Protein Modelling Highlighted Key Catalytic Sites Involved in Position-Specific Glycosylation of Isoflavonoids
Abstract
1. Introduction
2. Results
2.1. Multiple Sequence Alignment and Phylogenetic Analysis
2.2. Protein Modeling and Docking Analysis
3. Discussion
4. Materials and Methods
4.1. UGT Sequence Selection and Retrieval
4.2. Phylogenetic Analysis and Protein Modelling
4.3. Protein Docking Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Rehman, H.M.; Nawaz, M.A.; Bao, L.; Shah, Z.H.; Lee, J.-M.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-Wide Analysis of Family-1 UDP-Glycosyltransferases in Soybean Confirms Their Abundance and Varied Expression during Seed Development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Bulmer, G.S.; Mattey, A.P.; Parmeggiani, F.; Williams, R.; Ledru, H.; Marchesi, A.; Seibt, L.S.; Both, P.; Huang, K.; Galan, M.C. A Promiscuous Glycosyltransferase Generates Poly-β-1, 4-Glucan Derivatives That Facilitate Mass Spectrometry-Based Detection of Cellulolytic Enzymes. Org. Biomol. Chem. 2021, 19, 5529–5533. [Google Scholar] [CrossRef] [PubMed]
- Schwab, W.; Fischer, T.C.; Giri, A.; Wüst, M. Potential Applications of Glucosyltransferases in Terpene Glucoside Production: Impacts on the Use of Aroma and Fragrance. Appl. Microbiol. Biotechnol. 2015, 99, 165–174. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Bai, X.; Tan, Y.; Xie, W.; Feng, Y.; Yang, G.-Y. Glycosyltransferases: Mining, Engineering and Applications in Biosynthesis of Glycosylated Plant Natural Products. Synth. Syst. Biotechnol. 2022, 7, 602–620. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Stone, S.R.; Kaur, P. Recent Advances in Heterologous Synthesis Paving Way for Future Green-Modular Bioindustries—A Review with Special Reference to Isoflavonoids. Front. Bioeng. Biotechnol. 2021, 9, 532. [Google Scholar] [CrossRef]
- Sajid, M.; Stone, S.R.; Kaur, P. Phylogenetic Analysis and Protein Modelling of Isoflavonoid Synthase Highlights Key Catalytic Sites towards Realising New Bioengineering Endeavours. Bioengineering 2022, 9, 609. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant Flavonoids: Classification, Distribution, Biosynthesis, and Antioxidant Activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Sajid, M.; Channakesavula, C.N.; Stone, S.R.; Kaur, P. Synthetic Biology towards Improved Flavonoid Pharmacokinetics. Biomolecules 2021, 11, 754. [Google Scholar] [CrossRef]
- Xiao, J.; Muzashvili, T.S.; Georgiev, M.I. Advances in the Biotechnological Glycosylation of Valuable Flavonoids. Biotechnol. Adv. 2014, 32, 1145–1156. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Yang, J.; Kang, C.; Huang, L.; Guo, L. Glycosylation of Plant Secondary Metabolites: Regulating from Chaos to Harmony. Environ. Exp. Bot. 2022, 194, 104703. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Li, G.; Savolainen, O.; Chen, Y.; Nielsen, J. De Novo Biosynthesis of Bioactive Isoflavonoids by Engineered Yeast Cell Factories. Nat. Commun. 2021, 12, 6085. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-G. Biological Synthesis of Genistein in Escherichia coli. J. Microbiol. Biotechnol. 2020, 30, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Sordon, S.; Popłoński, J.; Tronina, T.; Huszcza, E. Microbial Glycosylation of Daidzein, Genistein and Biochanin A: Two New Glucosides of Biochanin A. Molecules 2017, 22, 81. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Liu, X.; Zhang, L.; Zhao, G.-R. Modular Engineering of Saccharomyces Cerevisiae for De Novo Biosynthesis of Genistein. Microorganisms 2022, 10, 1402. [Google Scholar] [CrossRef]
- Koirala, N.; Pandey, R.P.; Van Thang, D.; Jung, H.J.; Sohng, J.K. Glycosylation and Subsequent Malonylation of Isoflavonoids in E. Coli: Strain Development, Production and Insights into Future Metabolic Perspectives. J. Ind. Microbiol. Biotechnol. 2014, 41, 1647–1658. [Google Scholar] [CrossRef]
- Chu, L.; Li, S.; Dong, Z.; Zhang, Y.; Jin, P.; Ye, L.; Wang, X.; Xiang, W. Mining and Engineering Exporters for Titer Improvement of Macrolide Biopesticides in Streptomyces. Microb. Biotechnol. 2022, 15, 1120–1132. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, R.; Zhang, G.; Liu, Z.; Jiang, H.; Mao, X. Heterologous Expression of the Plant-Derived Astaxanthin Biosynthesis Pathway in Yarrowia Lipolytica for Glycosylated Astaxanthin Production. J. Agric. Food Chem. 2023, 71, 2943–2951. [Google Scholar] [CrossRef]
- Hofer, B. Recent Developments in the Enzymatic O-Glycosylation of Flavonoids. Appl. Microbiol. Biotechnol. 2016, 100, 4269–4281. [Google Scholar] [CrossRef]
- Lim, E.-K.; Bowles, D.J. A Class of Plant Glycosyltransferases Involved in Cellular Homeostasis. EMBO J. 2004, 23, 2915–2922. [Google Scholar] [CrossRef]
- Bowles, D.; Isayenkova, J.; Lim, E.-K.; Poppenberger, B. Glycosyltransferases: Managers of Small Molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Modolo, L.V.; Escamilla-Trevino, L.L.; Achnine, L.; Dixon, R.A.; Wang, X. Crystal Structure of Medicago Truncatula UGT85H2–Insights into the Structural Basis of a Multifunctional (Iso) Flavonoid Glycosyltransferase. J. Mol. Biol. 2007, 370, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, C.; Zhou, Z.; Zhang, Y. Identification of Three (Iso) Flavonoid Glucosyltransferases from Pueraria lobata. Front. Plant Sci. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Offen, W.; Martinez-Fleites, C.; Yang, M.; Kiat-Lim, E.; Davis, B.G.; Tarling, C.A.; Ford, C.M.; Bowles, D.J.; Davies, G.J. Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification. EMBO J. 2006, 25, 1396–1405. [Google Scholar] [CrossRef]
- Shao, H.; He, X.; Achnine, L.; Blount, J.W.; Dixon, R.A.; Wang, X. Crystal Structures of a Multifunctional Triterpene/Flavonoid Glycosyltransferase from Medicago Truncatula. Plant Cell 2005, 17, 3141–3154. [Google Scholar] [CrossRef]
- Modolo, L.V.; Li, L.; Pan, H.; Blount, J.W.; Dixon, R.A.; Wang, X. Crystal Structures of Glycosyltransferase UGT78G1 Reveal the Molecular Basis for Glycosylation and Deglycosylation of (Iso) Flavonoids. J. Mol. Biol. 2009, 392, 1292–1302. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A. Highly Accurate Protein Structure Prediction for the Human Proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Hameduh, T.; Haddad, Y.; Adam, V.; Heger, Z. Homology Modeling in the Time of Collective and Artificial Intelligence. Comput. Struct. Biotechnol. J. 2020, 18, 3494. [Google Scholar] [CrossRef]
- Chen, K.; Hu, Z.; Song, W.; Wang, Z.; He, J.; Shi, X.; Cui, Q.; Qiao, X.; Ye, M. Diversity of O-Glycosyltransferases Contributes to the Biosynthesis of Flavonoid and Triterpenoid Glycosides in Glycyrrhiza Uralensis. ACS Synth. Biol. 2019, 8, 1858–1866. [Google Scholar] [CrossRef]
- Yin, Q.; Shen, G.; Chang, Z.; Tang, Y.; Gao, H.; Pang, Y. Involvement of Three Putative Glucosyltransferases from the UGT72 Family in Flavonol Glucoside/Rhamnoside Biosynthesis in Lotus Japonicus Seeds. J. Exp. Bot. 2017, 68, 597–612. [Google Scholar] [PubMed]
- Funaki, A.; Waki, T.; Noguchi, A.; Kawai, Y.; Yamashita, S.; Takahashi, S.; Nakayama, T. Identification of a Highly Specific Isoflavone 7-O-Glucosyltransferase in the Soybean (Glycine max (L.) Merr.). Plant Cell Physiol. 2015, 56, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Z.; Li, C.; Gou, J.; Zhang, Y. Molecular Cloning and Characterization of an Isoflavone 7-O-Glucosyltransferase from Pueraria Lobata. Plant Cell Rep. 2014, 33, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Kramer, C.M.; Prata, R.T.; Willits, M.G.; De Luca, V.; Steffens, J.C.; Graser, G. Cloning and Regiospecificity Studies of Two Flavonoid Glucosyltransferases from Allium cepa. Phytochemistry 2003, 64, 1069–1076. [Google Scholar] [CrossRef]
- Noguchi, A.; Saito, A.; Homma, Y.; Nakao, M.; Sasaki, N.; Nishino, T.; Takahashi, S.; Nakayama, T. A UDP-Glucose: Isoflavone 7-O-Glucosyltransferase from the Roots of Soybean (Glycine max) Seedlings: Purification, Gene Cloning, Phylogenetics, and an Implication for an Alternative Strategy of Enzyme Catalysis. J. Biol. Chem. 2007, 282, 23581–23590. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.-Y.; Wang, J.; Zha, L.-P.; Peng, H.-S.; Zhao, Y.-P.; Yuan, Y.; Huang, L.-Q. Molecular Cloning and Functional Characterization of an Isoflavone Glucosyltransferase from Pueraria thomsonii. Chin. J. Nat. Med. 2022, 20, 133–138. [Google Scholar] [CrossRef]
- Nagashima, S.; Inagaki, R.; Kubo, A.; Hirotani, M.; Yoshikawa, T. CDNA Cloning and Expression of Isoflavonoid-Specific Glucosyltransferase from Glycyrrhiza echinata Cell-Suspension Cultures. Planta 2004, 218, 456–459. [Google Scholar] [CrossRef]
- Dhaubhadel, S.; Farhangkhoee, M.; Chapman, R. Identification and Characterization of Isoflavonoid Specific Glycosyltransferase and Malonyltransferase from Soybean Seeds. J. Exp. Bot. 2008, 59, 981–994. [Google Scholar] [CrossRef]
- Ko, J.H.; Kim, B.G.; Kim, J.H.; Kim, H.; Lim, C.E.; Lim, J.; Lee, C.; Lim, Y.; Ahn, J.-H. Four Glucosyltransferases from Rice: CDNA Cloning, Expression, and Characterization. J. Plant Physiol. 2008, 165, 435–444. [Google Scholar] [CrossRef]
- Modolo, L.V.; Escamilla-Treviño, L.L.; Dixon, R.A.; Wang, X. Single Amino Acid Mutations of Medicago Glycosyltransferase UGT85H2 Enhance Activity and Impart Reversibility. FEBS Lett. 2009, 583, 2131–2135. [Google Scholar] [CrossRef]
- Akere, A.; Chen, S.H.; Liu, X.; Chen, Y.; Dantu, S.C.; Pandini, A.; Bhowmik, D.; Haider, S. Structure-Based Enzyme Engineering Improves Donor-Substrate Recognition of Arabidopsis Thaliana Glycosyltransferases. Biochem. J. 2020, 477, 2791–2805. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ren, S.; Liu, X.; Liu, X.; Guo, F.; Sun, W.; Feng, X.; Li, C. Mining of UDP-glucosyltrfansferases in Licorice for Controllable Glycosylation of Pentacyclic Triterpenoids. Biotechnol. Bioeng. 2020, 117, 3651–3663. [Google Scholar] [CrossRef] [PubMed]
- Mameda, R.; Waki, T.; Kawai, Y.; Takahashi, S.; Nakayama, T. Involvement of Chalcone Reductase in the Soybean Isoflavone Metabolon: Identification of GmCHR5, Which Interacts with 2-Hydroxyisoflavanone Synthase. Plant J. 2018, 96, 56–74. [Google Scholar] [CrossRef]
- Lee, P.-G.; Kim, J.; Kim, E.-J.; Lee, S.-H.; Choi, K.-Y.; Kazlauskas, R.J.; Kim, B.-G. Biosynthesis of (−)-5-Hydroxy-Equol and 5-Hydroxy-Dehydroequol from Soy Isoflavone, Genistein Using Microbial Whole Cell Bioconversion. ACS Chem. Biol. 2017, 12, 2883–2890. [Google Scholar] [CrossRef] [PubMed]
- Lacchini, E.; Venegas-Molina, J.; Goossens, A. Structural and Functional Diversity in Plant Specialized Metabolism Signals and Products: The Case of Oxylipins and Triterpenes. Curr. Opin. Plant Biol. 2023, 74, 102371. [Google Scholar] [CrossRef]
- Xiao, H.-Y.; Chen, D.-L.; Lu, T.-T.; Yao, Y.-J.; Liu, N.-Y. The UDP-Glycosyltransferase Gene Family in Achelura Yunnanensis (Lepidoptera: Zygaenidae): Identification, Phylogeny, and Diverse Expression Patterns. Diversity 2022, 14, 407. [Google Scholar] [CrossRef]
- Wang, H.; Guo, H.; Wang, N.; Huo, Y.-X. Toward the Heterologous Biosynthesis of Plant Natural Products: Gene Discovery and Characterization. ACS Synth. Biol. 2021, 10, 2784–2795. [Google Scholar] [CrossRef]
- Wilson, A.E.; Tian, L. Phylogenomic Analysis of UDP-dependent Glycosyltransferases Provides Insights into the Evolutionary Landscape of Glycosylation in Plant Metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]


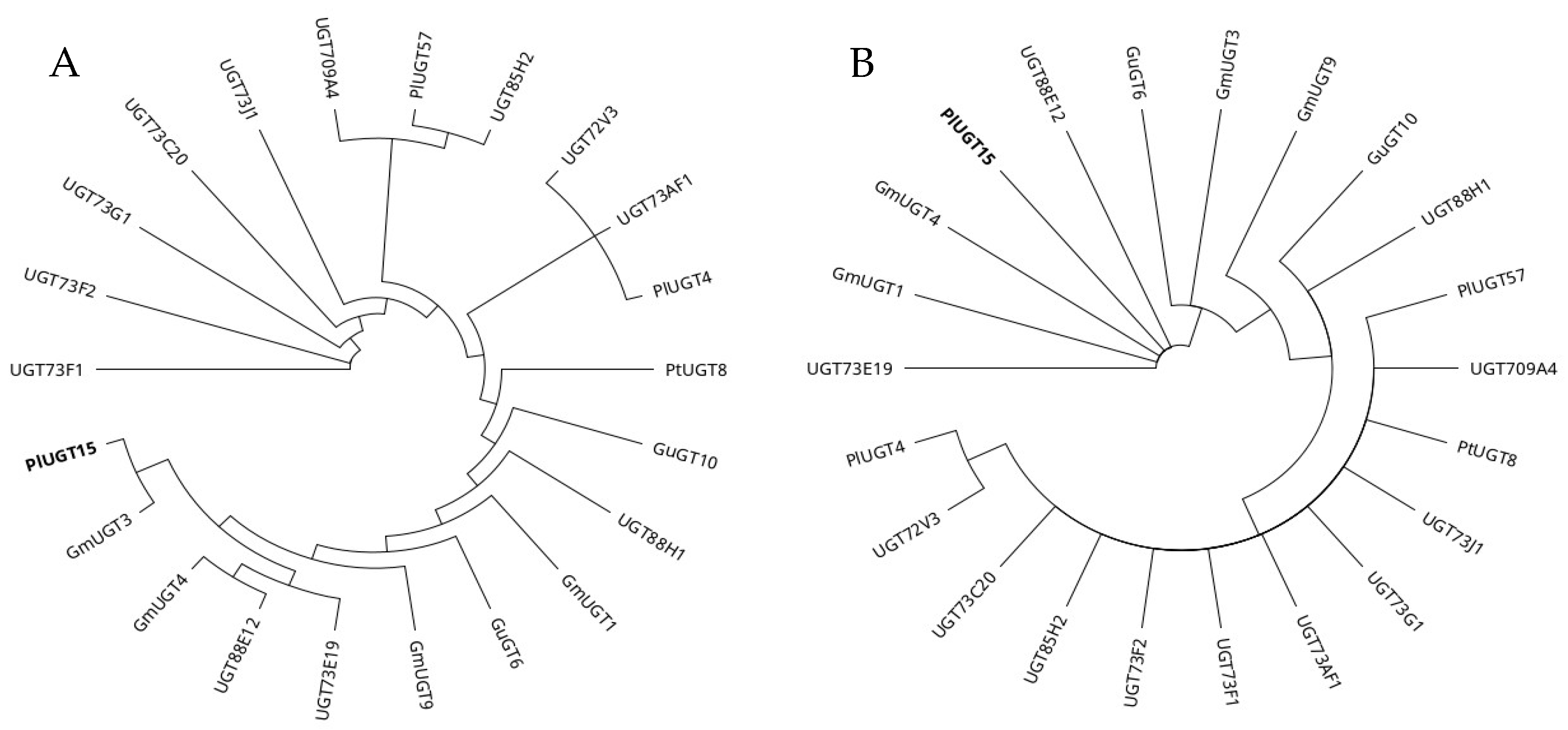
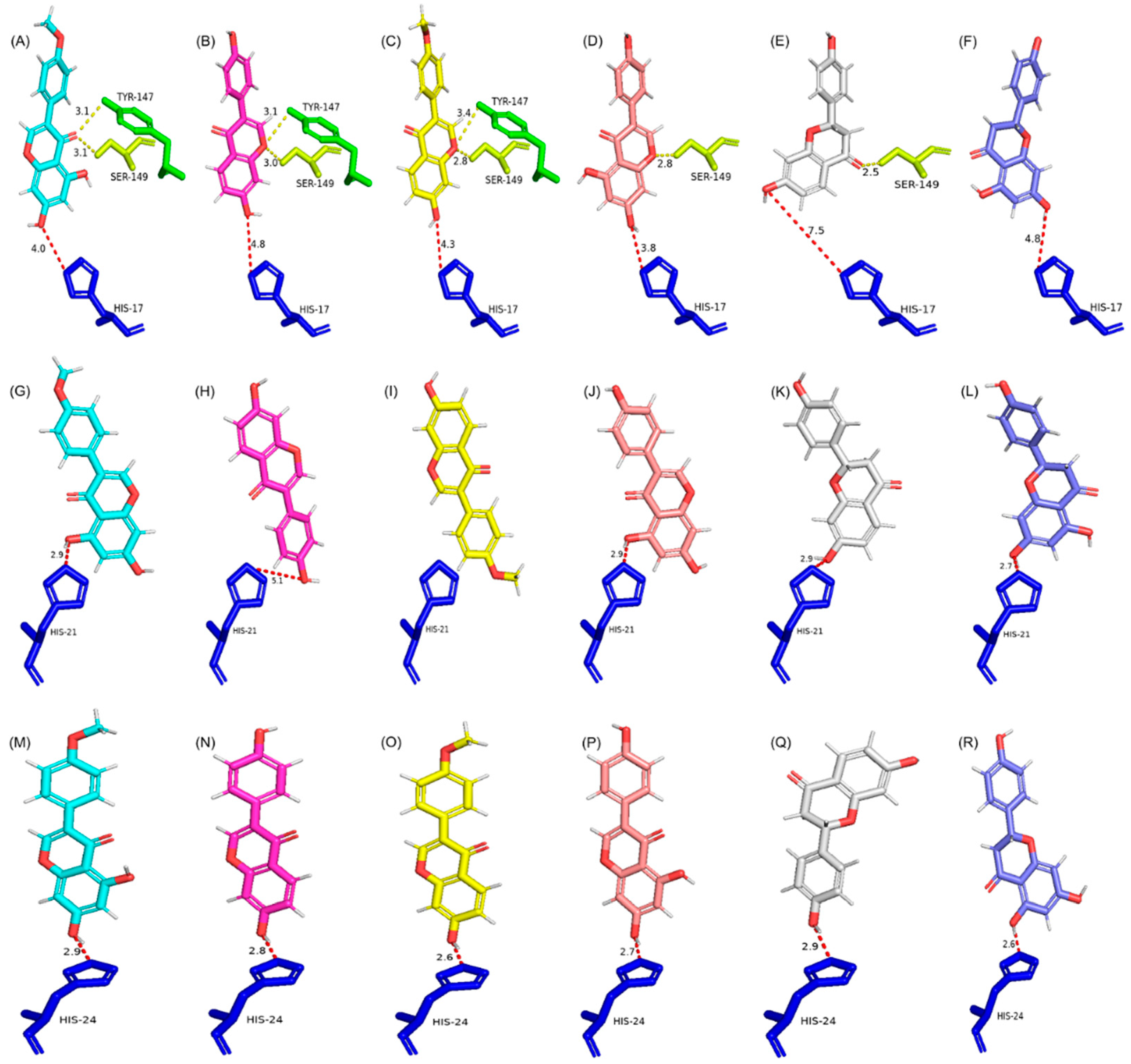
| Description | Total UGTs | Pairwise Identity (%) | Identical Site (%) | Preferred OH Sites |
|---|---|---|---|---|
| All UGTs | 22 | 33.2 | 4.5 | n.a. |
| N-terminus | 22 | 25.1 | 0.3 | n.a. |
| C-terminus | 22 | 44.1 | 9.1 | n.a. |
| PSPG motif | 22 | 67.9 | 28.3 | n.a. |
| Group 1 | 7 | 67.4 | 31.7 | 7-OH |
| Group 2 | 8 | 28.6 | 8.9 | All 3 OH |
| Group 3 | 5 | 28.3 | 8.2 | 7 and 4′ OH |
| Group | Name | Amino Acids | RMSD (a) | Mol. Weight | Isoelectric Point | Aliphatic Index | Instability Index | Subcellular Location | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| G1 | GuGT6 | 486 | 0.51 | 53.40 | 6.24 | 93.83 | Unstable | Cytoplasm | [30] |
| PlUGT15 | 472 | Model | 52.34 | 5.87 | 90.64 | Stable | Chloroplast | [23] | |
| UGT73E19 | 473 | 1.46 | 52.75 | 5.60 | 90.47 | Stable | Chloroplast | [31] | |
| GmUGT9 | 468 | 0.58 | 51.50 | 6.76 | 100.13 | Unstable | Chloroplast | [32] | |
| GmUGT3 | 473 | 0.30 | 52.29 | 5.51 | 91.52 | Stable | Chloroplast | [32] | |
| UGT88E12 | 471 | 0.62 | 52.20 | 5.48 | 92.72 | Unstable | Chloroplast | [33] | |
| UGT88H1 | 451 | 1.61 | 50.26 | 8.10 | 87.49 | Unstable | Chloroplast | [33] | |
| G2 | UGT72V3 | 474 | 1.95 | 52.37 | 5.14 | 97.64 | Unstable | Chloroplast | [31] |
| UGT72AF1 | 469 | 1.30 | 51.95 | 6.12 | 99.74 | Unstable | Chloroplast | [31] | |
| GuGT10 | 462 | 1.06 | 51.53 | 5.99 | 94.48 | Unstable | Chloroplast | [30] | |
| UGT73J1 | 469 | 1.75 | 52.95 | 5.36 | 83.94 | Stable | Chloroplast | [34] | |
| UGT73G1 | 479 | 1.42 | 53.58 | 6.08 | 93.01 | Unstable | Chloroplast | [34] | |
| GmUGT1 | 474 | 0.86 | 52.04 | 4.96 | 95.08 | Unstable | Chloroplast | [35] | |
| PlUGT4 | 465 | 1.61 | 51.08 | 5.85 | 102.90 | Unstable | Cytoplasm | [23] | |
| PtUGT8 | 426 | 1.81 | 47.71 | 5.86 | 99.74 | Unstable | Cytoplasm | [36] | |
| G3 | UGT73F1 | 482 | 1.73 | 54.32 | 5.64 | 86.37 | Unstable | Cytoplasm | [37] |
| UGT73F2 | 476 | 1.22 | 53.24 | 6.46 | 84.20 | Unstable | Chloroplast | [38] | |
| UGT709A4 | 480 | 1.21 | 50.87 | 6.29 | 94.79 | Unstable | Chloroplast | [39] | |
| UGT85H2 | 482 | 0.78 | 54.50 | 5.33 | 88.13 | Stable | Chloroplast | [22,40] | |
| UGT73C20 | 502 | 0.32 | 56.82 | 6.95 | 91.24 | Stable | Chloroplast | [31] | |
| GmUGT4 | 473 | 0.62 | 52.25 | 5.70 | 92.54 | Stable | Chloroplast | [32] | |
| PlUGT57 | 477 | 0.90 | 52.72 | 6.24 | 88.47 | Stable | Cytoplasm | [23] |
| Group | UGTs | GEN | BCN | DZN | FMN | LQN | NGN | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fitness Score | OH Site | Dis. to His (a) | Fitness Score | OH Site | Dis. to His | Fitness Score | OH Site | Dis. to His | Fitness Score | OH Site | Dis. to His | Fitness Score | OH Site | Dis. to His | Fitness Score | OH Site | Dis. to His | ||
| G1 | GuGT6 | 74.90 | 7 | 3.8 | 80.80 | 7 | 3.9 | 73.40 | 7 | 3.9 | 79.45 | 7 | 3.9 | 64.79 | 7 | 6.6 | 71.52 | 7 | 5.6 |
| PlUGT15 | 70.00 | 7 | 3.8 | 76.14 | 7 | 4.0 | 70.81 | 7 | 4.8 | 75.59 | 7 | 4.3 | 66.66 | 7 | 7.5 | 66.93 | 5 | 4.8 | |
| UGT73E19 | 74.76 | 7 | 4.3 | 73.80 | 7 | 3.8 | 73.91 | 7 | 4.3 | 71.33 | 7 | 3.6 | 66.45 | 7 | 6.8 | 7.40 | 7 | 5.6 | |
| GmUGT9 | 66.96 | 7 | 4.2 | 71.91 | (b) | - | 66.33 | 7 | 4.2 | 72.67 | 7 | 4.0 | 67.84 | 7 | 3.1 | 61.90 | 5 | 2.9 | |
| GmUGT3 | 67.18 | 7 | 4.4 | 69.39 | 7 | 3.9 | 65.69 | 7 | 4.0 | 67.79 | 7 | 3.9 | 57.57 | 4′ | 3.0 | 60.49 | 4′ | 2.8 | |
| UGT88E12 | 53.93 | 7 | 3.1 | 52.91 | (c) | - | 57.29 | 4′ | 2.8 | 58.75 | 7 | 3.1 | 49.20 | - | - | 51.52 | 5 | 2.9 | |
| UGT88H1 | 66.00 | 7 | 3.1 | 67.94 | 7 | 3.0 | 63.06 | 4′ | 2.7 | 61.90 | 7 | 3.0 | 63.43 | 7 | 2.9 | 61.19 | 7 | 2.8 | |
| G2 | UGT72V3 | 58.36 | 5 | 2.9 | 60.58 | 5 | 2.9 | 56.97 | 4′ | 5.1 | 61.24 | (b) | - | 54.44 | 7 | 2.9 | 52.42 | 7 | H1.8 |
| UGT72AF1 | 55.89 | 4′ | 3.6 | 56.84 | 7 | 2.1 | 55.47 | 4′ | 3.6 | 56.67 | 7 | 2.9 | 60.62 | 4′ | 3.1 | 58.24 | 4′ | 3.0 | |
| GuGT10 | 53.11 | (c) | - | 54.82 | (c) | - | 56.14 | - | - | 57.82 | (b) | - | 54.90 | - | - | 57.52 | 5 | 3.0 | |
| UGT73J1 | 52.73 | (c) | - | 54.37 | (c) | - | 54.30 | 4′ | 2.8 | 55.41 | (b) | - | 56.96 | 4′ | 2.6 | 55.10 | 5 | 2.8 | |
| UGT73G1 | 51.35 | 5 | 3.1 | 54.36 | 5 | 3.0 | 48.59 | 7 | 5.1 | 50.17 | 7 | 5.6 | 51.22 | 7 | 2.8 | 51.89 | 5 | 4.7 | |
| GmUGT1 | 58.89 | 4′ | 2.9 | 63.98 | 7 | 3.0 | 61.56 | 4′ | 3.0 | 64.84 | 7 | 2.7 | 60.58 | 4′ | 4.8 | 59.59 | 5 | 3.0 | |
| PlUGT4 | 54.34 | 7 | 2.6 | 50.35 | 7 | 2.9 | 54.91 | 7 | 2.6 | 52.74 | 7 | 2.5 | 55.22 | 4′ | 2.5 | 53.66 | 4′ | 2.9 | |
| PtUGT8 | 50.10 | 7 | 2.6 | 50.19 | 7 | 2.6 | 50.75 | 7 | 2.9 | 52.79 | 7 | 2.7 | 53.02 | 7 | 2.9 | 55.24 | 5 | 2.5 | |
| G3 | UGT73F1 | 47.13 | 7 | 2.7 | 50.30 | 7 | 2.9 | 47.25 | 7 | 2.8 | 50.39 | 7 | 2.6 | 46.00 | 4′ | 2.9 | 44.48 | 5 | 2.6 |
| UGT73C20 | 36.10 | 4′ | 4.4 | 38.81 | 5 | 4.3 | 32.31 | 7 | 4.4 | 40.60 | (b) | - | 38.31 | 4′ | 3.6 | 38.20 | 4′ | 4.3 | |
| UGT73F2 | 47.78 | 4′ | 3.0 | 48.46 | (c) | - | 47.05 | 4′ | 3.0 | 47.70 | 7 | 3.0 | 48.50 | 7 | 2.9 | 45.26 | 4′ | 3.2 | |
| UGT709A4 | 40.99 | 7 | 4.2 | 33.10 | 7 | 4.3 | 42.41 | 7 | 4.2 | 34.63 | 7 | 4.2 | 47.47 | - | - | 46.77 | - | - | |
| UGT85H2 | 51.52 | 7 | 5.9 | 42.08 | (c) | - | 52.41 | 7 | 6.0 | 49.39 | 7 | 7.2 | 54.69 | 7 | 5.7 | 49.46 | 7 | 6.0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sajid, M.; Kaur, P. Protein Modelling Highlighted Key Catalytic Sites Involved in Position-Specific Glycosylation of Isoflavonoids. Int. J. Mol. Sci. 2023, 24, 12356. https://doi.org/10.3390/ijms241512356
Sajid M, Kaur P. Protein Modelling Highlighted Key Catalytic Sites Involved in Position-Specific Glycosylation of Isoflavonoids. International Journal of Molecular Sciences. 2023; 24(15):12356. https://doi.org/10.3390/ijms241512356
Chicago/Turabian StyleSajid, Moon, and Parwinder Kaur. 2023. "Protein Modelling Highlighted Key Catalytic Sites Involved in Position-Specific Glycosylation of Isoflavonoids" International Journal of Molecular Sciences 24, no. 15: 12356. https://doi.org/10.3390/ijms241512356
APA StyleSajid, M., & Kaur, P. (2023). Protein Modelling Highlighted Key Catalytic Sites Involved in Position-Specific Glycosylation of Isoflavonoids. International Journal of Molecular Sciences, 24(15), 12356. https://doi.org/10.3390/ijms241512356







