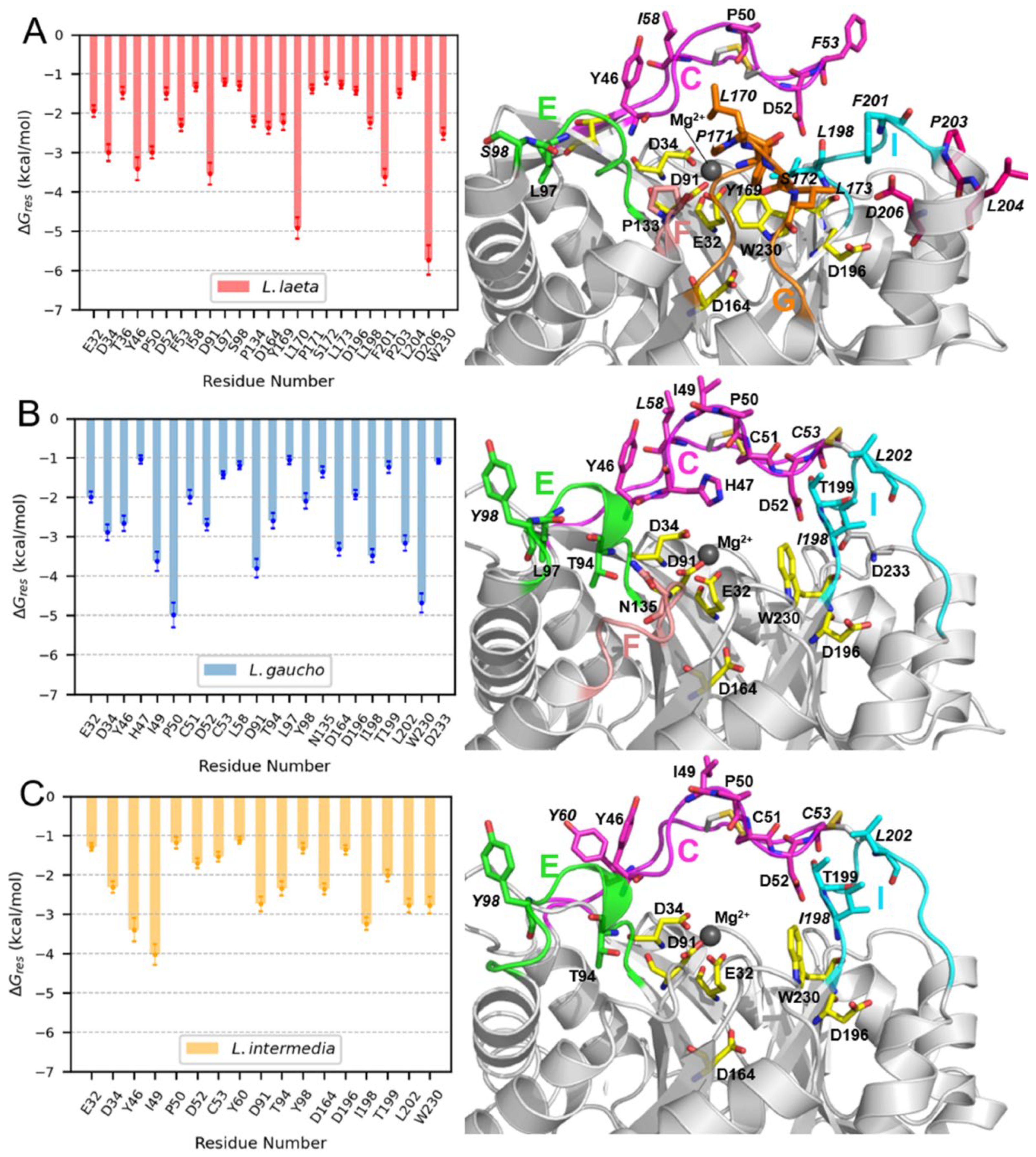
2.8. Molecular Dynamics (MD) Simulations of PLDs and LysoPC Micelles
3. Discussion
3.1. General Discussion
3.2. Sequence Identity/Structure
3.3. Membrane Binding Mode
4. Materials and Methods
4.1. Reagents
4.2. Recombinant Protein Expression and Purification
4.3. Immunological Cross-Reactivity
4.4. Circular Dichroism Spectroscopy (CD)
4.5. Differential Scanning Calorimetry (DSC)
4.6. Amplex RedTM Assay
4.7. HPTLC (High-Performance Thin-Layer Chromatography)
4.8. NMR Spectroscopy (Nuclear Magnetic Resonance)
4.9. Hemolysis Assay
4.10. Animals
4.11. Lethality
4.12. Molecular Dynamics (MD) Simulations of PLDs ad LysoPC Micelles
4.13. MM-GBSA Free Energy Calculations
4.14. Trajectory Analyses
4.15. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hogan, C.J.; Barbaro, K.C.; Winkel, K. Loxoscelism: Old obstacles, new directions. Ann. Emerg. Med. 2004, 44, 608–624. [Google Scholar] [CrossRef]
- Da Silva, P.H.; Da Silveira, R.B.; Helena Appel, M.; Mangili, O.C.; Gremski, W.; Veiga, S.S. Brown spiders and loxoscelism. Toxicon 2004, 44, 693–709. [Google Scholar] [CrossRef]
- Chaim, O.M.; Trevisan-Silva, D.; Chaves-Moreira, D.; Wille, A.C.M.; Ferrer, V.P.; Matsubara, F.H.; Mangili, O.C.; da Silveira, R.B.; Gremski, L.H.; Gremski, W.; et al. Brown spider (Loxosceles genus) venom toxins: Tools for biological purposes. Toxins 2011, 3, 309–344. [Google Scholar] [CrossRef]
- Gremski, L.H.; Trevisan-Silva, D.; Ferrer, V.P.; Matsubara, F.H.; Meissner, G.O.; Wille, A.C.M.; Vuitika, L.; Dias-Lopes, C.; Ullah, A.; De Moraes, F.R.; et al. Recent advances in the understanding of brown spider venoms: From the biology of spiders to the molecular mechanisms of toxins. Toxicon 2014, 83, 91–120. [Google Scholar] [CrossRef]
- Panorama dos Acidentes Causados por Aranhas No Brasil, de 2017 a 2021. Available online: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/epidemiologicos/edicoes/2022/boletim-epidemiologico-vol-53-no31/view (accessed on 25 April 2023).
- World Spider Catalog. Available online: https://wsc.nmbe.ch/ (accessed on 15 March 2023).
- Cordeiro, F.A.; Amorim, F.G.; Anjolette, F.A.P.; Arantes, E.C. Arachnids of medical importance in Brazil: Main active compounds present in scorpion and spider venoms and tick saliva. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 24. [Google Scholar] [CrossRef]
- Futrell, J.M. Loxoscelism. Am. J. Med. Sci. 1992, 304, 261–267. [Google Scholar] [CrossRef]
- Chaim, O.M.; Sade, Y.B.; Da Silveira, R.B.; Toma, L.; Kalapothakis, E.; Chávez-Olórtegui, C.; Mangili, O.C.; Gremski, W.; Von Dietrich, C.P.; Nader, H.B.; et al. Brown spider dermonecrotic toxin directly induces nephrotoxicity. Toxicol. Appl. Pharmacol. 2006, 211, 64–77. [Google Scholar] [CrossRef]
- Magalhães, G.S.; Caporrino, M.C.; Della-Casa, M.S.; Kimura, L.F.; Prezotto-Neto, J.P.; Fukuda, D.A.; Portes-Junior, J.A.; Neves-Ferreira, A.G.C.; Santoro, M.L.; Barbaro, K.C. Cloning, expression and characterization of a phospholipase D from Loxosceles gaucho venom gland. Biochimie 2013, 95, 1773–1783. [Google Scholar] [CrossRef]
- Kusma, J.; Chaim, O.M.; Wille, A.C.M.; Ferrer, V.P.; Sade, Y.B.; Donatti, L.; Gremski, W.; Mangili, O.C.; Veiga, S.S. Nephrotoxicity caused by brown spider venom phospholipase-D (dermonecrotic toxin) depends on catalytic activity. Biochimie 2008, 90, 1722–1736. [Google Scholar] [CrossRef]
- Chaves-Moreira, D.; Chaim, O.M.; Sade, Y.B.; Paludo, K.S.; Gremski, L.H.; Donatti, L.; De Moura, J.; Mangili, O.C.; Gremski, W.; Da Silveira, R.B.; et al. Identification of a direct hemolytic effect dependent on the catalytic activity induced by phospholipase-D (dermonecrotic toxin) from brown spider venom. J. Cell. Biochem. 2009, 107, 655–666. [Google Scholar] [CrossRef]
- Fernandes Pedrosa, M.D.F.; Junqueira de Azevedo, I.D.L.M.; Gonçalves-de-Andrade, R.M.; Van Den Berg, C.W.; Ramos, C.R.R.; Lee Ho, P.; Tambourgi, D.V. Molecular cloning and expression of a functional dermonecrotic and haemolytic factor from Loxosceles laeta venom. Biochem. Biophys. Res. Commun. 2002, 298, 638–645. [Google Scholar] [CrossRef]
- Lajoie, D.M.; Zobel-Thropp, P.A.; Kumirov, V.K.; Bandarian, V.; Binford, G.J.; Cordes, M.H.J. Phospholipase D Toxins of Brown Spider Venom Convert Lysophosphatidylcholine and Sphingomyelin to Cyclic Phosphates. PLoS ONE 2013, 8, e72372. [Google Scholar] [CrossRef]
- Gremski, L.H.; Da Justa, H.C.; Da Silva, T.P.; Polli, N.L.C.; Antunes, B.C.; Minozzo, J.C.; Wille, A.C.M.; Senff-Ribeiro, A.; Arni, R.K.; Veiga, S.S. Forty years of the description of brown spider venom phospholipases-D. Toxins 2020, 12, 164. [Google Scholar] [CrossRef]
- Murakami, M.T.; Fernandes-Pedrosa, M.F.; Tambourgi, D.V.; Arni, R.K. Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases D. J. Biol. Chem. 2005, 280, 13658–13664. [Google Scholar] [CrossRef]
- Murakami, M.T.; Freitas Fernandes-Pedrosa, M.; De Andrade, S.A.; Gabdoulkhakov, A.; Betzel, C.; Tambourgi, D.V.; Arni, R.K. Structural insights into the catalytic mechanism of sphingomyelinases D and evolutionary relationship to glycerophosphodiester phosphodiesterases. Biochem. Biophys. Res. Commun. 2006, 342, 323–329. [Google Scholar] [CrossRef]
- Coronado, M.A.; Ullah, A.; Silva, L.S.; Chaves-Moreira, D.; Vuitika, L.; Chaim, O.M.; Veiga, S.S.; Chahine, J.; Murakami, M.T.; Arni, R.K. Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D. Curr. Protein Pept. Sci. 2015, 16, 768–774. [Google Scholar] [CrossRef]
- De Giuseppe, P.O.; Ullah, A.; Silva, D.T.; Gremski, L.H.; Wille, A.C.M.; Chaves Moreira, D.; Ribeiro, A.S.; Chaim, O.M.; Murakami, M.T.; Veiga, S.S.; et al. Structure of a novel class II phospholipase D: Catalytic cleft is modified by a disulphide bridge. Biochem. Biophys. Res. Commun. 2011, 409, 622–627. [Google Scholar] [CrossRef]
- Dias-Lopes, C.; Neshich, I.A.P.; Neshich, G.; Ortega, J.M.; Granier, C.; Chávez-Olortegui, C.; Molina, F.; Felicori, L. Identification of new sphingomyelinases D in pathogenic fungi and other pathogenic organisms. PLoS ONE 2013, 8, e79240. [Google Scholar] [CrossRef]
- Vuitika, L.; Chaves-Moreira, D.; Caruso, I.; Lima, M.A.; Matsubara, F.H.; Murakami, M.T.; Takahashi, H.K.; Toledo, M.S.; Coronado, M.A.; Nader, H.B.; et al. Active site mapping of Loxosceles phospholipases D: Biochemical and biological features. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Van Meeteren, L.A.; Frederiks, F.; Giepmans, B.N.G.; Fernandes Pedrosa, M.F.; Billington, S.J.; Jost, B.H.; Tambourgi, D.V.; Moolenaar, W.H. Spider and Bacterial Sphingomyelinases D Target Cellular Lysophosphatidic Acid Receptors by Hydrolyzing Lysophosphatidylcholine. J. Biol. Chem. 2004, 279, 10833–10836. [Google Scholar] [CrossRef]
- Lee, S.; Lynch, K.R. Brown recluse spider (Loxosceles reclusa) venom phospholipase D (PLD) generates lysophosphatidic acid (LPA). Biochem. J. 2005, 391, 317–323. [Google Scholar] [CrossRef]
- Chaim, O.M.; Da Silveira, R.B.; Trevisan-Silva, D.; Ferrer, V.P.; Sade, Y.B.; Bóia-Ferreira, M.; Gremski, L.H.; Gremski, W.; Senff-Ribeiro, A.; Takahashi, H.K.; et al. Phospholipase-D activity and inflammatory response induced by brown spider dermonecrotic toxin: Endothelial cell membrane phospholipids as targets for toxicity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2011, 1811, 84–96. [Google Scholar] [CrossRef]
- Van Meeteren, L.A.; Stortelers, C.; Moolenaar, W.H. Upregulation of cytokine expression in fibroblasts exposed to Loxosceles sphingomyelinase D: What is the trigger? J. Investig. Dermatol. 2007, 127, 1266–1267. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, D.M.; Cordes, M.H.J. Spider, bacterial and fungal phospholipase D toxins make cyclic phosphate products. Toxicon 2015, 108, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, D.M.; Roberts, S.A.; Zobel-Thropp, P.A.; Delahaye, J.L.; Bandarian, V.; Binford, G.J.; Cordes, M.H.J. Variable substrate preference among phospholipase D toxins from sicariid spiders. J. Biol. Chem. 2015, 290, 10994–11007. [Google Scholar] [CrossRef]
- Zobel-Thropp, P.A.; Kerins, A.E.; Binford, G.J. Sphingomyelinase D in sicariid spider venom is a potent insecticidal toxin. Toxicon 2012, 60, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Pedrosa, M.d.F.; Junqueira-de-Azevedo, I.d.L.M.; Gonçalves-de-Andrade, R.M.; Kobashi, L.S.; Almeida, D.D.; Ho, P.L.; Tambourgi, D. V Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genom. 2008, 9, 279. [Google Scholar] [CrossRef]
- Gremski, L.H.; Da Silveira, R.B.; Chaim, O.M.; Probst, C.M.A.; Ferrer, V.P.; Nowatzki, J.; Weinschutz, H.C.; Madeira, H.M.I.; Gremski, W.; Nader, H.B.; et al. A novel expression profile of the Loxosceles intermedia spider venomous gland revealed by transcriptome analysis. Mol. Biosyst. 2010, 6, 2403–2416. [Google Scholar] [CrossRef]
- Dantas, A.E.; Carmo, A.O.; Horta, C.C.R.; Leal, H.G.; Oliveira-Mendes, B.B.R.; Martins, A.P.V.; Chávez-Olórtegui, C.; Kalapothakis, E. Description of Loxtox protein family and identification of a new group of Phospholipases D from Loxosceles similis venom gland. Toxicon 2016, 120, 97–106. [Google Scholar] [CrossRef]
- Medina-Santos, R.; Fernandes Costa, T.G.; Silva de Assis, T.C.; Kalapothakis, Y.; de Almeida Lima, S.; do Carmo, A.O.; Gonzalez-Kozlova, E.E.; Kalapothakis, E.; Chávez-Olórtegui, C.; Guerra-Duarte, C. Analysis of NGS data from Peruvian Loxosceles laeta spider venom gland reveals toxin diversity. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 43, 101017. [Google Scholar] [CrossRef]
- Dos Santos, L.D.; Dias, N.B.; Roberto, J.; Pinto, A.S.; Palma, M.S. Brown recluse spider venom: Proteomic analysis and proposal of a putative mechanism of action. Protein Pept. Lett. 2009, 16, 933–943. [Google Scholar] [CrossRef]
- Machado, L.F.; Laugesen, S.; Botelho, E.D.; Ricart, C.A.O.; Fontes, W.; Barbaro, K.C.; Roepstorff, P.; Valle De Sousa, M. Proteome analysis of brown spider venom: Identification of loxnecrogin isoforms in Loxosceles gaucho venom. Proteomics 2005, 5, 2167–2176. [Google Scholar] [CrossRef]
- Trevisan-Silva, D.; Bednaski, A.V.; Fischer, J.S.G.; Veiga, S.S.; Bandeira, N.; Guthals, A.; Marchini, F.K.; Leprevost, F.V.; Barbosa, V.C.; Senff-Ribeiro, A.; et al. A multi-protease, multi-dissociation, bottom-up-to-top-down proteomic view of the Loxosceles intermedia venom. Sci. Data 2017, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dragulev, B.; Bao, Y.; Ramos-Cerrillo, B.; Vazquez, H.; Olvera, A.; Stock, R.; Algaron, A.; Fox, J.W. Upregulation of IL-6, IL-8, CXCL1, and CXCL2 dominates gene expression in human fibroblast cells exposed to Loxosceles reclusa sphingomyelinase D: Insights into spider venom dermonecrosis. J. Investig. Dermatol. 2007, 127, 1264–1266. [Google Scholar] [CrossRef] [PubMed]
- Horta, C.C.R.; Oliveira-Mendes, B.B.R.; Do Carmo, A.O.; Siqueira, F.F.; Barroca, T.M.; Dos Santos Nassif Lacerda, S.M.; De Almeida Campos, P.H.; De França, L.R.; Ferreira, R.L.; Kalapothakis, E. Lysophosphatidic acid mediates the release of cytokines and chemokines by human fibroblasts treated with loxosceles spider venom. J. Investig. Dermatol. 2013, 133, 1682–1685. [Google Scholar] [CrossRef]
- Barbaro, K.C.; Lira, M.S.; Araújo, C.A.; Pareja-Santos, A.; Távora, B.C.L.F.; Prezotto-Neto, J.P.; Kimura, L.F.; Lima, C.; Lopes-Ferreira, M.; Santoro, M.L. Inflammatory mediators generated at the site of inoculation of Loxosceles gaucho spider venom. Toxicon 2010, 56, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Rojas, J.M.; Arán-Sekul, T.; Cortés, E.; Jaldín, R.; Ordenes, K.; Orrego, P.R.; González, J.; Araya, J.E.; Catalán, A. Phospholipase d from loxosceles laeta spider venom induces IL-6, IL-8, CXCL1/GRO-α, and CCL2/MCP-1 production in human skin fibroblasts and stimulates monocytes migration. Toxins 2017, 9, 125. [Google Scholar] [CrossRef]
- Manzoni-de-Almeida, D.; Squaiella-Baptistão, C.C.; Lopes, P.H.; van den Berg, C.W.; Tambourgi, D.V. Loxosceles venom Sphingomyelinase D activates human blood leukocytes: Role of the complement system. Mol. Immunol. 2018, 94, 45–53. [Google Scholar] [CrossRef]
- De Oliveira, K.C.; Gonçalves De Andrade, R.M.; Piazza, R.M.F.; Ferreira, J.M.C.; Van Den Berg, C.W.; Tambourgi, D.V. Variations in Loxosceles spider venom composition and toxicity contribute to the severity of envenomation. Toxicon 2005, 45, 421–429. [Google Scholar] [CrossRef]
- Catalán, A.; Cortés, W.; Muñoz, C.; Araya, J.E. Tryptophan and aspartic acid residues present in the glycerophosphoryl diester phosphodiesterase (GDPD) domain of the Loxosceles laeta phospholipase D are essential for substrate recognition. Toxicon 2014, 81, 43–47. [Google Scholar] [CrossRef]
- Buch, D.R.; Souza, F.N.; Meissner, G.O.; Morgon, A.M.; Gremski, L.H.; Ferrer, V.P.; Trevisan-Silva, D.; Matsubara, F.H.; Boia-Ferreira, M.; Sade, Y.B.; et al. Brown spider (Loxosceles genus) venom toxins: Evaluation of biological conservation by immune cross-reactivity. Toxicon 2015, 108, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Tavares, F.L.; Peichoto, M.E.; Rangel, D.D.M.; Barbaro, K.C.; Cirillo, M.C.; Santoro, M.L.; Sano-Martins, I.S. Loxosceles gaucho spider venom and its sphingomyelinase fraction trigger the main functions of human and rabbit platelets. Hum. Exp. Toxicol. 2011, 30, 1567–1574. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, D.A.; Caporrino, M.C.; Barbaro, K.C.; Della-Casa, M.S.; Faquim-Mauro, E.L.; Magalhaes, G.S. Recombinant phospholipase D from Loxosceles gaucho binds to platelets and promotes phosphatidylserine exposure. Toxins 2017, 9, 191. [Google Scholar] [CrossRef] [PubMed]
- Ingraham, L.; Boxer, L.; Haak, R.; Baehner, R. Membrane fluidity changes accompanying phagocytosis in normal and in chronic granulomatous disease polymorphonuclear leukocytes. Blood 1981, 58, 830–835. [Google Scholar] [CrossRef]
- Harlow, E.; Lane, D. Antibodies: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1988. [Google Scholar]
- Da Silva, T.P.; de Castro, F.J.; Vuitika, L.; Polli, N.L.C.; Antunes, B.C.; Bóia-Ferreira, M.; Minozzo, J.C.; Mariutti, R.B.; Matsubara, F.H.; Arni, R.K.; et al. Brown Spiders’ Phospholipases-D with Potential Therapeutic Applications: Functional Assessment of Mutant Isoforms. Biomedicines 2021, 9, 320. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Perez-Iratxeta, C.; Andrade-Navarro, M.A. K2D2: Estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 2008, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Louis-Jeune, C.; Andrade-Navarro, M.A.; Perez-Iratxeta, C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins Struct. Funct. Bioinforma. 2012, 80, 374–381. [Google Scholar] [CrossRef]
- Exnowitz, F.; Meyer, B.; Hackl, T. NMR for direct determination of Km and Vmax of enzyme reactions based on the Lambert W function-analysis of progress curves. Biochim. Biophys. Acta Proteins Proteom. 2012, 1824, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.J.; Marshall, C.B.; Theillet, F.X.; Binolfi, A.; Selenko, P.; Ikura, M. Real-time NMR monitoring of biological activities in complex physiological environments. Curr. Opin. Struct. Biol. 2015, 32, 39–47. [Google Scholar] [CrossRef]
- Hwang, T.-L.; Shaka, A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Waveforms and Pulsed Field Gradients. J. Magn. Reson. 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Cheng, X.; Jo, S.; Lee, H.S.; Klauda, J.B.; Im, W. CHARMM-GUI micelle builder for pure/mixed micelle and protein/micelle complex systems. J. Chem. Inf. Model. 2013, 53, 2171–2180. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cisneros, G.A.; Cruzeiro, V.W.D.; et al. Amber 22; University of California: San Francisco, CA, USA, 2022; pp. 13–945. [Google Scholar]
- Dickson, C.J.; Walker, R.C.; Gould, I.R. Lipid21: Complex Lipid Membrane Simulations with AMBER. J. Chem. Theory Comput. 2022, 18, 1726–1736. [Google Scholar] [CrossRef] [PubMed]
- Schott-Verdugo, S.; Gohlke, H. PACKMOL-Memgen: A Simple-To-Use, Generalized Workflow for Membrane-Protein-Lipid-Bilayer System Building. J. Chem. Inf. Model. 2019, 59, 2522–2528. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Åqvist, J.; Wennerström, P.; Nervall, M.; Bjelic, S.; Brandsdal, B.O. Molecular dynamics simulations of water and biomolecules with a Monte Carlo constant pressure algorithm. Chem. Phys. Lett. 2004, 384, 288–294. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005, 33, 368–371. [Google Scholar] [CrossRef]
- Lilkova, E. The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Izadi, S.; Anandakrishnan, R.; Onufriev, A. V Building Water Models: A Different Approach. J. Phys. Chem. Lett. 2014, 5, 3863–3871. [Google Scholar] [CrossRef]
- Hopkins, C.W.; Le Grand, S.; Walker, R.C.; Roitberg, A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015, 11, 1864–1874. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; Mcgee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Mongan, J.; Simmerling, C.; McCammon, J.A.; Case, D.A.; Onufriev, A. Generalized Born model with a simple, robust molecular volume correction. J. Chem. Theory Comput. 2007, 3, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
| Control | LiRecDT1 | LgRecDT1 | LlRecDT1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| - | - | 12.5 µg | 25 µg | 50 µg | 12.5 µg | 25 µg | 50 µg | 12.5 µg | 25 µg | 50 µg |
| 16 h | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) |
| 20 h | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 2/5 (40%) | 4/5 (80%) | 0/5 (0%) | 0/5 (0%) | 3/5 (60%) |
| 24 h | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 2/5 (40%) | 3/5 (60%) | 5/5 (100%) | 0/5 (0%) | 1/5 (20%) | 3/5 (60%) |
| 30 h | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 3/5 (60%) * | 3/5 (60%) * | 5/5 (100%) | - | 0/5 (0%) | 3/5 (60%) * | 5/5 (100%) |
| 36 h | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 3/5 (60%) | 3/5 (60%) | - | - | 0/5 (0%) | 3/5 (60%) | - |
| 42 h | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) | 4/5 (80%) | 5/5 (100%) | - | - | 0/5 (0%) | 3/5 (60%) | - |
| PLD | ∆GEl+GB b (kcal/mol) | ∆Gvdw c (kcal/mol) | ∆Gsurf d (kcal/mol) | ∆Geff e (kcal/mol) |
|---|---|---|---|---|
| LgRecDT1 | 76.3 ± 4.0 | −129.7 ± 6.4 | −17.2 ± 0.8 | −70.6 ± 4.0 |
| LlRecDT1 | 117.1 ± 5.3 | −179.5 ± 8.0 | −24.1 ± 1.0 | −86.5 ± 4.1 |
| LiRecDT1 | 98.3 ± 4.8 | −136.3 ± 6.8 | −18.8 ± 0.9 | −56.7 ± 3.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Justa, H.C.; Hernández González, J.E.; Vuitika, L.; Mariutti, R.B.; Magnago, P.A.M.; de Moraes, F.R.; Senff-Ribeiro, A.; Gremski, L.H.; Arni, R.K.; Veiga, S.S. Comparative Biochemical, Structural, and Functional Analysis of Recombinant Phospholipases D from Three Loxosceles Spider Venoms. Int. J. Mol. Sci. 2023, 24, 12006. https://doi.org/10.3390/ijms241512006
da Justa HC, Hernández González JE, Vuitika L, Mariutti RB, Magnago PAM, de Moraes FR, Senff-Ribeiro A, Gremski LH, Arni RK, Veiga SS. Comparative Biochemical, Structural, and Functional Analysis of Recombinant Phospholipases D from Three Loxosceles Spider Venoms. International Journal of Molecular Sciences. 2023; 24(15):12006. https://doi.org/10.3390/ijms241512006
Chicago/Turabian Styleda Justa, Hanna Câmara, Jorge Enrique Hernández González, Larissa Vuitika, Ricardo Barros Mariutti, Pedro Augusto Martinho Magnago, Fábio Rogério de Moraes, Andrea Senff-Ribeiro, Luiza Helena Gremski, Raghuvir Krishnaswamy Arni, and Silvio Sanches Veiga. 2023. "Comparative Biochemical, Structural, and Functional Analysis of Recombinant Phospholipases D from Three Loxosceles Spider Venoms" International Journal of Molecular Sciences 24, no. 15: 12006. https://doi.org/10.3390/ijms241512006
APA Styleda Justa, H. C., Hernández González, J. E., Vuitika, L., Mariutti, R. B., Magnago, P. A. M., de Moraes, F. R., Senff-Ribeiro, A., Gremski, L. H., Arni, R. K., & Veiga, S. S. (2023). Comparative Biochemical, Structural, and Functional Analysis of Recombinant Phospholipases D from Three Loxosceles Spider Venoms. International Journal of Molecular Sciences, 24(15), 12006. https://doi.org/10.3390/ijms241512006







