The Potential of Bile Acids as Biomarkers for Metabolic Disorders
Abstract
:1. Introduction
2. An Overview of BA Metabolism and Physiology
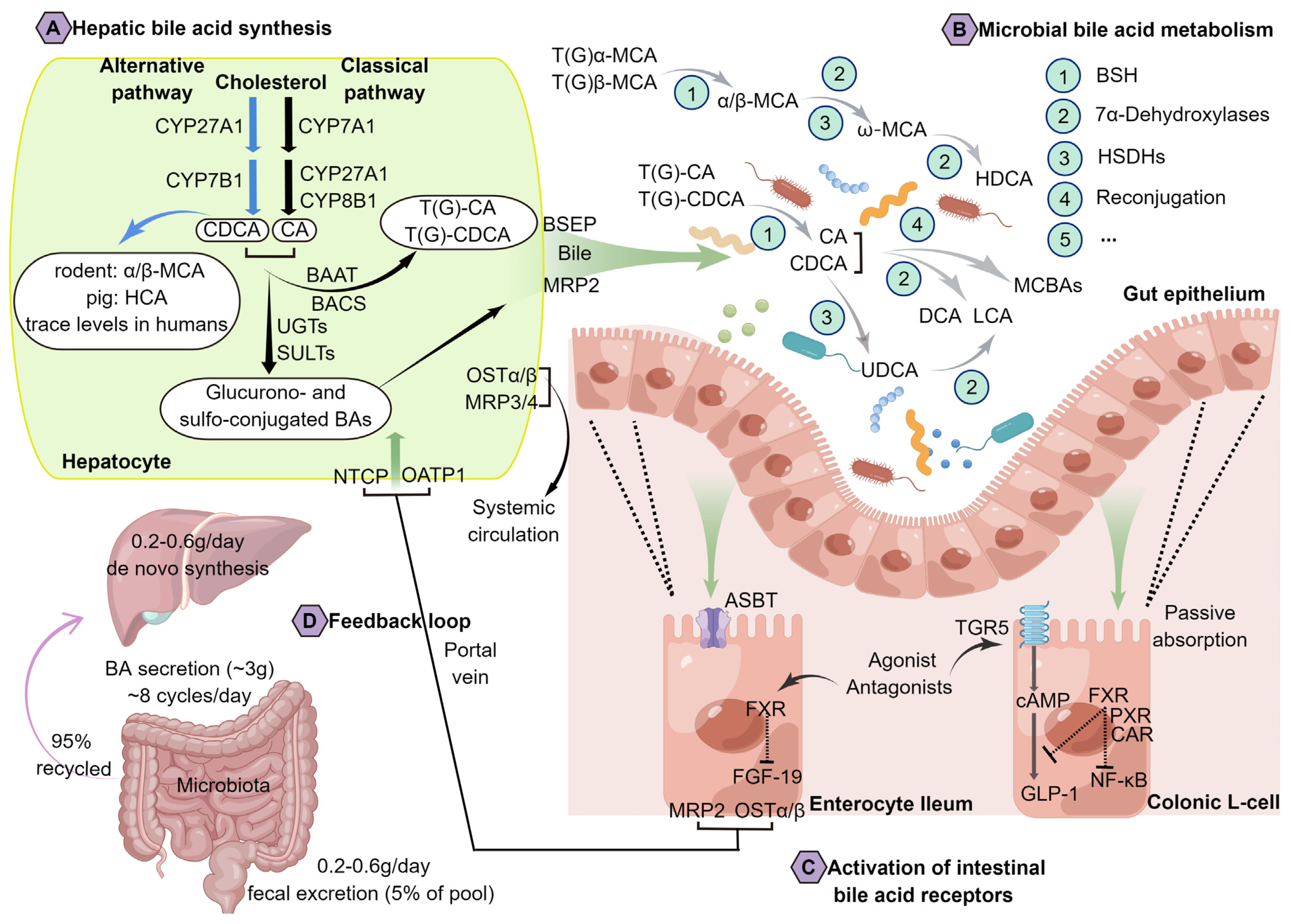
3. Bacterial Metabolism of BAs Has Been Recognized as an Important Actor within the Metabolic Homeostasis of the Host
4. Major Factors Associated with Alterations in BA Metabolism
4.1. Antibiotics
4.2. Diet
4.3. Disease
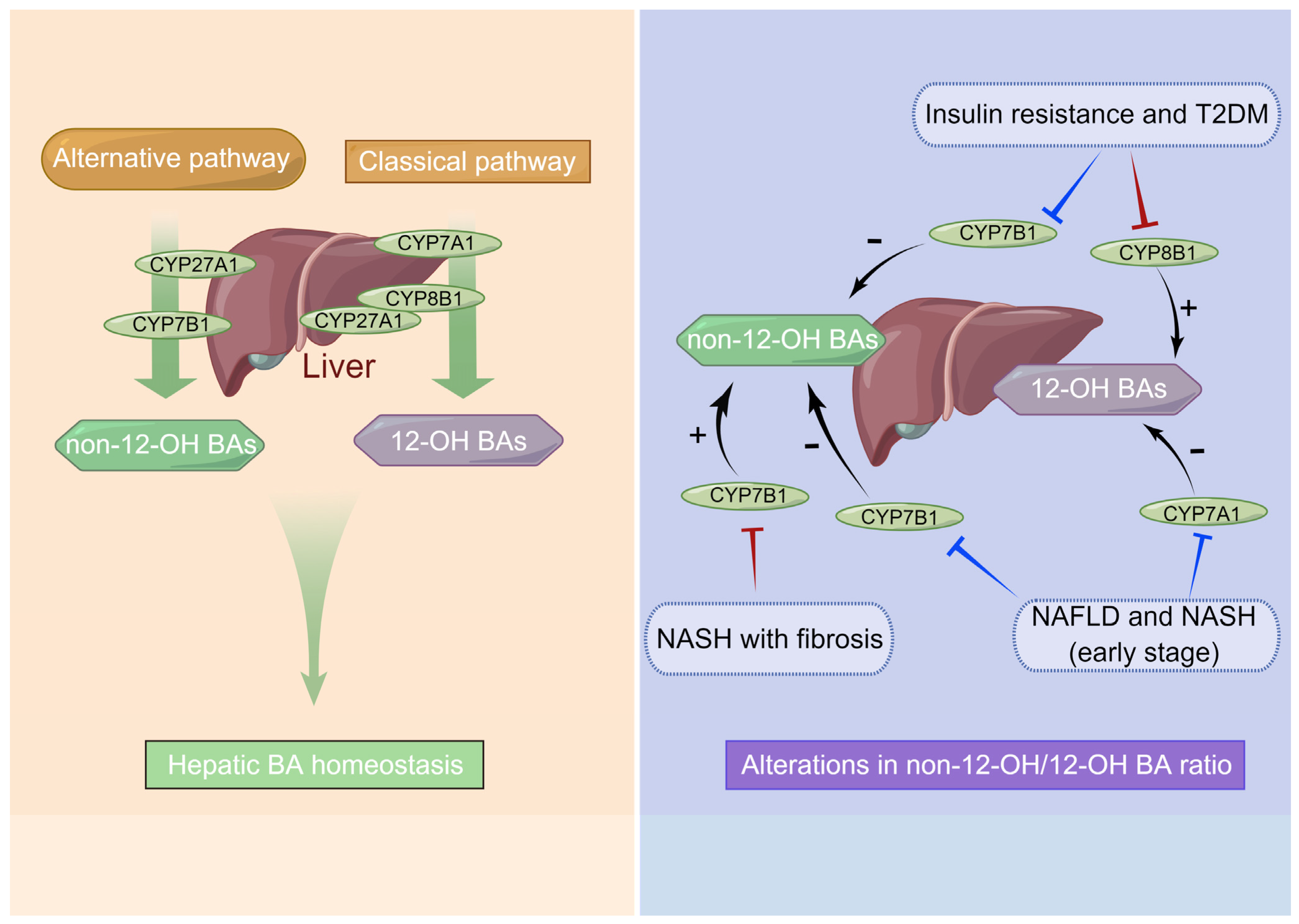
4.3.1. Chronic Liver Diseases Such as NAFLD and NASH
4.3.2. T2DM
4.3.3. Gastric and Hepatocellular Cancer
4.3.4. Enteric Diseases
4.3.5. Cholestatic Liver Diseases
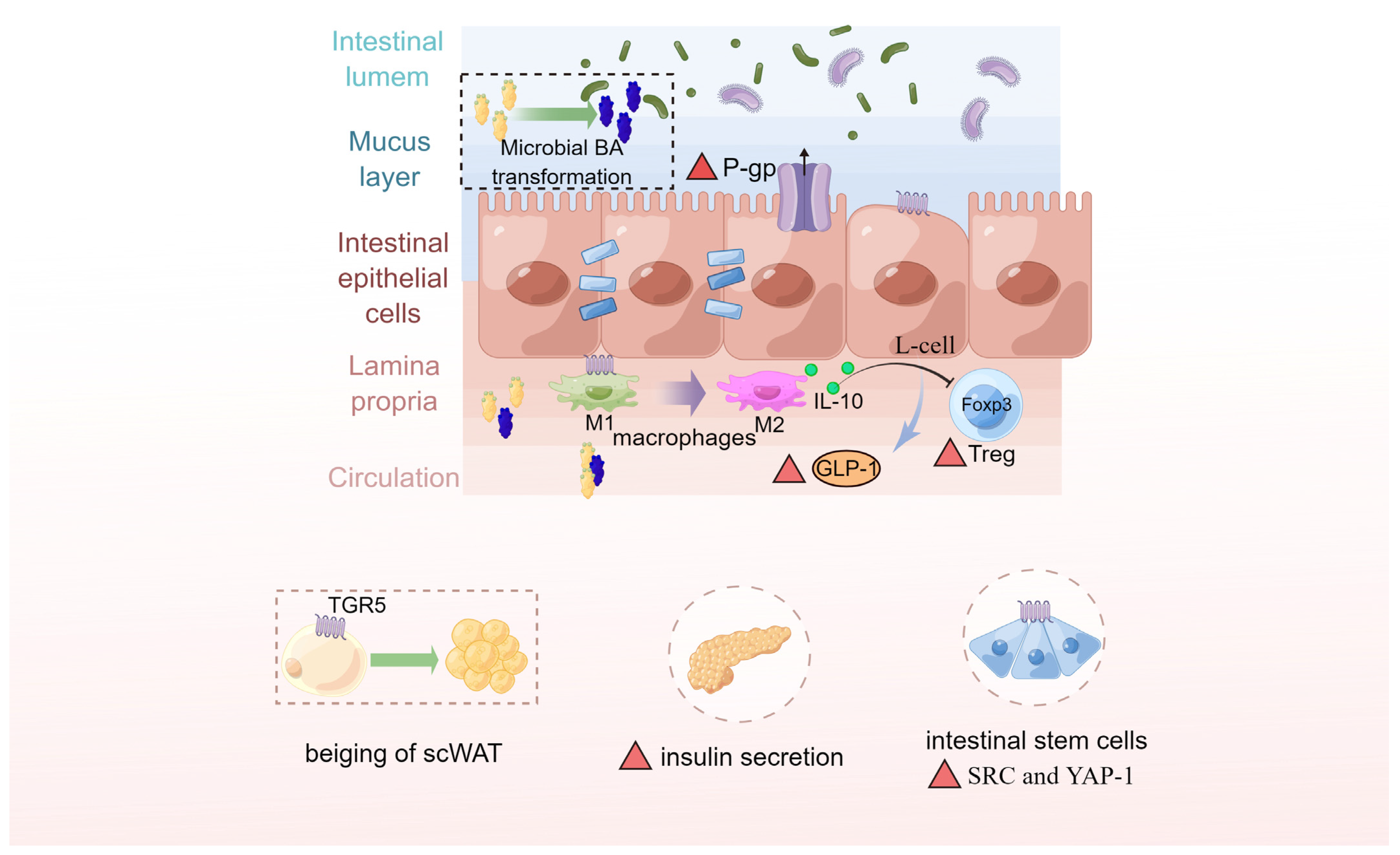
5. BAs Shape the Gut Microbiota
6. BAs Are Closely Associated with Metabolic Disorders
6.1. Dysregulation of BA Synthesis Contributes to Metabolic Disorders
6.2. Gut Microbiota Interacting with BAs and Altering BA Metabolism Are Critical in Metabolic Disorders
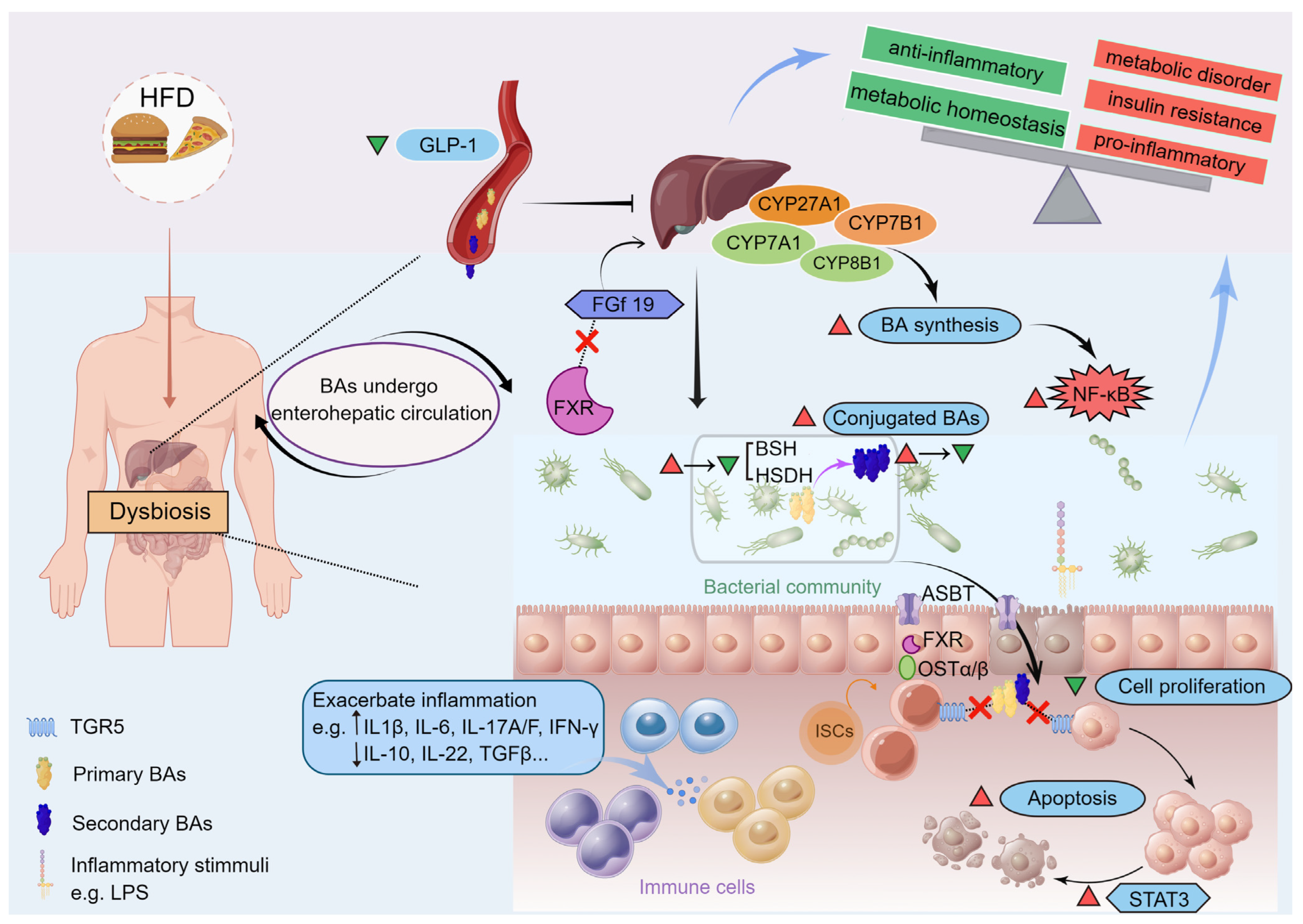
6.3. Microbiota-Derived BAs Serve as the Chemical Signals and Biomarkers in Gut–Liver Axis
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Wang, D.Q.; Carey, M.C. Therapeutic uses of animal biles in traditional Chinese medicine: An ethnopharmacological, biophysical chemical and medicinal review. World J. Gastroenterol. 2014, 20, 9952–9975. [Google Scholar] [CrossRef] [PubMed]
- Kritchevsky, D.; Nair, P.P. (Eds.) Chemistry of the bile acids. In The Bile Acids Chemistry, Physiology, and Metabolism, 1st ed; Springer: Boston, MA, USA, 1971; pp. 1–10. [Google Scholar] [CrossRef]
- Midtvedt, T. Microbial bile acid transformation. Am. J. Clin. Nutr. 1974, 27, 1341–1347. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R.; Forrest, E.K. Bile acid content of human serum. I. Serum bile acids in patients with hepatic disease. J. Clin. Investig. 1957, 36, 530–537. [Google Scholar] [CrossRef] [Green Version]
- Holzbach, R.T.; Marsh, M.E.; Freedman, M.R.; Fazio, V.W.; Lavery, I.; Jagelman, D.A. Portal vein bile acids in patients with severe inflammatory bowel disease. Gut 1980, 21, 428–435. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorakova, K.; Garewal, H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 2005, 589, 47–65. [Google Scholar] [CrossRef]
- Li, J.; Dawson, P.A. Animal models to study bile acid metabolism. Biochim. Biophys. Acta Mol. Basis. Dis. 2019, 1865, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Kootte, R.S.; Levin, E.; Salojärvi, J.; Smits, L.P.; Hartstra, A.V.; Udayappan, S.D.; Hermes, G.; Bouter, K.E.; Koopen, A.M.; Holst, J.J.; et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017, 26, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Li, T.; Chiang, J.Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 2014, 66, 948–983. [Google Scholar] [CrossRef] [Green Version]
- Wahlstrom, A.; Sayin, S.I.; Marschall, H.U.; Backhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, N.; Baker, S.S.; Chapa-Rodriguez, A.; Liu, W.; Nugent, C.A.; Tsompana, M.; Mastrandrea, L.; Buck, M.J.; Baker, R.D.; Genco, R.J.; et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 2018, 67, 1881–1891. [Google Scholar] [CrossRef]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019, 26, 222–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.; Pellicciari, R.; Pruzanski, M.; Auwerx, J.; Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 2008, 7, 678–693. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.Q.; Xu, C.F.; Chen, H.T.; Liu, S.; Teng, X.D.; Xu, G.Y.; Yu, C.H. Association of caveolin-3 and cholecystokinin A receptor with cholesterol gallstone disease in mice. World J. Gastroenterol. 2014, 20, 9513–9518. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef] [Green Version]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.; Gioiello, A.; Noriega, L.; Strehle, A.; Oury, J.; Rizzo, G.; Macchiarulo, A.; Yamamoto, H.; Mataki, C.; Pruzanski, M.; et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009, 10, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, S.; Li, Q.; Cai, L.; Wang, C.; Lei, X. Chemoproteomic profiling of bile acid interacting proteins. ACS Cent. Sci. 2017, 3, 501–509. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, A.A.; Seegar, T.C.M.; Ficarro, S.B.; McCurry, M.D.; Ramachandran, D.; Yao, L.; Chaudhari, S.N.; Ndousse-Fetter, S.; Banks, A.S.; Marto, J.A.; et al. Development of a covalent inhibitor of gut bacterial bile salt hydrolases. Nat. Chem. Biol. 2020, 16, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Chen, T.; Jiang, R.; Zhao, A.; Wu, Q.; Kuang, J.; Sun, D.; Ren, Z.; Li, M.; Zhao, M.; et al. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab. 2021, 33, 791–803 e797. [Google Scholar] [CrossRef] [PubMed]
- Martinot, E.; Sèdes, L.; Baptissart, M.; Lobaccaro, J.M.; Caira, F.; Beaudoin, C.; Volle, D.H. Bile acids and their receptors. Mol. Aspects Med. 2017, 56, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, H.; Kolodziejczyk, A.A.; Halstuch, D.; Elinav, E. Bile acids in glucose metabolism in health and disease. J. Exp. Med. 2018, 215, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Albaugh, V.L.; Banan, B.; Antoun, J.; Xiong, Y.; Guo, Y.; Ping, J.; Alikhan, M.; Clements, B.A.; Abumrad, N.N.; Flynn, C.R. Role of Bile Acids and GLP-1 in Mediating the Metabolic Improvements of Bariatric Surgery. Gastroenterology 2019, 156, 1041–1051. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Ye, J.; Zhang, F.; Su, H.; Yang, X.; He, H.; Liu, F.; Zhu, X.; Wang, L.; Gao, P.; et al. Chenodeoxycholic Acid (CDCA) protects against the lipopolysaccharide-induced impairment of the intestinal epithelial barrier function via the FXR-MLCK pathway. J. Agric. Food Chem. 2019, 67, 8868–8874. [Google Scholar] [CrossRef]
- Xie, Q.; Khaoustov, V.I.; Chung, C.C.; Sohn, J.; Krishnan, B.; Lewis, D.E.; Yoffe, B. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress-induced caspase-12 activation. Hepatology 2002, 36, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [Green Version]
- Monte, M.J.; Marin, J.J.; Antelo, A.; Vazquez-Tato, J. Bile acids: Chemistry, physiology, and pathophysiology. World J. Gastroenterol. 2009, 15, 804–816. [Google Scholar] [CrossRef]
- Dumas, M.E.; Barton, R.H.; Toye, A.; Cloarec, O.; Blancher, C.; Rothwell, A.; Fearnside, J.; Tatoud, R.; Blanc, V.; Lindon, J.C.; et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. USA 2006, 103, 12511–12516. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.; Kinross, J.; Gibson, G.R.; Burcelin, R.; Jia, W.; Pettersson, S.; Nicholson, J.K. Therapeutic modulation of microbiota-host metabolic interactions. Sci. Transl. Med. 2012, 4, 137rv6. [Google Scholar] [CrossRef] [PubMed]
- Gérard, P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, R.A.; Melnik, A.V.; Vrbanac, A.; Fu, T.; Patras, K.A.; Christy, M.P.; Bodai, Z.; Belda-Ferre, P.; Tripathi, A.; Chung, L.K.; et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 2020, 579, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Swann, J.R.; Want, E.J.; Geier, F.M.; Spagou, K.; Wilson, I.D.; Sidaway, J.E.; Nicholson, J.K.; Holmes, E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4523–4530. [Google Scholar] [CrossRef]
- Kuno, T.; Hirayama-Kurogi, M.; Ito, S.; Ohtsuki, S. Reduction in hepatic secondary bile acids caused by short-term antibiotic-induced dysbiosis decreases mouse serum glucose and triglyceride levels. Sci. Rep. 2018, 8, 1253. [Google Scholar] [CrossRef] [Green Version]
- Yokota, A.; Fukiya, S.; Islam, K.B.; Ooka, T.; Ogura, Y.; Hayashi, T.; Hagio, M.; Ishizuka, S. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes 2012, 3, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Degirolamo, C.; Rainaldi, S.; Bovenga, F.; Murzilli, S.; Moschetta, A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 2014, 7, 12–18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wu, W.; Lee, Y.-K.; Xie, J.; Zhang, H. Spatial heterogeneity and co-occurrence of mucosal and luminal microbiome across swine intestinal tract. Front. Microbiol. 2018, 9, 48. [Google Scholar] [CrossRef] [Green Version]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Arifuzzaman, M.; Won, T.H.; Li, T.T.; Yano, H.; Digumarthi, S.; Heras, A.F.; Zhang, W.; Parkhurst, C.N.; Kashyap, S.; Jin, W.B.; et al. Inulin fibre promotes microbiota-derived bile acids and type 2 inflammation. Nature 2022, 611, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, J.; Li, J.; Lei, X.; Xu, D.; Wang, Y.; Li, C.; Li, X.; Mao, Y. Turnover of bile acids in liver, serum and caecal content by high-fat diet feeding affects hepatic steatosis in rats. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhou, K.; Sun, T.; Sang, C.; Jia, W.; Xie, G. Altered bile acid glycine: Taurine ratio in the progression of chronic liver disease. J. Gastroenterol. Hepatol. 2022, 37, 208–215. [Google Scholar] [CrossRef]
- Xie, G.; Jiang, R.; Wang, X.; Liu, P.; Zhao, A.; Wu, Y.; Huang, F.; Liu, Z.; Rajani, C.; Zheng, X.; et al. Conjugated secondary 12α-hydroxylated bile acids promote liver fibrogenesis. EBioMedicine 2021, 66, 103290. [Google Scholar] [CrossRef]
- Drzymała-Czyż, S.; Dziedzic, K.; Szwengiel, A.; Krzyżanowska-Jankowska, P.; Nowak, J.K.; Nowicka, A.; Aringazina, R.; Drzymała, S.; Kashirskaya, N.; Walkowiak, J. Serum bile acids in cystic fibrosis patients-glycodeoxycholic acid as a potential marker of liver disease. Dig. Liver Dis. 2022, 54, 111–117. [Google Scholar] [CrossRef]
- Rajani, C.; Jia, W. Disruptions in gut microbial-host co-metabolism and the development of metabolic disorders. Clin. Sci. 2018, 132, 791–811. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, T.; Zhao, A.; Ning, Z.; Kuang, J.; Wang, S.; You, Y.; Bao, Y.; Ma, X.; Yu, H.; et al. Hyocholic acid species as novel biomarkers for metabolic disorders. Nat. Commun. 2021, 12, 1487. [Google Scholar] [CrossRef]
- Wang, S.; Kuang, J.; Zhang, H.; Chen, W.; Zheng, X.; Wang, J.; Huang, F.; Ge, K.; Li, M.; Zhao, M.; et al. Bile acid-microbiome interaction promotes gastric carcinogenesis. Adv. Sci. 2022, 9, e2200263. [Google Scholar] [CrossRef]
- Jia, W.; Wei, M.; Rajani, C.; Zheng, X. Targeting the alternative bile acid synthetic pathway for metabolic diseases. Protein Cell 2021, 12, 411–425. [Google Scholar] [CrossRef]
- Liu, N.; Feng, J.; Lv, Y.; Liu, Q.; Deng, J.; Xia, Y.; Guo, C.; Zhou, Y. Role of bile acids in the diagnosis and progression of liver cirrhosis: A prospective observational study. Exp. Ther. Med. 2019, 18, 4058–4066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Qin, W.X.; Li, Z.L.; Xu, A.J.; Xing, H.; Wu, H.; Zhang, H.; Wang, M.D.; Li, C.; Liang, L.; et al. Tissue and serum metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin. Chim. Acta 2019, 488, 68–75. [Google Scholar] [CrossRef]
- Funabashi, M.; Grove, T.L.; Wang, M.; Varma, Y.; McFadden, M.E.; Brown, L.C.; Guo, C.; Higginbottom, S.; Almo, S.C.; Fischbach, M.A. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 2020, 582, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Paik, D.; Yao, L.; Zhang, Y.; Bae, S.; D’Agostino, G.D.; Zhang, M.; Kim, E.; Franzosa, E.A.; Avila-Pacheco, J.; Bisanz, J.E.; et al. Human gut bacteria produce Τ(H)17-modulating bile acid metabolites. Nature 2022, 603, 907–912. [Google Scholar] [CrossRef]
- Attili, A.F.; Angelico, M.; Cantafora, A.; Alvaro, D.; Capocaccia, L. Bile acid-induced liver toxicity: Relation to the hydrophobic-hydrophilic balance of bile acids. Med. Hypotheses 1986, 19, 57–69. [Google Scholar] [CrossRef]
- Boonstra, K.; Beuers, U.; Ponsioen, C.Y. Epidemiology of primary sclerosing cholangitis and primary biliary cirrhosis: A systematic review. J. Hepatol. 2012, 56, 1181–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Wei, Y.; Xiong, A.; Li, Y.; Guan, H.; Wang, Q.; Miao, Q.; Bian, Z.; Xiao, X.; Lian, M.; et al. Comprehensive Analysis of Serum and Fecal Bile Acid Profiles and Interaction with Gut Microbiota in Primary Biliary Cholangitis. Clin. Rev. Allergy Immunol. 2020, 58, 25–38. [Google Scholar] [CrossRef]
- Núñez, F.P.; Quera, P.R.; Gomollón, F. Primary sclerosing cholangitis and inflammatory bowel disease: Intestine-liver interrelation. Gastroenterol. Hepatol. 2019, 42, 316–325. [Google Scholar] [CrossRef] [Green Version]
- Mousa, O.Y.; Juran, B.D.; McCauley, B.M.; Vesterhus, M.N.; Folseraas, T.; Turgeon, C.T.; Ali, A.H.; Schlicht, E.M.; Atkinson, E.J.; Hu, C.; et al. Bile acid profiles in primary sclerosing cholangitis and their ability to predict hepatic decompensation. Hepatology 2021, 74, 281–295. [Google Scholar] [CrossRef]
- Trottier, J.; Białek, A.; Caron, P.; Straka, R.J.; Heathcote, J.; Milkiewicz, P.; Barbier, O. Metabolomic profiling of 17 bile acids in serum from patients with primary biliary cirrhosis and primary sclerosing cholangitis: A pilot study. Dig. Liver Dis. 2012, 44, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.M.; Candels, L.S.; Hov, J.R.; Myllys, M.; Hassan, R.; Schneider, C.V.; Wahlstrom, A.; Mohs, A.; Zuhlke, S.; Liao, L.; et al. Gut microbiota depletion exacerbates cholestatic liver injury via loss of FXR signalling. Nat. Metab. 2021, 3, 1228–1241. [Google Scholar] [CrossRef]
- Song, F.; Chen, Y.; Chen, L.; Li, H.; Cheng, X.; Wu, W. Association of elevated maternal serum total bile acids with low birth weight and intrauterine fetal growth restriction. JAMA Netw. Open 2021, 4, e2117409. [Google Scholar] [CrossRef] [PubMed]
- Min, H.K.; Kapoor, A.; Fuchs, M.; Mirshahi, F.; Zhou, H.; Maher, J.; Kellum, J.; Warnick, R.; Contos, M.J.; Sanyal, A.J. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012, 15, 665–674. [Google Scholar] [CrossRef] [Green Version]
- Haeusler, R.A.; Astiarraga, B.; Camastra, S.; Accili, D.; Ferrannini, E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes 2013, 62, 4184–4191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biddinger, S.B.; Haas, J.T.; Yu, B.B.; Bezy, O.; Jing, E.; Zhang, W.; Unterman, T.G.; Carey, M.C.; Kahn, C.R. Hepatic insulin resistance directly promotes formation of cholesterol gallstones. Nat. Med. 2008, 14, 778–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Hu, B.; Wu, T.; Zhang, Y.; Xu, Y.; Feng, Y.; Jiang, H. Bile acid profiles in diabetic (db/db) mice and their wild type littermates. J. Pharm. Biomed. Anal. 2016, 131, 473–481. [Google Scholar] [CrossRef]
- Joyce, S.A.; Gahan, C.G. Disease-associated changes in bile acid profiles and links to altered gut microbiota. Dig. Dis. 2017, 35, 169–177. [Google Scholar] [CrossRef]
- Labbé, A.; Ganopolsky, J.G.; Martoni, C.J.; Prakash, S.; Jones, M.L. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. PLoS ONE 2014, 9, e115175. [Google Scholar] [CrossRef]
- Bingham, S.A.; Day, N.E.; Luben, R.; Ferrari, P.; Slimani, N.; Norat, T.; Clavel-Chapelon, F.; Kesse, E.; Nieters, A.; Boeing, H.; et al. Dietary fibre in food and protection against colorectal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC): An observational study. Lancet 2003, 361, 1496–1501. [Google Scholar] [CrossRef]
- Tian, Y.; Gui, W.; Koo, I.; Smith, P.B.; Allman, E.L.; Nichols, R.G.; Rimal, B.; Cai, J.; Liu, Q.; Patterson, A.D. The microbiome modulating activity of bile acids. Gut Microbes 2020, 11, 979–996. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [Green Version]
- Solbach, P.; Chhatwal, P.; Woltemate, S.; Tacconelli, E.; Buhl, M.; Gerhard, M.; Thoeringer, C.K.; Vehreschild, M.; Jazmati, N.; Rupp, J.; et al. BaiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PLoS ONE 2018, 13, e0196977. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.K.; Yun, B.S.; Matsuzaki, K.; Furukawa, M.; Min, H.K.; Bajaj, J.S.; et al. Bile acid 7α-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: Role of secondary bile acids. Cell Chem. Biol. 2019, 26, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Johnston, M.P.; Patel, J.; Byrne, C.D. Causes of mortality in non-alcoholic fatty liver disease (NAFLD) and alcohol related fatty liver disease (AFLD). Curr. Pharm. Des. 2020, 26, 1079–1092. [Google Scholar] [CrossRef]
- Koutoukidis, D.A.; Astbury, N.M.; Tudor, K.E.; Morris, E.; Henry, J.A.; Noreik, M.; Jebb, S.A.; Aveyard, P. Association of weight loss interventions with changes in biomarkers of nonalcoholic fatty liver disease: A systematic review and meta-analysis. JAMA Intern. Med. 2019, 179, 1262–1271. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Patti, M.E. Bile acids, obesity, and the metabolic syndrome. Best Pract. Res. Clin. Gastroenterol. 2014, 28, 573–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertaggia, E.; Jensen, K.K.; Castro-Perez, J.; Xu, Y.; Di Paolo, G.; Chan, R.B.; Wang, L.; Haeusler, R.A. Cyp8b1 ablation prevents Western diet-induced weight gain and hepatic steatosis because of impaired fat absorption. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E121–E133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Ni, Y.; Bao, Y.; Zhang, P.; Zhao, A.; Chen, T.; Xie, G.; Tu, Y.; Zhang, L.; Su, M.; et al. Chenodeoxycholic acid as a potential prognostic marker for Roux-en-Y gastric bypass in Chinese obese patients. J. Clin. Endocrinol. Metab. 2015, 100, 4222–4230. [Google Scholar] [CrossRef] [Green Version]
- Lake, A.D.; Novak, P.; Shipkova, P.; Aranibar, N.; Robertson, D.; Reily, M.D.; Lu, Z.; Lehman-McKeeman, L.D.; Cherrington, N.J. Decreased hepatotoxic bile acid composition and altered synthesis in progressive human nonalcoholic fatty liver disease. Toxicol. Appl. Pharmacol. 2013, 268, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Nojima, K.; Sugimoto, K.; Ueda, H.; Babaya, N.; Ikegami, H.; Rakugi, H. Analysis of hepatic gene expression profile in a spontaneous mouse model of type 2 diabetes under a high sucrose diet. Endocr. J. 2013, 60, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Natividad, J.M.; Lamas, B.; Pham, H.P.; Michel, M.L.; Rainteau, D.; Bridonneau, C.; da Costa, G.; van Hylckama Vlieg, J.; Sovran, B.; Chamignon, C.; et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 2018, 9, 2802. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Houten, S.M.; Wang, L.; Moschetta, A.; Mangelsdorf, D.J.; Heyman, R.A.; Moore, D.D.; Auwerx, J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Investig. 2004, 113, 1408–1418. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, E.; Fu, L.; John, L.; Hultgren, B.; Huang, X.; Renz, M.; Stephan, J.P.; Tsai, S.P.; Powell-Braxton, L.; French, D.; et al. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology 2002, 143, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Wahlström, A.; Marschall, H.U. Role of Bile Acids in Metabolic Control. Trends Endocrinol. Metab. 2018, 29, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe 2020, 27, 659–670. [Google Scholar] [CrossRef]
- Dawson, P.A.; Lan, T.; Rao, A. Bile acid transporters. J. Lipid. Res. 2009, 50, 2340–2357. [Google Scholar] [CrossRef] [Green Version]
- Ridlon, J.M.; Alves, J.M.; Hylemon, P.B.; Bajaj, J.S. Cirrhosis, bile acids and gut microbiota: Unraveling a complex relationship. Gut Microbes 2013, 4, 382–387. [Google Scholar] [CrossRef] [Green Version]
- Chiang, J.Y. Bile acid metabolism and signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Xie, G.; Wang, X.; Fan, J.; Qiu, Y.; Zheng, X.; Qi, X.; Cao, Y.; Su, M.; Wang, X.; et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol. Cell Proteom. 2011, 10, M110.004945. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, X.; Xie, G.; Zhou, M.; Yu, H.; Lin, Y.; Du, G.; Luo, G.; Jia, W.; Liu, P. Urinary metabolite variation is associated with pathological progression of the post-hepatitis B cirrhosis patients. J. Proteome Res. 2012, 11, 3838–3847. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Wang, X.; Huang, F.; Zhao, A.; Chen, W.; Yan, J.; Zhang, Y.; Lei, S.; Ge, K.; Zheng, X.; et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int. J. Cancer 2016, 139, 1764–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadaleta, R.M.; van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.; Renooij, W.; Murzilli, S.; Klomp, L.W.; Siersema, P.D.; Schipper, M.E.; Danese, S.; et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef]
- De Gottardi, A.; Touri, F.; Maurer, C.A.; Perez, A.; Maurhofer, O.; Ventre, G.; Bentzen, C.L.; Niesor, E.J.; Dufour, J.F. The bile acid nuclear receptor FXR and the bile acid binding protein IBABP are differently expressed in colon cancer. Dig. Dis. Sci. 2004, 49, 982–989. [Google Scholar] [CrossRef]
- Modica, S.; Murzilli, S.; Salvatore, L.; Schmidt, D.R.; Moschetta, A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008, 68, 9589–9594. [Google Scholar] [CrossRef] [Green Version]
- Chávez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile Acid Control of Metabolism and Inflammation in Obesity, Type 2 Diabetes, Dyslipidemia, and Nonalcoholic Fatty Liver Disease. Gastroenterology 2017, 152, 1679–1694. [Google Scholar] [CrossRef]
- Honda, A.; Miyazaki, T.; Iwamoto, J.; Hirayama, T.; Morishita, Y.; Monma, T.; Ueda, H.; Mizuno, S.; Sugiyama, F.; Takahashi, S.; et al. Regulation of bile acid metabolism in mouse models with hydrophobic bile acid composition. J. Lipid Res. 2020, 61, 54–69. [Google Scholar] [CrossRef]
- Ward, J.B.J.; Lajczak, N.K.; Kelly, O.B.; O’Dwyer, A.M.; Giddam, A.K.; Ní Gabhann, J.; Franco, P.; Tambuwala, M.M.; Jefferies, C.A.; Keely, S.; et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G550–G558. [Google Scholar] [CrossRef] [Green Version]
- da Silva, J.A., Jr.; Figueiredo, L.S.; Chaves, J.O.; Oliveira, K.M.; Carneiro, E.M.; Abreu, P.A.; Ribeiro, R.A. Effects of tauroursodeoxycholic acid on glucose homeostasis: Potential binding of this bile acid with the insulin receptor. Life Sci. 2021, 285, 120020. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E.; Carino, A.; Zampella, A.; Biagioli, M. Bile acids and their receptors in metabolic disorders. Prog. Lipid Res. 2021, 82, 101094. [Google Scholar] [CrossRef]
- Chen, X.; Lou, G.; Meng, Z.; Huang, W. TGR5: A novel target for weight maintenance and glucose metabolism. Exp. Diabetes Res. 2011, 2011, 853501. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Nakamura, T.; Itadani, H.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Villegas, L.A.; Perino, A.; Lemos, V.; Zietak, M.; Nomura, M.; Pols, T.W.H.; Schoonjans, K. TGR5 signalling promotes mitochondrial fission and beige remodelling of white adipose tissue. Nat. Commun. 2018, 9, 245. [Google Scholar] [CrossRef] [Green Version]
- Foley, S.E.; Tuohy, C.; Dunford, M.; Grey, M.J.; De Luca, H.; Cawley, C.; Szabady, R.L.; Maldonado-Contreras, A.; Houghton, J.M.; Ward, D.V.; et al. Gut microbiota regulation of P-glycoprotein in the intestinal epithelium in maintenance of homeostasis. Microbiome 2021, 9, 183. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, G.; Perino, A.; Yildiz, E.; El Alam, G.; Bou Sleiman, M.; Gioiello, A.; Pellicciari, R.; Schoonjans, K. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology 2020, 159, 956–968.e958. [Google Scholar] [CrossRef]
- Sun, R.; Xu, C.; Feng, B.; Gao, X.; Liu, Z. Critical roles of bile acids in regulating intestinal mucosal immune responses. Therap. Adv. Gastroenterol. 2021, 14, 17562848211018098. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.F.; Bailly, C. Ursodeoxycholic acid and cancer: From chemoprevention to chemotherapy. Pharmacol. Ther. 2019, 203, 107396. [Google Scholar] [CrossRef]
- Eaton, J.E.; Silveira, M.G.; Pardi, D.S.; Sinakos, E.; Kowdley, K.V.; Luketic, V.A.; Harrison, M.E.; McCashland, T.; Befeler, A.S.; Harnois, D.; et al. High-dose ursodeoxycholic acid is associated with the development of colorectal neoplasia in patients with ulcerative colitis and primary sclerosing cholangitis. Am. J. Gastroenterol. 2011, 106, 1638–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenman, L.K.; Holma, R.; Korpela, R. High-fat-induced intestinal permeability dysfunction associated with altered fecal bile acids. World J. Gastroenterol. 2012, 18, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.D.; Mohammed, Z.; Roland, M.M.; Chatzistamou, I.; Jolly, A.; Schoettmer, L.M.; Arroyo, M.; Kakar, K.; Tian, Y.; Patterson, A.; et al. Defective humoral immunity disrupts bile acid homeostasis which promotes inflammatory disease of the small bowel. Nat. Commun. 2022, 13, 525. [Google Scholar] [CrossRef]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Cusi, K. Role of obesity and lipotoxicity in the development of nonalcoholic steatohepatitis: Pathophysiology and clinical implications. Gastroenterology 2012, 142, 711–725.e716. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Zhao, L.; Yu, H.; Ma, X.; Bao, Y.; Rajani, C.; Loo, L.W.; Shvetsov, Y.B.; Yu, H.; Chen, T.; et al. Circulating unsaturated fatty acids delineate the metabolic status of obese individuals. EBioMedicine 2015, 2, 1513–1522. [Google Scholar] [CrossRef] [Green Version]
- Hamer, M.; Stamatakis, E. Metabolically healthy obesity and risk of all-cause and cardiovascular disease mortality. J. Clin. Endocrinol. Metab. 2012, 97, 2482–2488. [Google Scholar] [CrossRef] [Green Version]
- St-Onge, M.P.; Janssen, I.; Heymsfield, S.B. Metabolic syndrome in normal-weight Americans: New definition of the metabolically obese, normal-weight individual. Diabetes Care 2004, 27, 2222–2228. [Google Scholar] [CrossRef] [Green Version]
- Stefan, N.; Häring, H.U.; Hu, F.B.; Schulze, M.B. Metabolically healthy obesity: Epidemiology, mechanisms, and clinical implications. Lancet Diabetes Endocrinol. 2013, 1, 152–162. [Google Scholar] [CrossRef]
- Chávez-Talavera, O.; Wargny, M.; Pichelin, M.; Descat, A.; Vallez, E.; Kouach, M.; Bigot-Corbel, E.; Joliveau, M.; Goossens, J.F.; Le May, C.; et al. Bile acids associate with glucose metabolism, but do not predict conversion from impaired fasting glucose to diabetes. Metabolism 2020, 103, 154042. [Google Scholar] [CrossRef]
- Schramm, C. Bile acids, the microbiome, immunity, and liver tumors. N. Engl. J. Med. 2018, 379, 888–890. [Google Scholar] [CrossRef]
| Disease | Bile Acid Alterations | Ref. |
|---|---|---|
| Non-alcoholic fatty liver disease (NAFLD) | ↑Serum GCA:TCA ratio, GDCA:TDCA ratio, GCDCA:TCDCA ratio; ↓Serum non-12α-OH BAs, such as UDCA and THDCA. | [44,45] |
| Non-alcoholic steatohepatitis (NASH) | ↑Serum conjugated 12α-OH-BAs such as TDCA and GDCA; ↑Serum 12α-OH BAs (patients with fibrosis); ↓Serum non-12α-OH BAs (patients without fibrosis). | [46,47,65] |
| Type 2 diabetes mellitus (T2DM) | ↑Serum 12α-OH BAs; ↓Serum non-12α-OH BAs, including HCA species. | [24,49,66,67,68] |
| Hepatocellular carcinoma (HCC) | ↑Serum TBA, mainly in conjugated PBAs including GCA, GCDCA, TCA, TCDCA, and TUDCA (early-stage HCC patients with cirrhosis); ↓Serum 12α-OH BAs:non-12α-OH BAs ratio (patients with advanced HCC). | [51,52,53] |
| Inflammatory bowel diseases (IBD) | ↑Fecal conjugated BA; Conjugated PBAs; 3α-OH-sulfated SBAs; ↓Fecal SBA, especially TGR5-specific BAs; 3-oxoLCA and isoalloLCA. | [16,55,69,70] |
| Colorectal cancer (CRC) | ↑Fecal SBA, mainly in DCA and LCA in early-stage | [54,57,71] |
| Primary biliary cholangitis (PBC) | ↑Serum PBA, such as CA, CDCA, and their conjugates; ↓Serum SBA, such as DCA and its conjugates, LCA and TLCA; ↑Fecal PBA, such as Fecal CA, TCA, CDCA and TCDCA; ↓Fecal LCA and GDCA. | [59,62] |
| Primary sclerosing cholangitis (PSC) | ↑Serum TBA; ↑Serum CA, CDCA and their conjugates, UDCA, TDCA; ↓Serum LCA, TLCA, DCA and GDCA. | [61,62,63] |
| Intrahepatic cholestasis of pregnancy (ICP) | ↑Maternal serum TBA | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, C.; Zhong, R.; Zhang, W.; Liu, L.; Chen, L.; Zhang, H. The Potential of Bile Acids as Biomarkers for Metabolic Disorders. Int. J. Mol. Sci. 2023, 24, 12123. https://doi.org/10.3390/ijms241512123
Yin C, Zhong R, Zhang W, Liu L, Chen L, Zhang H. The Potential of Bile Acids as Biomarkers for Metabolic Disorders. International Journal of Molecular Sciences. 2023; 24(15):12123. https://doi.org/10.3390/ijms241512123
Chicago/Turabian StyleYin, Chang, Ruqing Zhong, Weidong Zhang, Lei Liu, Liang Chen, and Hongfu Zhang. 2023. "The Potential of Bile Acids as Biomarkers for Metabolic Disorders" International Journal of Molecular Sciences 24, no. 15: 12123. https://doi.org/10.3390/ijms241512123





