Characterization of Dof Transcription Factors and the Heat-Tolerant Function of PeDof-11 in Passion Fruit (Passiflora edulis)
Abstract
1. Introduction
2. Results
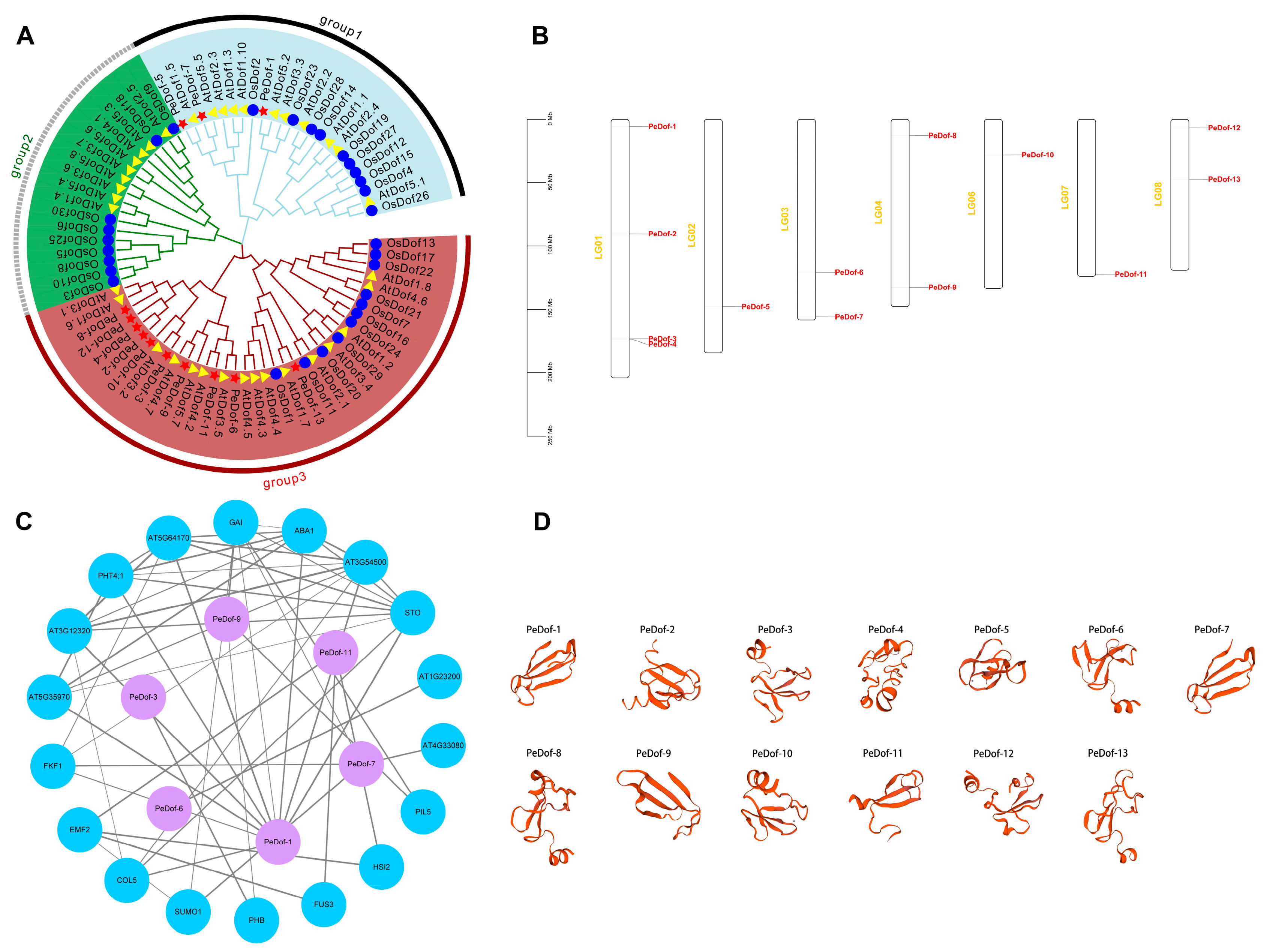
2.1. Identification of Passion Fruit Dof Members and Information Analysis
2.2. Analysis of the Structures of PeDof Motifs and Promoters
2.3. Collinearity Analysis of PeDofs
2.4. Expression of PeDof in Different Tissues of Passion Fruit and under High-Temperature Stress
2.5. Analysis of PeDof-11 in Different Tissues of Passion Fruit and under High-Temperature Stress
3. Discussion
3.1. Identification and Characteristics of PeDof Genes
3.2. Cis-Elements of PeDof Genes
3.3. Potential Role of PeDof genes in Different Tissues
3.4. The Response of Dof Genes in Abiotic Stresses
4. Materials and Methods
4.1. Identification of Dofs and Information in Passion Fruit
4.2. Plant Materials, Transcriptome, and qRT-PCR Analysis
4.3. Cloning and Vector Construction of PeDof-11 and the Promoter
4.4. Functional Complementation of PeDof-11 in Yeast (Saccharomyces cerevisiae)
4.5. Plant Transformation and High-Temperature Treatment
4.6. Detection of GUS Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Costa, J.L.; Jesus, O.N.D.; Oliverira, G.A.F.; Oliverira, E.J.D. Effect of selection on genetic variability in yellow passion fruit. Crop Breed. Appl. Biotechnol. 2014, 12, 253–260. [Google Scholar] [CrossRef]
- Huang, D.; Xu, Y.; Wu, B.; Ma, F.N.; Song, S. Comparative analysis of basic quality of passion fruits (Passiflora edulis Sims) in Guangxi, Guizhou and Fujian, China. Bangladesh J. Bot. 2019, 48, 901–906. [Google Scholar]
- Song, S.; Zhang, D.; Ma, F.; Xing, W.; Huang, D.; Wu, B.; Chen, J.; Chen, D.; Xu, B.; Xu, Y. Genome-wide identification and expression analyses of the aquaporin gene family in passion fruit (Passiflora edulis), revealing PeTIP3-2 to be involved in drought stress. Int. J. Mol. Sci. 2022, 23, 5720. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, P.; Ma, F.; Huang, D.; Xing, W.; Wu, B.; Sun, P.; Xu, B.; Song, S. Characterization of the NAC Transcription Factor in Passion Fruit (Passiflora edulis) and Functional Identification of PeNAC-19 in Cold Stress. Plants 2023, 12, 1393. [Google Scholar] [CrossRef]
- Corrales, A.R.; Nebauer, S.G.; Carrillo, L.; Fernández-Nohales, P.; Marqués, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R.-V.; et al. Characterization of tomato cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic tress responses. J. Exp. Bot. 2014, 65, 995–1012. [Google Scholar] [CrossRef]
- Ewas, M.; Khames, E.; Ziaf, K.; Shahzad, R.; Nishawy, E.; Ali, F.; Subthain, H.; Amar, M.H.; Ayaad, M.; Ghaly, O.; et al. The Tomato DOF Daily Fluctuations 1, TDDF1 acts as flowering accelerator and protector against various stresses. Sci. Rep. 2017, 7, 10299. [Google Scholar] [CrossRef]
- Ruta, V.; Longo, C.; Lepri, A.; Angelis, V.D.; Occhigrossi, S.; Costantino, P.; Vittorioso, P. The DOF Transcription Factors in Seed and Seedling Development. Plants 2020, 9, 218. [Google Scholar] [CrossRef]
- Blair, E.J.; Goralogia, G.S.; Lincoln, M.J.; Imaizumi, T.; Nagel, D.H. Clock-Controlled and Cold-Induced CYCLING DOF FACTOR6 Alters Growth and Development in Arabidopsis. Front. Plant Sci. 2022, 13, 919676. [Google Scholar] [CrossRef]
- Ramírez Gonzales, L.; Shi, L.; Bergonzi, S.B.; Oortwijn, M.; Franco-Zorrilla, J.M.; Solano-Tavira, R.; Visser, R.G.F.; Abelenda, J.A.; Bachem, C.W.B. Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 2021, 105, 855–869. [Google Scholar] [CrossRef]
- Da Silva, D.C.; da Silveira Falavigna, V.; Fasoli, M.; Buffon, V.; Porto, D.D.; Pappas, G.J., Jr.; Pezzotti, M.; Pasquali, G.; Revers, L.F. Transcriptome analyses of the Dof-like gene family in grapevine reveal its involvement in berry, flower and seed development. Hortic. Res. 2016, 3, 16042. [Google Scholar] [CrossRef]
- Rojas-Gracia, P.; Roque, E.; Medina, M.; López-Martín, M.J.; Cañas, L.A.; Beltrán, J.P.; Gómez-Mena, C. The DOF Transcription Factor SlDOF10 Regulates Vascular Tissue Formation during Ovary Development in Tomato. Front. Plant Sci. 2019, 10, 216. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Diego, L.; Pilar, C.; Jesús, V.C. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17. [Google Scholar]
- Hariom, K.; Shubhra, G.; Kumar, S.V.; Smita, R.; Dinesh, Y. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol. Biol. Rep. 2011, 38, 5037–5053. [Google Scholar]
- Venkatesh, J.; Park, S.W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Izui, K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 1993, 268, 16028–16036. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Y.; Ma, Y.Y.; Lu, Y.J.; Yue, J.J.; Ming, R. Expression profiling of the Dof gene family under abiotic stresses in spinach. Sci. Rep. 2021, 11, 14429. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, S.; Gao, Y.; Yang, J. Characterization of Dof transcription factors and their responses to osmotic stress in poplar (Populus trichocarpa). PLoS ONE 2017, 12, e0170210. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.; Xian, J.; Jia, Z.; Hou, X.; Zhang, L. Genome-wide identification of Dof transcription factors possibly associated with internal browning of postharvest pineapple fruits. Sci. Hortic. 2019, 251, 80–87. [Google Scholar] [CrossRef]
- Wen, C.L.; Cheng, Q.; Zhao, L.; Mao, A.; Yang, J.; Yu, S.; Weng, Y.; Xu, Y. Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 2016, 6, 23072. [Google Scholar] [CrossRef]
- Koralewski, T.; Krutovsky, K. Evolution of exon–intron structure and alternative splicing. PLoS ONE 2011, 6, e18055. [Google Scholar] [CrossRef] [PubMed]
- Malviya, N.; Gupta, S.; Singh, V.K.; Yadav, M.K.; Bisht, N.C.; Sarangi, B.K.; Yadav, D. Genome wide in silico characterization of Dof gene families of pigeonpea (Cajanus cajan (L) Millsp.). Mol. Biol. Rep. 2015, 42, 535–552. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N. Dissecting cis-regulation of FLOWERING LOCUS T. Plant Cell 2010, 22, 1422. [Google Scholar] [CrossRef][Green Version]
- Yanagisawa, S.; Sheen, J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 1998, 10, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Neeta, L.; Saeid, B.; Mohan, B.S.; Prem, L.B. Genome-Wide In Silico Identification and Comparative Analysis of Dof Gene Family in Brassica napus. Plants 2021, 10, 709. [Google Scholar]
- Gandass, N.; Kajal Salvi, P. Intrinsically disordered protein, DNA binding with one finger transcription factor (OsDOF27) implicates thermotolerance in yeast and rice. Front. Plant Sci. 2022, 13, 956299. [Google Scholar] [CrossRef]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Morata, B.R.; Pollmann, S.; Granell, A.; Molina, R.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, W.; Ma, F.; Huang, D.; Xing, W.; Wu, B.; Sun, P.; Chen, D.; Xu, B.; Song, S. Characterization of the Passion Fruit (Passiflora edulis Sim) bHLH Family in Fruit Development and Abiotic Stress and Functional Analysis of PebHLH56 in Cold Stress. Horticulturae 2023, 9, 272. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Xu, Y.; Xing, W.T.; Wu, B.; Huang, D.M.; Ma, F.N. Identification of the passion fruit (Passiflora edulis Sims) MYB family in fruit development and abiotic stress, and functional analysis of PeMYB87 in abiotic stresses. Front. Plant Sci. 2023, 14, 1124351. [Google Scholar] [CrossRef]
- Xu, Y.; Jin, Z.Q.; Xu, B.Y.; Li, J.Y.; Li, Y.J.; Wang, X.Y.; Wang, A.B.; Hu, W.; Huang, D.M.; Wei, Q.; et al. Identification of transcription factors interacting with a 1274bp promoter of MaPIP1;1 which confers high-level gene expression and drought stress Inducibility in transgenic Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 278. [Google Scholar] [CrossRef]







| Gene | Gene ID | CDS Length (bp) | Protein Length (aa) | Molecular Formula | MW (Da) | pI | Number of Phosphate Sites | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| PeDof-1 | P_edulia010000233.g | 1434 | 477 | C2245H3535N647O725S22 | 51,894.98 | 6.72 | Ser:56 Thr:22 Tyr:19 | Nucleus |
| PeDof-2 | P_edulia010001122.g | 852 | 283 | C1289H1984N386O407S11 | 29,752.93 | 9.25 | Ser:39 Thr:13 Tyr:9 | Nucleus |
| PeDof-3 | P_edulia010002592.g | 891 | 296 | C1396H2158N410O463S18 | 32,670.04 | 6.31 | Ser:35 Thr:21 Tyr:8 | Nucleus |
| PeDof-4 | P_edulia010002622.g | 849 | 282 | C1274H1983N375O408S14 | 29,529.87 | 8.96 | Ser:42 Thr:13 Tyr:6 | Nucleus |
| PeDof-5 | P_edulia020006400.g | 1083 | 360 | C1701H2678N490O553S19 | 39,450.07 | 8.07 | Ser:51 Thr:24 Tyr:13 | Nucleus |
| PeDof-6 | P_edulia030008409.g | 1014 | 337 | C1564H2429N459O502S14 | 36,143.10 | 9.31 | Ser:49 Thr:17 Tyr:11 | Nucleus |
| PeDof-7 | P_edulia030009124.g | 1503 | 500 | C2359H3647N679O782S23 | 54,769.37 | 5.63 | Ser:59 Thr:30 Tyr:22 | Nucleus |
| PeDof-8 | P_edulia040010004.g | 1038 | 345 | C1599H2471N457O511S14 | 36,721.80 | 8.85 | Ser:51 Thr:18 Tyr:15 | Nucleus |
| PeDof-9 | P_edulia040010672.g | 738 | 245 | C1179H1839N333O367S10 | 26,871.18 | 9.01 | Ser:27 Thr:19 Tyr:10 | Nucleus |
| PeDof-10 | P_edulia060015094.g | 906 | 301 | C1437H2240N404O482S20 | 33,529.21 | 4.75 | Ser:44 Thr:14 Tyr:13 | Nucleus |
| PeDof-11 | P_edulia070018540.g | 750 | 249 | C1138H1749N339O373S11 | 26,500.11 | 8.14 | Ser:32 Thr:12 Tyr:6 | Nucleus |
| PeDof-12 | P_edulia080018881.g | 804 | 267 | C1237H1945N369O399S12 | 28,755.00 | 9.28 | Ser:40 Thr:15 Tyr:7 | Nucleus |
| PeDof-13 | P_edulia080019069.g | 960 | 319 | C1501H2312N466O486S13 | 35,078.48 | 8.57 | Ser:44 Thr:14 Tyr:11 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Xu, Y.; Gui, J.; Huang, Y.; Ma, F.; Wu, W.; Han, T.; Qiu, W.; Yang, L.; Song, S. Characterization of Dof Transcription Factors and the Heat-Tolerant Function of PeDof-11 in Passion Fruit (Passiflora edulis). Int. J. Mol. Sci. 2023, 24, 12091. https://doi.org/10.3390/ijms241512091
Chen G, Xu Y, Gui J, Huang Y, Ma F, Wu W, Han T, Qiu W, Yang L, Song S. Characterization of Dof Transcription Factors and the Heat-Tolerant Function of PeDof-11 in Passion Fruit (Passiflora edulis). International Journal of Molecular Sciences. 2023; 24(15):12091. https://doi.org/10.3390/ijms241512091
Chicago/Turabian StyleChen, Ge, Yi Xu, Jie Gui, Yongcai Huang, Funing Ma, Wenhua Wu, Te Han, Wenwu Qiu, Liu Yang, and Shun Song. 2023. "Characterization of Dof Transcription Factors and the Heat-Tolerant Function of PeDof-11 in Passion Fruit (Passiflora edulis)" International Journal of Molecular Sciences 24, no. 15: 12091. https://doi.org/10.3390/ijms241512091
APA StyleChen, G., Xu, Y., Gui, J., Huang, Y., Ma, F., Wu, W., Han, T., Qiu, W., Yang, L., & Song, S. (2023). Characterization of Dof Transcription Factors and the Heat-Tolerant Function of PeDof-11 in Passion Fruit (Passiflora edulis). International Journal of Molecular Sciences, 24(15), 12091. https://doi.org/10.3390/ijms241512091






