Serum NMR-Based Metabolomics Profiling Identifies Lipoprotein Subfraction Variables and Amino Acid Reshuffling in Myeloma Development and Progression
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics of Study Populations
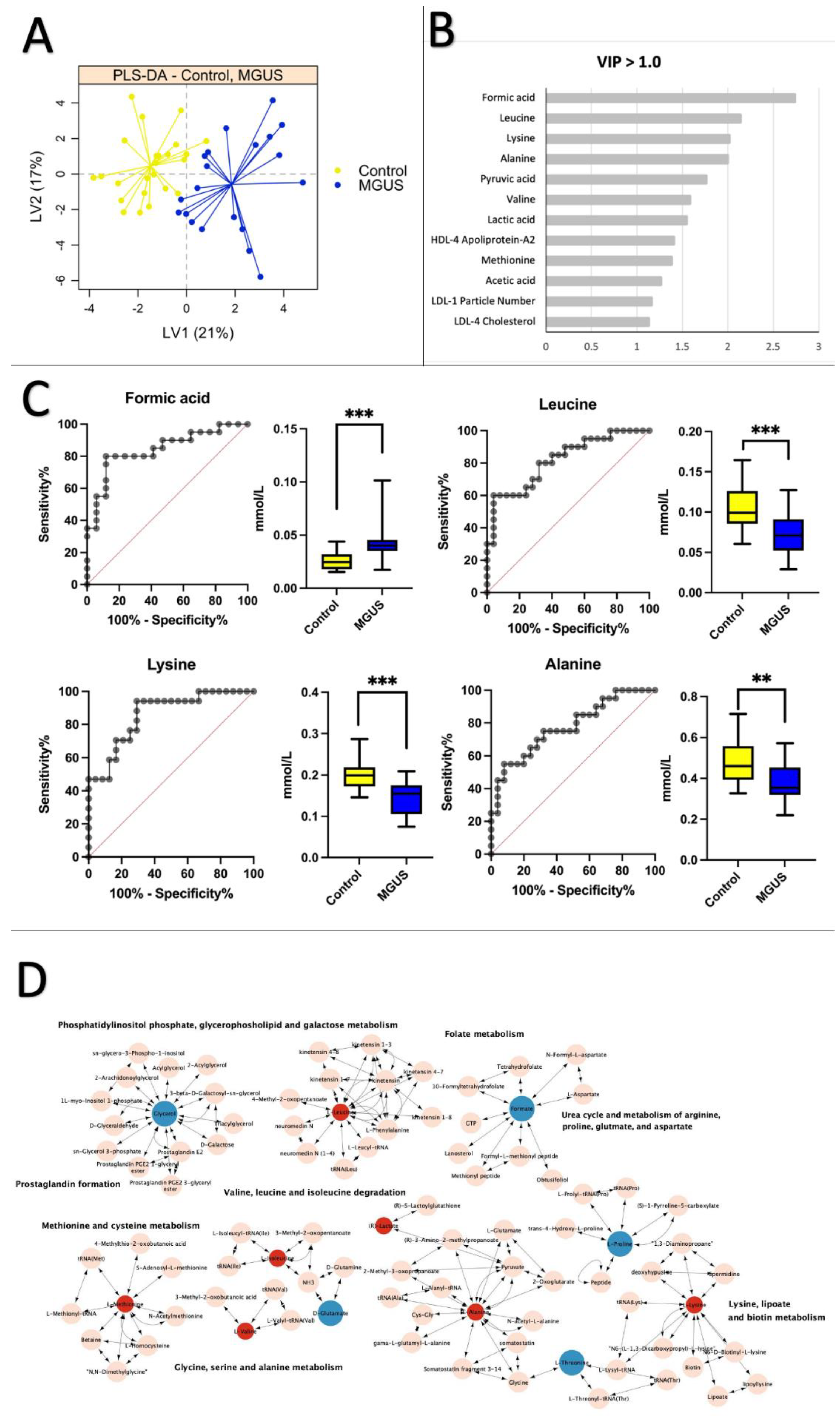
2.2. Healthy Control vs. MGUS: Progression of MGUS Associated with Imbalanced Amino Acid Metabolism
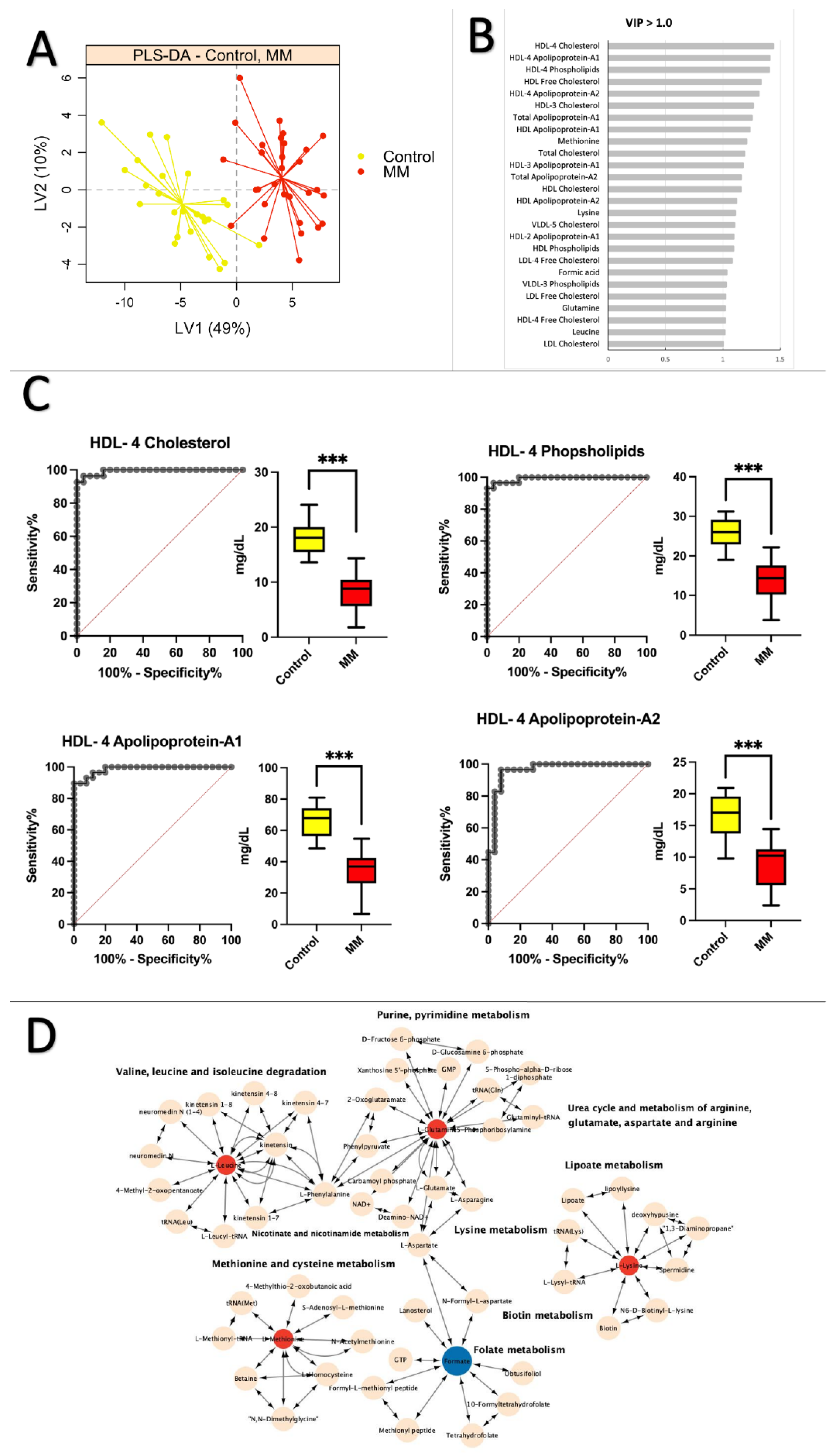
2.3. Healthy Control vs. MM: Low Levels of Apolipoprotein and Cholesterol Are Prevalent in MM Patients
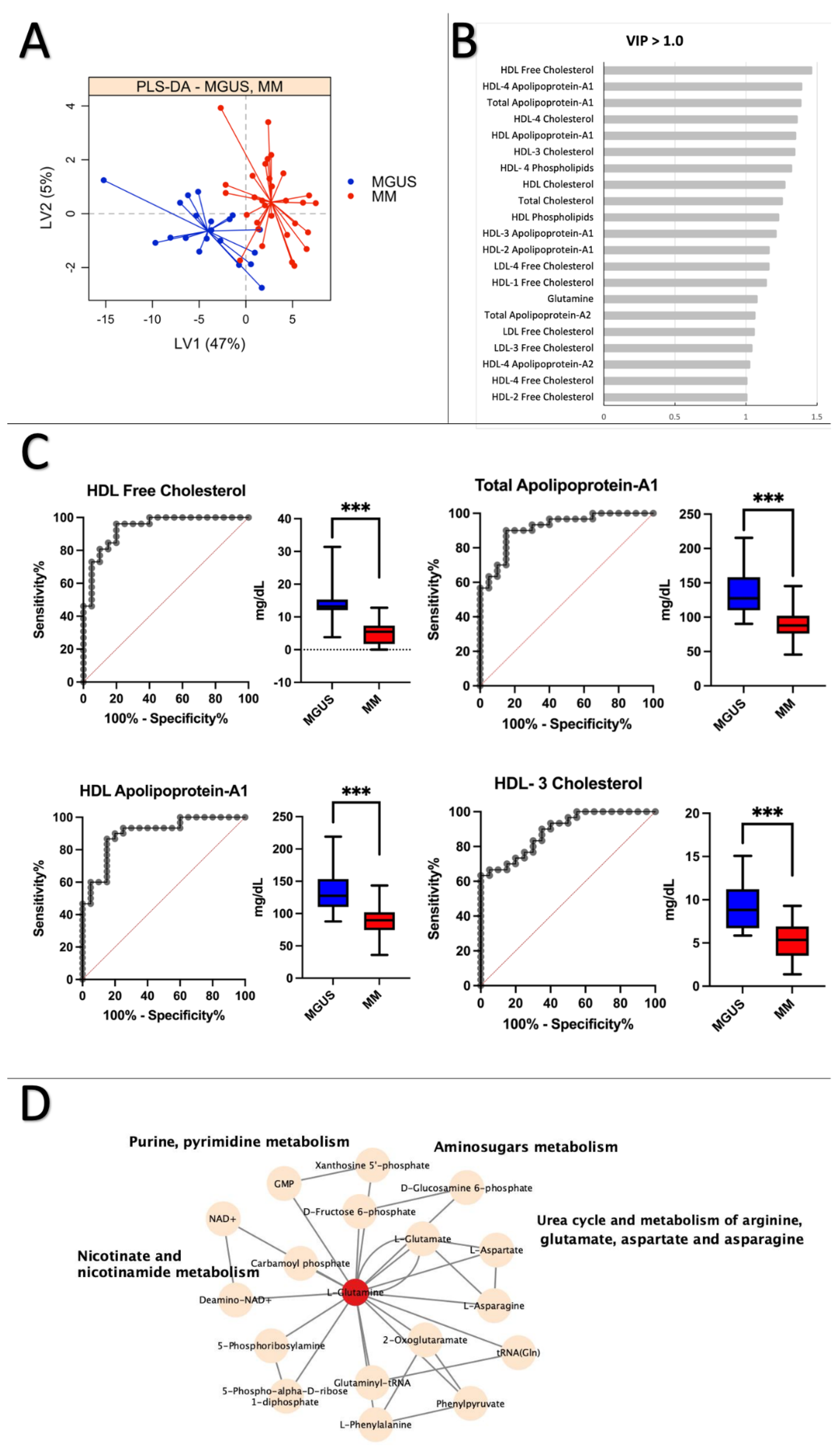
2.4. MGUS vs. MM: Lipoprotein Subfractions Alterations in MGUS Contribute to Symptomatic MM
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. Sample Collection and Processing
4.3. Biochemical Analysis
4.4. Nuclear Magnetic Resonance Spectroscopy
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajkumar, S.V. Updated Diagnostic Criteria and Staging System for Multiple Myeloma. Am. Soc. Clin. Oncol. Educ. Book 2016, 35, e418–e423. [Google Scholar] [CrossRef] [PubMed]
- Koshiaris, C.; Oke, J.; Abel, L.; Nicholson, B.D.; Ramasamy, K.; Bruel, A.V.D. Quantifying intervals to diagnosis in myeloma: A systematic review and meta-analysis. BMJ Open 2018, 8, e019758. [Google Scholar] [CrossRef] [PubMed]
- van de Donk, N.W.C.J.; Pawlyn, C.; Yong, K.L. Multiple myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; van Duin, M.; Sonneveld, P.; Mateos, M.-V.; Gay, F.; Anderson, K.C. Multiple myeloma. Nat. Rev. Dis. Prim. 2017, 3, 17046. [Google Scholar] [CrossRef] [PubMed]
- Bird, S.A.; Boyd, K. Multiple myeloma: An overview of management. Palliat. Care Soc. Pract. 2019, 13, 1178224219868235. [Google Scholar] [CrossRef]
- Ludwig, H.; Novis Durie, S.; Meckl, A.; Hinke, A.; Durie, B. Multiple Myeloma Incidence and Mortality Around the Globe; Interrelations Between Health Access and Quality, Economic Resources, and Patient Empowerment. Oncologist 2020, 25, e1406–e1413. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Li, W.; Lin, M.; Li, T. Serum Protein Electrophoresis and Immunofixation Electrophoresis Detection in Multiple Myeloma. J. Coll. Physicians Surg. Pak. 2021, 31, 864–867. [Google Scholar] [CrossRef]
- Steiner, N.; Müller, U.; Hajek, R.; Sevcikova, S.; Borjan, B.; Jöhrer, K.; Göbel, G.; Pircher, A.; Gunsilius, E. The metabolomic plasma profile of myeloma patients is considerably different from healthy subjects and reveals potential new therapeutic targets. PLoS ONE 2018, 13, e0202045. [Google Scholar] [CrossRef]
- Hideshima, T.; Mitsiades, C.; Tonon, G.; Richardson, P.G.; Anderson, K.C. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat. Rev. Cancer 2007, 7, 585–598. [Google Scholar] [CrossRef]
- Fairfield, H.; Falank, C.; Avery, L.; Reagan, M.R. Multiple myeloma in the marrow: Pathogenesis and treatments. Ann. N. Y. Acad. Sci. 2016, 1364, 32–51. [Google Scholar] [CrossRef] [Green Version]
- Chiecchio, L.; Dagrada, G.P.; White, H.E.; Towsend, M.R.; Protheroe, R.K.M.; Cheung, K.L.; Stockley, D.M.; Orchard, K.H.; Cross, N.C.P.; Harrison, C.J.; et al. Frequent upregulation of MYC in plasma cell leukemia. Genes Chromosom. Cancer 2009, 48, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Puchades-Carrasco, L.; Lecumberri, R.; Martínez-López, J.; Lahuerta, J.-J.; Mateos, M.-V.; Prósper, F.; San-Miguel, J.F.; Pineda-Lucena, A. Multiple Myeloma Patients Have a Specific Serum Metabolomic Profile That Changes after Achieving Complete Remission. Clin. Cancer Res. 2013, 19, 4770–4779. [Google Scholar] [CrossRef] [Green Version]
- Schraw, J.M.; Junco, J.; Brown, A.L.; Scheurer, M.; Rabin, K.R.; Lupo, P.J. Metabolomic profiling identifies pathways associated with minimal residual disease in childhood acute lymphoblastic leukaemia. Ebiomedicine 2019, 48, 49–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Peng, X.-C.; Li, B. The Metabolic Profiles in Hematological Malignancies. Indian J. Hematol. Blood Transfus. 2019, 35, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef]
- Emwas, A.H. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Moco, S. Studying Metabolism by NMR-Based Metabolomics. Front. Mol. Biosci. 2022, 9, 882487. [Google Scholar] [CrossRef]
- Dieterle, F.; Riefke, B.; Schlotterbeck, G.; Ross, A.; Senn, H.; Amberg, A. NMR and MS Methods for Metabonomics. Methods Mol. Biol. 2011, 691, 385–415. [Google Scholar] [CrossRef]
- Bajpai, R.; Matulis, S.M.; Wei, C.; Nooka, A.K.; Von Hollen, H.E.; Lonial, S.; Boise, L.H.; Shanmugam, M. Targeting glutamine metabolism in multiple myeloma enhances BIM binding to BCL-2 eliciting synthetic lethality to venetoclax. Oncogene 2016, 35, 3955–3964. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, C.; Williams, D.S.; Bartlett, D.B.; Essex, S.J.; McNee, G.; Allwood, J.W.; Jewell, E.; Barkhuisen, A.; Parry, H.; Anandram, S.; et al. Alterations in bone marrow metabolism are an early and consistent feature during the development of MGUS and multiple myeloma. Blood Cancer J. 2015, 5, e359. [Google Scholar] [CrossRef] [PubMed]
- Fei, F.; Ma, T.; Zhou, X.; Zheng, M.; Cao, B.; Li, J. Metabolic markers for diagnosis and risk-prediction of multiple myeloma. Life Sci. 2021, 265, 118852. [Google Scholar] [CrossRef]
- Du, H.; Wang, L.; Liu, B.; Wang, J.; Su, H.; Zhang, T.; Huang, Z. Analysis of the Metabolic Characteristics of Serum Samples in Patients with Multiple Myeloma. Front. Pharmacol. 2018, 9, 884. [Google Scholar] [CrossRef]
- Lodi, A.; Tiziani, S.; Khanim, F.L.; Günther, U.L.; Viant, M.R.; Morgan, G.J.; Bunce, C.M.; Drayson, M.T. Proton NMR-Based Metabolite Analyses of Archived Serial Paired Serum and Urine Samples from Myeloma Patients at Different Stages of Disease Activity Identifies Acetylcarnitine as a Novel Marker of Active Disease. PLoS ONE 2013, 8, e56422. [Google Scholar] [CrossRef] [Green Version]
- Gonsalves, W.I.; Broniowska, K.; Jessen, E.; Petterson, X.-M.; Bush, A.G.; Gransee, J.; Lacy, M.Q.; Hitosugi, T.; Kumar, S.K. Metabolomic and Lipidomic Profiling of Bone Marrow Plasma Differentiates Patients with Monoclonal Gammopathy of Undetermined Significance from Multiple Myeloma. Sci. Rep. 2020, 10, 10250. [Google Scholar] [CrossRef]
- Medina, E.A.; Oberheu, K.; Polusani, S.R.; Ortega, V.; Velagaleti, G.V.N.; Oyajobi, B.O. PKA/AMPK signaling in relation to adiponectin’s antiproliferative effect on multiple myeloma cells. Leukemia 2014, 28, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Kuliszkiewicz-Janus, M.; Baczynski, S. Chemotherapy-associated changes in31P MRS spectra of sera from patients with multiple myeloma. NMR Biomed. 1995, 8, 127–132. [Google Scholar] [CrossRef]
- Hachem, H.; Favre, G.; Raynal, G.; Soula, G. Plasma lipoproteins and multiple myeloma. Variations of lipid constituents of HDL and apolipoproteins A1 and B. Ann. Biol. Clin. 1983, 41, 181–185. [Google Scholar]
- Papachristou, N.I.; Blair, H.C.; Kypreos, K.; Papachristou, D.J. High-density lipoprotein (HDL) metabolism and bone mass. J. Endocrinol. 2017, 233, R95–R107. [Google Scholar] [CrossRef]
- Lazaris, V.; Hatziri, A.; Symeonidis, A.; Kypreos, K.E. The Lipoprotein Transport System in the Pathogenesis of Multiple Myeloma: Advances and Challenges. Front. Oncol. 2021, 11, 638288. [Google Scholar] [CrossRef]
- Liang, L.; Li, J.; Fu, H.; Liu, X.; Liu, P. Identification of High Serum Apolipoprotein A1 as a Favorable Prognostic Indicator in Patients with Multiple Myeloma. J. Cancer 2019, 10, 4852–4859. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, X. Prognostic Significance of Pretreatment Apolipoprotein A-I as a Noninvasive Biomarker in Cancer Survivors: A Meta-Analysis. Dis. Markers 2018, 2018, 1034037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, T.; Kristensen, S.R.; Gregersen, H.; Teodorescu, E.M.; Pedersen, S. Prothrombotic abnormalities in patients with multiple myeloma and monoclonal gammopathy of undetermined significance. Thromb. Res. 2021, 202, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Mayers, J.R.; Heiden, M.G.V. Nature and Nurture: What Determines Tumor Metabolic Phenotypes? Cancer Res. 2017, 77, 3131–3134. [Google Scholar] [CrossRef] [Green Version]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef]
- Yavasoglu, I.; Tombuloglu, M.; Kadikoylu, G.; Donmez, A.; Cagırgan, S.; Bolaman, Z. Cholesterol levels in patients with multiple myeloma. Ann. Hematol. 2008, 87, 223–228. [Google Scholar] [CrossRef]
- Tabet, F.; Rye, K.-A. High-density lipoproteins, inflammation and oxidative stress. Clin. Sci. 2009, 116, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-T.; Tian, E.-B.; Chen, Y.-L.; Deng, H.-T.; Wang, Q.-T. Proteomic Analysis for Finding Serum Pathogenic Factors and Potential Biomarkers in Multiple Myeloma. Chin. Med. J. 2015, 128, 1108–1113. [Google Scholar] [CrossRef]
- Mangaraj, M.; Nanda, R.; Panda, S. Apolipoprotein A-I: A Molecule of Diverse Function. Indian J. Clin. Biochem. 2016, 31, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Hungria, V.T.M.; Latrilha, M.C.; Rodrigues, D.G.; Bydlowski, S.P.; Chiattone, C.S.; Maranhão, R.C. Metabolism of a cholesterol-rich microemulsion (LDE) in patients with multiple myeloma and a preliminary clinical study of LDE as a drug vehicle for the treatment of the disease. Cancer Chemother. Pharmacol. 2004, 53, 51–60. [Google Scholar] [CrossRef]
- Scolozzi, R.; Boccafogli, A.; Salmi, R.; Furlani, M.R.; Guidoboni, C.A.; Vicentini, L.; Coletti, M.; Tocchetto, M. Hypocholesterolemia in multiple myeloma. Inverse relation to the component M and the clinical stage. Minerva Med. 1983, 74, 2359–2364. [Google Scholar] [PubMed]
- Edwards, C.M.; Zhuang, J.; Mundy, G.R. The pathogenesis of the bone disease of multiple myeloma. Bone 2008, 42, 1007–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Corral, L.; Gutiérrez, N.C.; Vidriales, M.B.; Mateos, M.V.; Rasillo, A.; García-Sanz, R.; Paiva, B.; Miguel, J.F.S. The Progression from MGUS to Smoldering Myeloma and Eventually to Multiple Myeloma Involves a Clonal Expansion of Genetically Abnormal Plasma Cells. Clin. Cancer Res. 2011, 17, 1692–1700. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Xu, P.; Wang, L.; Zhang, C.; Wang, M.; Ouyang, J.; Chen, B. Cholesterol Levels Provide Prognostic Information in Patients with Multiple Myeloma. Clin. Lab. 2020, 66. [Google Scholar] [CrossRef]
- Misselwitz, B.; Goede, J.S.; Pestalozzi, B.C.; Schanz, U.; Seebach, J.D. Hyperlipidemic myeloma: Review of 53 cases. Ann. Hematol. 2010, 89, 569–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMartin, K.E.; Ambre, J.J.; Tephly, T.R. Methanol poisoning in human subjects: Role for formic acid accumulation in the metabolic acidosis. Am. J. Med. 1980, 68, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Ghadiany, M.; Tabarraee, M.; Salari, S.; Haghighi, S.; Rezvani, H.; Ghasemi, S.N.; Karimi-Sari, H. Adding Oral Pioglitazone to Standard Induction Chemotherapy of Acute Myeloid Leukemia: A Randomized Clinical Trial. Clin. Lymphoma Myeloma Leuk. 2019, 19, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, V.; Yermiahu, T.; Gorodischer, R. Renal pathology and retinol status in multiple myeloma patients. Kidney Int. 2006, 69, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Batuman, V.; Sastrasinh, M.; Sastrasinh, S. Light chain effects on alanine and glucose uptake by renal brush border membranes. Kidney Int. 1986, 30, 662–665. [Google Scholar] [CrossRef] [Green Version]
- Hellman, R.N.; Decker, B.S.; Murray, M. Elevated Serum Creatinine and a Normal Urinalysis: A Short Differential Diagnosis in the Etiology of Renal Failure. Ren. Fail. 2006, 28, 389–394. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Koo, E.H.; Shin, J.-H.; Jang, H.R.; Park, H.-D.; Kwon, G.-Y.; Huh, W.; Kim, D.J.; Kim, Y.-G.; Oh, H.Y.; Lee, J.E. Diagnostic performances of M-protein tests according to the clinical presentations of kidney disease. Eur. J. Intern. Med. 2016, 33, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Derman, B.A.; Langerman, S.S.; Maric, M.; Jakubowiak, A.; Zhang, W.; Chiu, B.C. Sex differences in outcomes in multiple myeloma. Br. J. Haematol. 2021, 192, e66–e69. [Google Scholar] [CrossRef]
- Palumbo, A.; Avet-Loiseau, H.; Oliva, S.; Lokhorst, H.M.; Goldschmidt, H.; Rosinol, L.; Richardson, P.; Caltagirone, S.; Lahuerta, J.J.; Facon, T.; et al. Revised International Staging System for Multiple Myeloma: A Report from International Myeloma Working Group. J. Clin. Oncol. 2015, 33, 2863–2869. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.; Hansen, J.B.; Maltesen, R.G.; Szejniuk, W.M.; Andreassen, T.; Falkmer, U.; Kristensen, S.R. Identifying metabolic alterations in newly diagnosed small cell lung cancer patients. Metab. Open 2021, 12, 100127. [Google Scholar] [CrossRef]
- Dona, A.C.; Jiménez, B.; Schäfer, H.; Humpfer, E.; Spraul, M.; Lewis, M.R.; Pearce, J.T.M.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Precision High-Throughput Proton NMR Spectroscopy of Human Urine, Serum, and Plasma for Large-Scale Metabolic Phenotyping. Anal. Chem. 2014, 86, 9887–9894. [Google Scholar] [CrossRef]
- Jiménez, B.; Holmes, E.; Heude, C.; Tolson, R.F.; Harvey, N.; Lodge, S.L.; Chetwynd, A.J.; Cannet, C.; Fang, F.; Pearce, J.T.M.; et al. Quantitative Lipoprotein Subclass and Low Molecular Weight Metabolite Analysis in Human Serum and Plasma by 1H NMR Spectroscopy in a Multilaboratory Trial. Anal. Chem. 2018, 90, 11962–11971. [Google Scholar] [CrossRef]
- Ma, H.; Sorokin, A.; Mazein, A.; Selkov, A.; Selkov, E.; Demin, O.; Goryanin, I. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol. Syst. Biol. 2007, 3, 135. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Salem, K.B.; Abdelaziz, A.B. Principal Component Analysis (PCA). Tunis Med. 2021, 99, 383–389. [Google Scholar] [PubMed]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopinath, P.P.; Parsad, R.; Joseph, B.; Adarsh, V.S. grapesAgri1: Collection of Shiny Applications for Data Analysis in Agriculture-Part 1. R Package Version 1.1.0. 2021. Available online: https://cran.r-project.org/package=grapesAgri1 (accessed on 22 October 2022).
| MGUS (n = 20) | MM (n = 30) | Between Groups, p-Value | Reference Range, Male/Female | |
|---|---|---|---|---|
| Demographics | ||||
| Age in years (mean ± SD) * | 70.35 ± 11 | 70.7 ± 10 | 0.996 | |
| Male gender | 10 (50%) | 14 (47%) | ||
| Clinical and Biochemical characteristics | ||||
| ISS stage (%) | ||||
| I | 4 (13%) | |||
| II | 16 (53%) | |||
| III | 10 (33%) | |||
| Bone changes (%) | ||||
| None | 8 (27%) | |||
| Halisteresis | 0 (0%) | |||
| Localized | 3 (10%) | |||
| Spread | 19 (63%) | |||
| M-protein, isotype (%) | ||||
| IgG | 15 (50%) | 22 (73%) | ||
| Kappa | 8 (53%) | 17 (77%) | ||
| Lambda | 7 (47%) | 5 (23%) | ||
| IgA | 4 (20%) | 8 (27%) | ||
| Kappa | 2 (50%) | 6 (75%) | ||
| Lambda | 2 (50%) | 2 (25%) | ||
| Plasma cells in bone marrow (%) | 6.0 ± 2.3 | 41 ± 19.4 | <0.001 | |
| M-protein (g/L) | 7.4 ± 6.6 | 42.9 ± 22.4 | <0.001 | |
| κ-Chain, free (mg/L) | 128.5 ± 355.3 | 1179.1 ± 3434.8 | 0.080 | 3.3–19.4 |
| λ-Chain, free (mg/L) | 26.3 ± 35.0 | 225.6 ± 652.5 | 0.014 | 5.7–26.3 |
| Creatinine (µmol/L) | 74.4 ± 26.6 | 120.2 ± 94.1/87.4 ± 35.6 | 0.199 | 60–105/45–90 |
| CRP (mg/L) | 7.4 ± 10.9 | 12.3 ± 25.0 | 0.812 | <8.0 |
| Protein (g/L) | 77.2 ± 7.2 | 107.8 ± 20.0 | <0.001 | 62–78 |
| Albumin (g/L) | 36.8 ± 3.1 | 29.5 ± 4.9 | <0.001 | 34–45 |
| Fibrinogen (μM) | 11.4 ± 3.4 | 10.6 ± 3.9 | 0.156 | 5–12 |
| Hemoglobin (M/F) (mmol/L) | 8.6 ± 1.3\7.7 ± 0.8 | 6.4 ± 1.4\5.8 ± 0.7 | <0.001 | 8.3–10.5/7.3–9.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedersen, S.; Mikkelstrup, M.F.; Kristensen, S.R.; Anwardeen, N.R.; Elrayess, M.A.; Andreassen, T. Serum NMR-Based Metabolomics Profiling Identifies Lipoprotein Subfraction Variables and Amino Acid Reshuffling in Myeloma Development and Progression. Int. J. Mol. Sci. 2023, 24, 12275. https://doi.org/10.3390/ijms241512275
Pedersen S, Mikkelstrup MF, Kristensen SR, Anwardeen NR, Elrayess MA, Andreassen T. Serum NMR-Based Metabolomics Profiling Identifies Lipoprotein Subfraction Variables and Amino Acid Reshuffling in Myeloma Development and Progression. International Journal of Molecular Sciences. 2023; 24(15):12275. https://doi.org/10.3390/ijms241512275
Chicago/Turabian StylePedersen, Shona, Morten Faarbæk Mikkelstrup, Søren Risom Kristensen, Najeha Rizwana Anwardeen, Mohamed A. Elrayess, and Trygve Andreassen. 2023. "Serum NMR-Based Metabolomics Profiling Identifies Lipoprotein Subfraction Variables and Amino Acid Reshuffling in Myeloma Development and Progression" International Journal of Molecular Sciences 24, no. 15: 12275. https://doi.org/10.3390/ijms241512275
APA StylePedersen, S., Mikkelstrup, M. F., Kristensen, S. R., Anwardeen, N. R., Elrayess, M. A., & Andreassen, T. (2023). Serum NMR-Based Metabolomics Profiling Identifies Lipoprotein Subfraction Variables and Amino Acid Reshuffling in Myeloma Development and Progression. International Journal of Molecular Sciences, 24(15), 12275. https://doi.org/10.3390/ijms241512275







