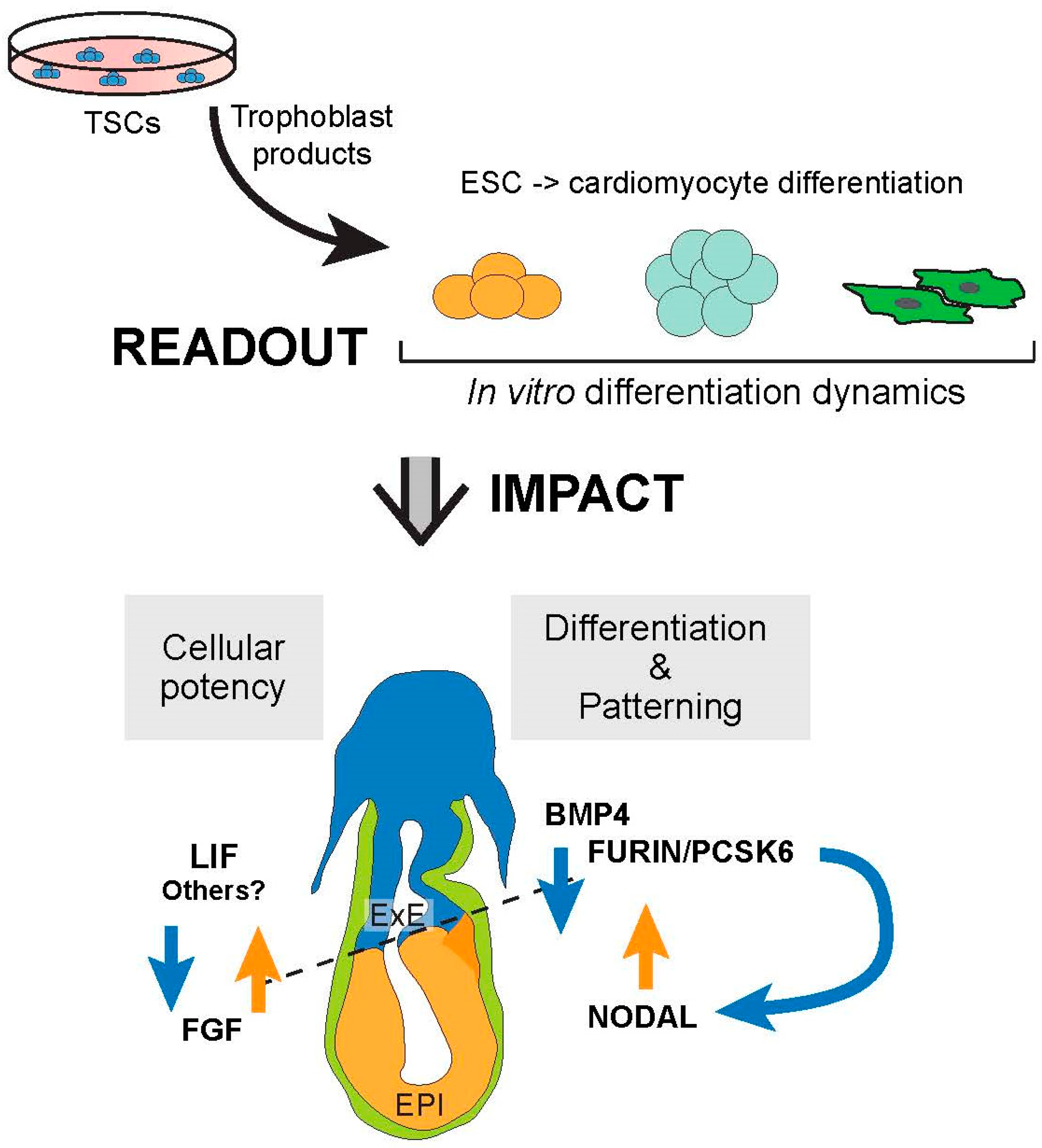
The Trophoblast Compartment Helps Maintain Embryonic Pluripotency and Delays Differentiation towards Cardiomyocytes
Abstract
1. Introduction
2. Results
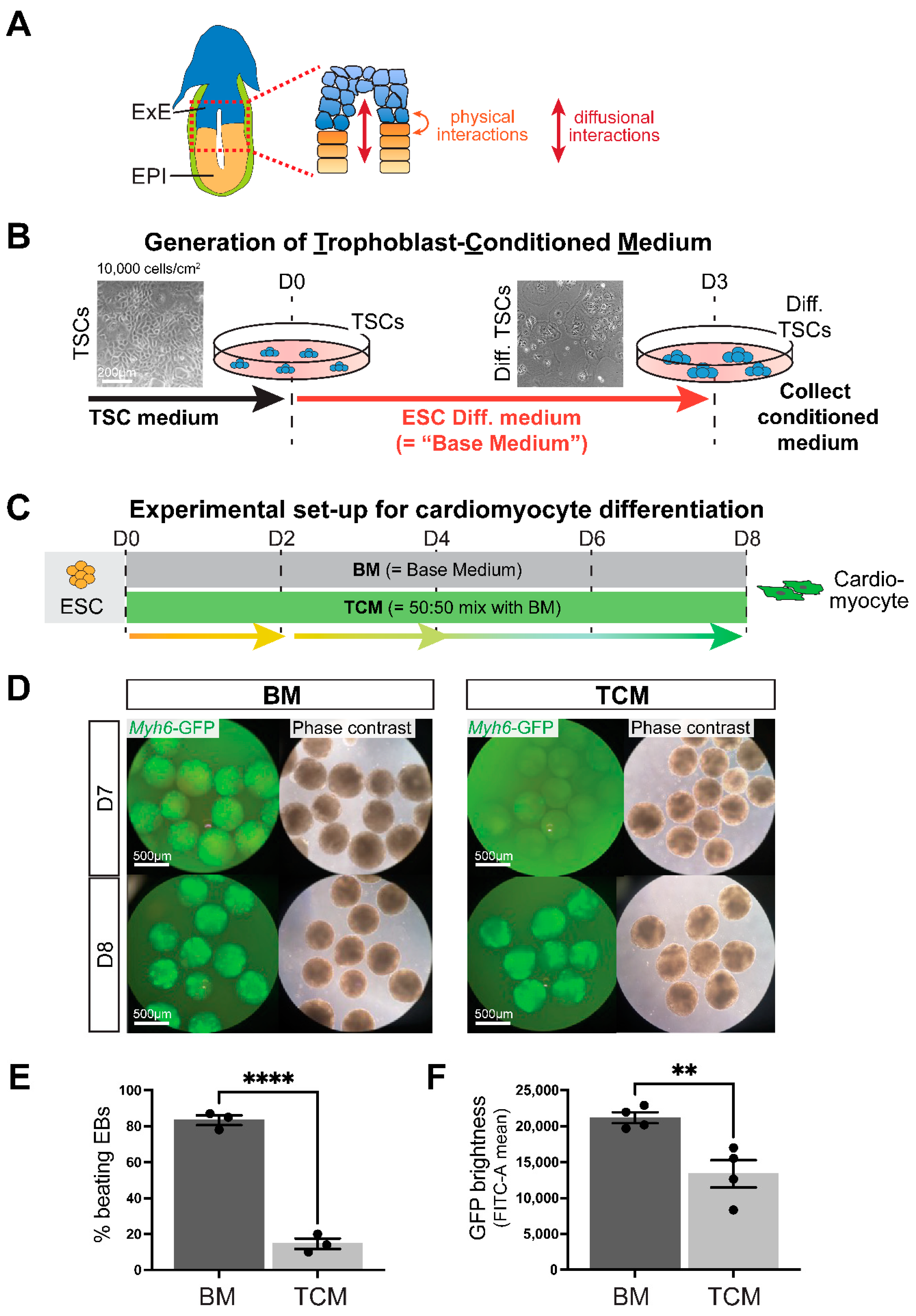
2.1. Establishment of a Robust Differentiation Protocol of Murine ESCs into Cardiomyocytes
2.2. Generation of Trophoblast-Conditioned Medium (TCM)
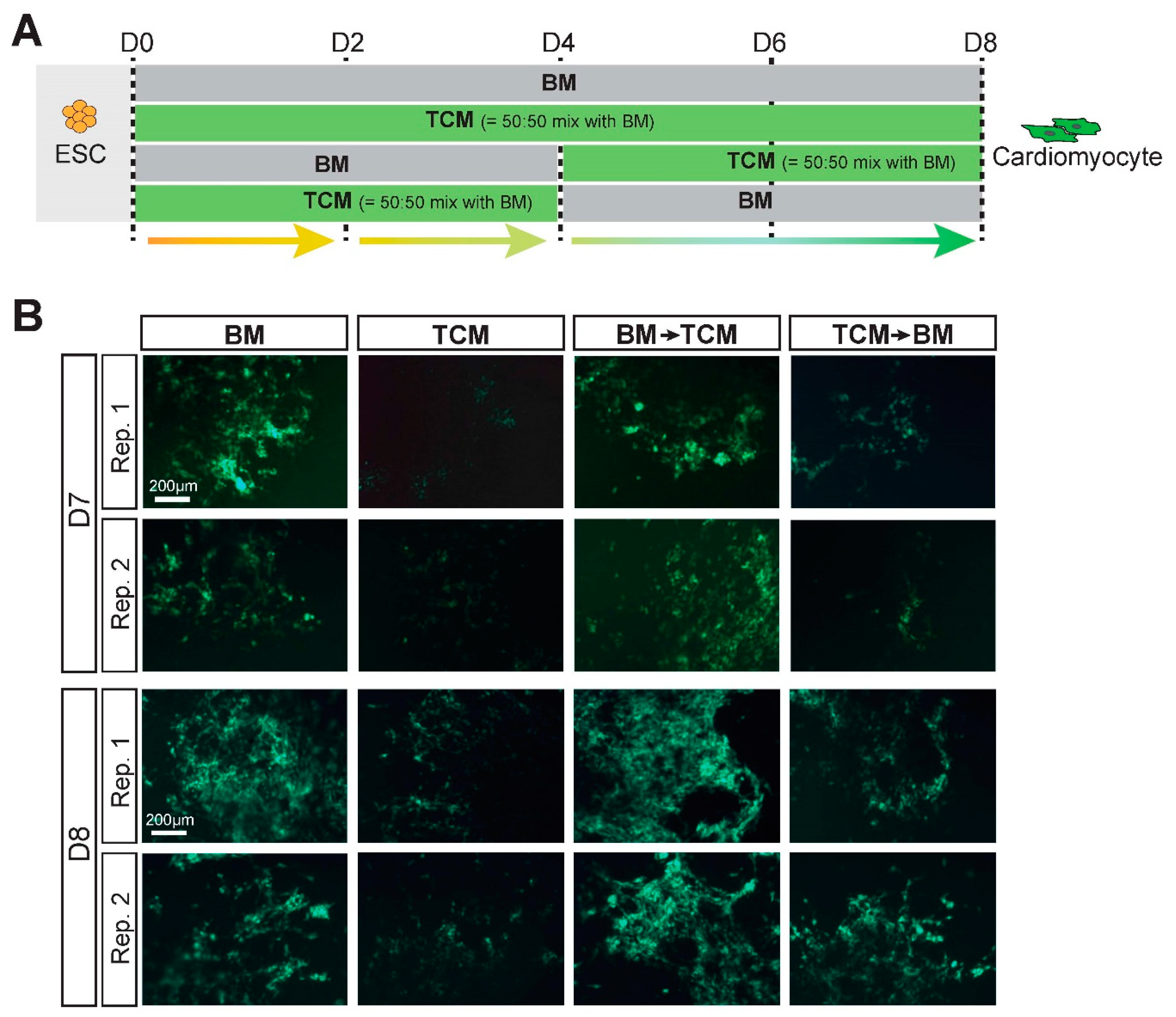
2.3. TCM Slows down Cardiomyocyte Differentiation Dynamics
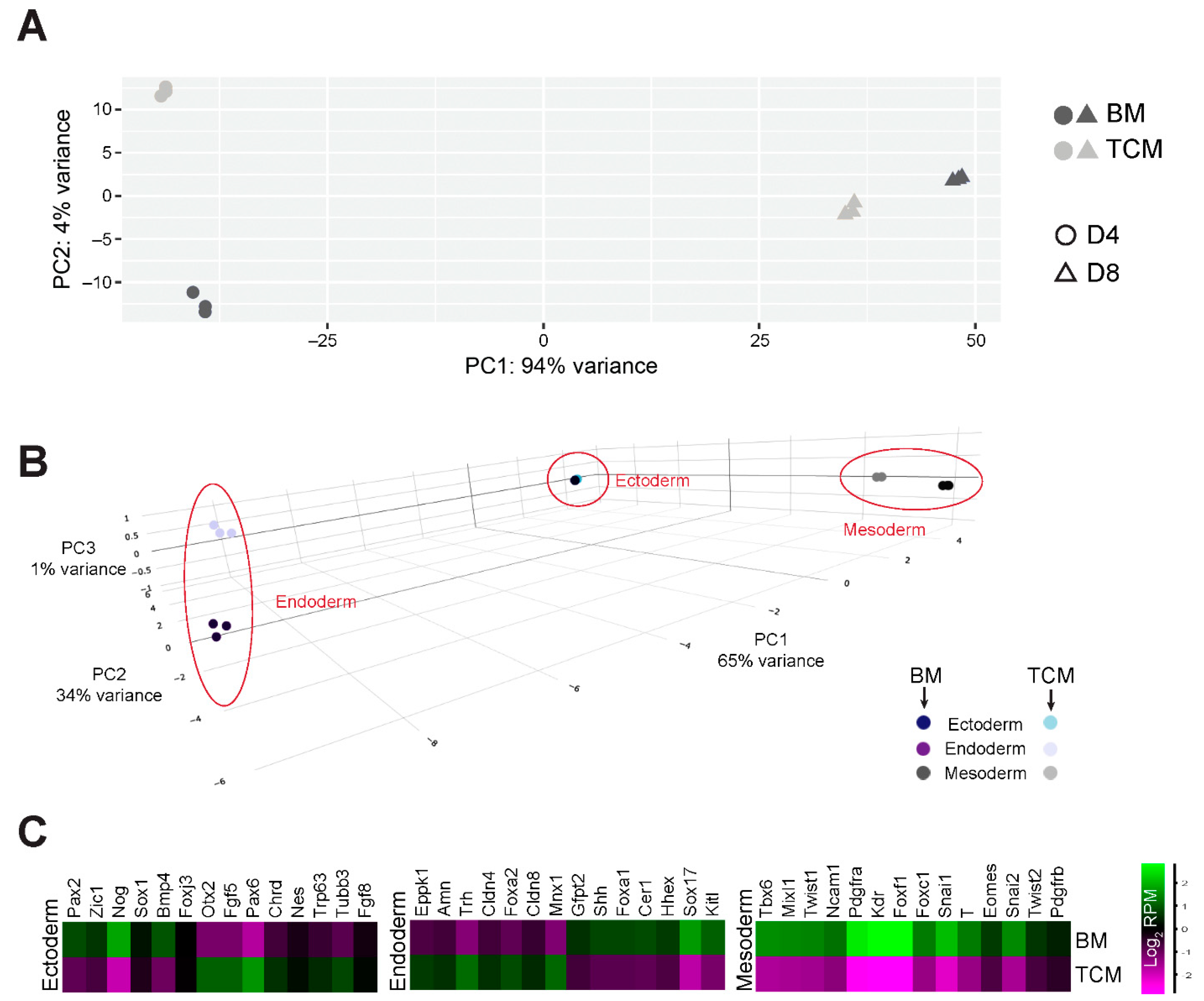
2.4. TCM Interferes Specifically with the Early Phases of Mesoderm and Cardiac Mesoderm Formation
2.5. Expression Profiling Underscores the Impact of TCM on Mesoderm Differentiation
2.6. TCM from Mutant Trophoblast Stem Cells Differentially Affects Cardiac Differentiation Dynamics
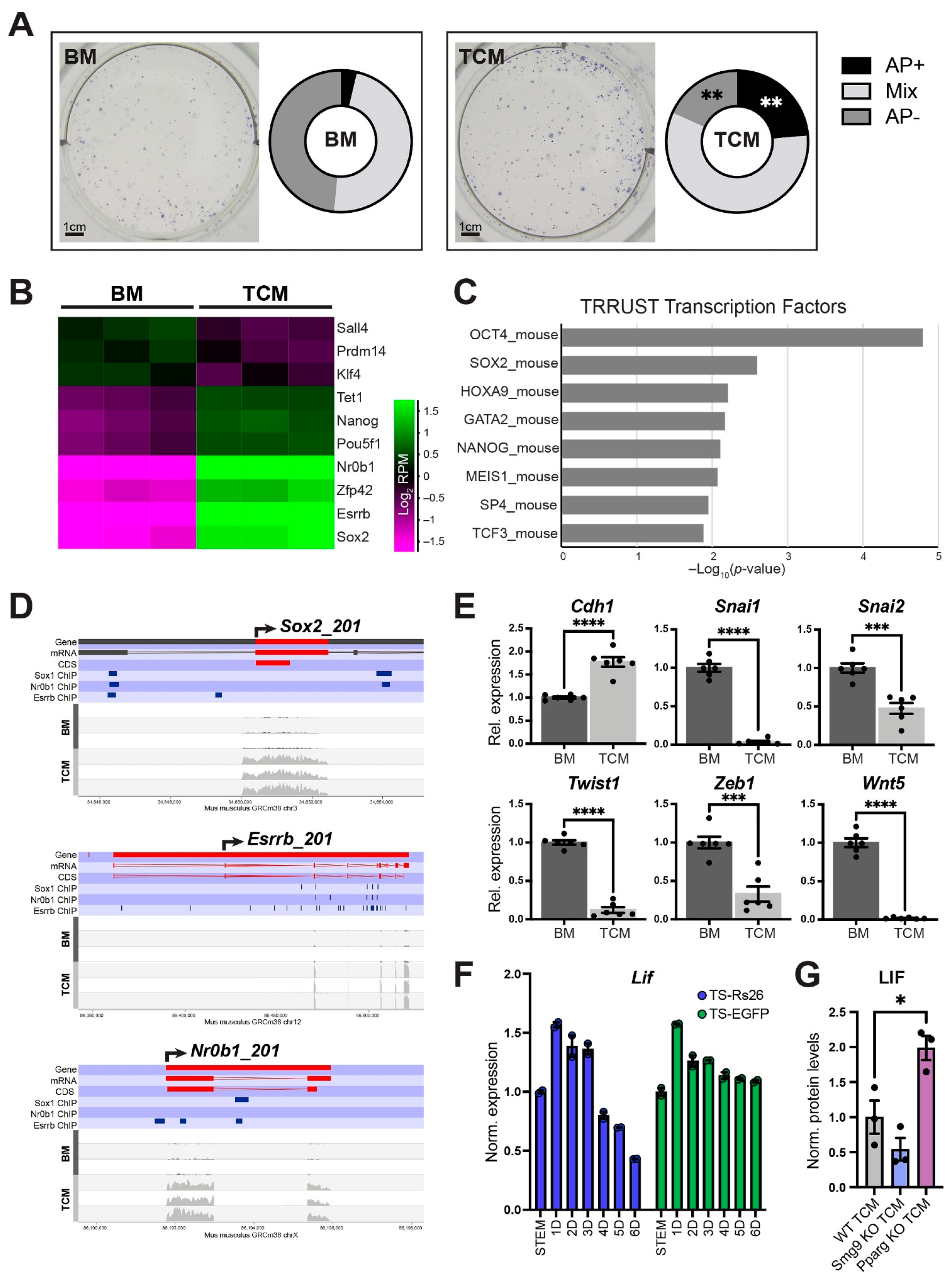
2.7. TCM Promotes ESC Pluripotency
3. Discussion
4. Materials and Methods
4.1. ESC and TSC Lines Used and Standard Culture Conditions
4.2. Conditioned Medium Generation
4.3. Embryoid Body Differentiation
4.4. Alkaline Phosphatase Staining
4.5. RNA Extraction and RT-qPCRs
4.6. RNA-Sequencing
4.7. Determination of LIF Protein Levels in TCM
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwon, G.S.; Viotti, M.; Hadjantonakis, A.K. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 2008, 15, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Rossant, J.; Tam, P.P. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 2009, 136, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Lawson, K.A.; Dunn, N.R.; Roelen, B.A.; Zeinstra, L.M.; Davis, A.M.; Wright, C.V.; Korving, J.P.; Hogan, B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo [see comments]. Genes Dev. 1999, 13, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Guzman-Ayala, M.; Ben-Haim, N.; Beck, S.; Constam, D.B. Nodal protein processing and fibroblast growth factor 4 synergize to maintain a trophoblast stem cell microenvironment. Proc. Natl. Acad. Sci. USA 2004, 101, 15656–15660. [Google Scholar] [CrossRef] [PubMed]
- Simmons, D.G.; Cross, J.C. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol. 2005, 284, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Ralston, A.; Rossant, J. How signaling promotes stem cell survival: Trophoblast stem cells and Shp2. Dev. Cell 2006, 10, 275–276. [Google Scholar] [CrossRef]
- Tanaka, S.; Kunath, T.; Hadjantonakis, A.K.; Nagy, A.; Rossant, J. Promotion of trophoblast stem cell proliferation by FGF4. Science 1998, 282, 2072–2075. [Google Scholar] [CrossRef]
- Brade, T.; Pane, L.S.; Moretti, A.; Chien, K.R.; Laugwitz, K.L. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a013847. [Google Scholar] [CrossRef]
- Barak, Y.; Nelson, M.C.; Ong, E.S.; Jones, Y.Z.; Ruiz-Lozano, P.; Chien, K.R.; Koder, A.; Evans, R.M. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell 1999, 4, 585–595. [Google Scholar] [CrossRef]
- Adams, R.H.; Porras, A.; Alonso, G.; Jones, M.; Vintersten, K.; Panelli, S.; Valladares, A.; Perez, L.; Klein, R.; Nebreda, A.R. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol. Cell 2000, 6, 109–116. [Google Scholar] [CrossRef]
- Hatano, N.; Mori, Y.; Oh-hora, M.; Kosugi, A.; Fujikawa, T.; Nakai, N.; Niwa, H.; Miyazaki, J.; Hamaoka, T.; Ogata, M. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells 2003, 8, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, E.O.; Lin, H.; Chiu, S.Y.; Yu, H.M.; Porter, G.A.; Hsu, W. Extraembryonic but not embryonic SUMO-specific protease 2 is required for heart development. Sci. Rep. 2016, 6, 20999. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.; Scheinast, M.; Pasteiner, W.; Lagger, S.; Hofner, M.; Hoellrigl, A.; Schultheis, M.; Weitzer, G. Self-organization phenomena in embryonic stem cell-derived embryoid bodies: Axis formation and breaking of symmetry during cardiomyogenesis. Cells Tissues Organs 2012, 195, 377–391. [Google Scholar] [CrossRef]
- Robbins, J.; Gulick, J.; Sanchez, A.; Howles, P.; Doetschman, T. Mouse embryonic stem cells express the cardiac myosin heavy chain genes during development in vitro. J. Biol. Chem. 1990, 265, 11905–11909. [Google Scholar] [CrossRef]
- Bader, A.; Gruss, A.; Hollrigl, A.; Al-Dubai, H.; Capetanaki, Y.; Weitzer, G. Paracrine promotion of cardiomyogenesis in embryoid bodies by LIF modulated endoderm. Differentiation 2001, 68, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Kibschull, M. Differentiating Mouse Embryonic Stem Cells into Embryoid Bodies in AggreWell Plates. Cold Spring Harb. Protoc. 2017, 2017, pdb.prot094169. [Google Scholar] [CrossRef] [PubMed]
- Bauwens, C.L.; Toms, D.; Ungrin, M. Aggregate Size Optimization in Microwells for Suspension-based Cardiac Differentiation of Human Pluripotent Stem Cells. J. Vis. Exp. 2016, 115, 54308. [Google Scholar] [CrossRef]
- Leahy, A.; Xiong, J.W.; Kuhnert, F.; Stuhlmann, H. Use of developmental marker genes to define temporal and spatial patterns of differentiation during embryoid body formation. J. Exp. Zool. 1999, 284, 67–81. [Google Scholar] [CrossRef]
- Liu, Y.; Kaneda, R.; Leja, T.W.; Subkhankulova, T.; Tolmachov, O.; Minchiotti, G.; Schwartz, R.J.; Barahona, M.; Schneider, M.D. Hhex and Cer1 mediate the Sox17 pathway for cardiac mesoderm formation in embryonic stem cells. Stem Cells 2014, 32, 1515–1526. [Google Scholar] [CrossRef]
- Radford, B.N.; Zhao, X.; Glazer, T.; Eaton, M.; Blackwell, D.; Mohammad, S.; Lo Vercio, L.D.; Devine, J.; Shalom-Barak, T.; Hallgrimsson, B.; et al. Defects in placental syncytiotrophoblast cells are a common cause of developmental heart disease. Nat. Commun. 2023, 14, 1174. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; Paraiso, K.D.; Zhou, J.J.; Blitz, I.L.; Fish, M.B.; Charney, R.M.; Cho, J.S.; Yasuoka, Y.; Sudou, N.; Bright, A.R.; et al. Uncovering the mesendoderm gene regulatory network through multi-omic data integration. Cell Rep. 2022, 38, 110364. [Google Scholar] [CrossRef] [PubMed]
- Hahnel, A.C.; Rappolee, D.A.; Millan, J.L.; Manes, T.; Ziomek, C.A.; Theodosiou, N.G.; Werb, Z.; Pedersen, R.A.; Schultz, G.A. Two alkaline phosphatase genes are expressed during early development in the mouse embryo. Development 1990, 110, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Matsui, Y.; Zsebo, K.; Hogan, B.L. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 1992, 70, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, A.P.; Choo, S.H.; Mistri, T.K.; Rahmani, M.; Woon, C.T.; Ng, C.K.; Jauch, R.; Robson, P. Co-motif discovery identifies an Esrrb-Sox2-DNA ternary complex as a mediator of transcriptional differences between mouse embryonic and epiblast stem cells. Stem Cells 2013, 31, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Uranishi, K.; Akagi, T.; Sun, C.; Koide, H.; Yokota, T. Dax1 associates with Esrrb and regulates its function in embryonic stem cells. Mol. Cell. Biol. 2013, 33, 2056–2066. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Nikaido, I.; Ohta, H.; Ohtsuka, S.; Ura, H.; Kadota, M.; Wakayama, T.; Ueda, H.R.; Niwa, H. Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol. Cell 2013, 52, 380–392. [Google Scholar] [CrossRef]
- Latos, P.A.; Goncalves, A.; Oxley, D.; Mohammed, H.; Turro, E.; Hemberger, M. Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat. Commun. 2015, 6, 7776. [Google Scholar] [CrossRef]
- Kim, Y.S.; Yi, B.R.; Kim, N.H.; Choi, K.C. Role of the epithelial-mesenchymal transition and its effects on embryonic stem cells. Exp. Mol. Med. 2014, 46, e108. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; You, Y.; Jiang, H.; Wang, Z.Z. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J. Cell. Physiol. 2017, 232, 3261–3272. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Warmflash, A. Self-organized signaling in stem cell models of embryos. Stem Cell Rep. 2021, 16, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; Doss, M.X.; Legros, S.; Artus, J.; Hadjantonakis, A.K.; Foley, A.C. eXtraembryonic ENdoderm (XEN) stem cells produce factors that activate heart formation. PLoS ONE 2010, 5, e13446. [Google Scholar] [CrossRef] [PubMed]
- Arman, E.; Haffner-Krausz, R.; Chen, Y.; Heath, J.K.; Lonai, P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl. Acad. Sci. USA 1998, 95, 5082–5087. [Google Scholar] [CrossRef] [PubMed]
- Feldman, B.; Poueymirou, W.; Papaioannou, V.E.; DeChaira, T.M.; Goldfarb, M. Requirement of FGF-4 for postimplantation mouse development. Science 1995, 267, 246–249. [Google Scholar] [CrossRef]
- Cheng, A.M.; Saxton, T.M.; Sakai, R.; Kulkarni, S.; Mbamalu, G.; Vogel, W.; Totorice, C.G.; Cardiff, R.D.; Cross, J.C.; Muller, W.J.; et al. Mammalian Grb2 regulates multiple steps in embryonic development, lineage commitment and malignant transformation. Cell 1998, 95, 793–803. [Google Scholar] [CrossRef]
- Gotoh, N.; Manova, K.; Tanaka, S.; Murohashi, M.; Hadari, Y.; Lee, A.; Hamada, Y.; Hiroe, T.; Ito, M.; Kurihara, T.; et al. The docking protein FRS2alpha is an essential component of multiple fibroblast growth factor responses during early mouse development. Mol. Cell. Biol. 2005, 25, 4105–4116. [Google Scholar] [CrossRef]
- Saba-El-Leil, M.K.; Vella, F.D.; Vernay, B.; Voisin, L.; Chen, L.; Labrecque, N.; Ang, S.L.; Meloche, S. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 2003, 4, 964–968. [Google Scholar] [CrossRef]
- Saxton, T.M.; Cheng, A.M.; Ong, S.H.; Lu, Y.; Sakai, R.; Cross, J.C.; Pawson, T. Gene dosage-dependent functions for phosphotyrosine-Grb2 signaling during mammalian tissue morphogenesis. Curr. Biol. 2001, 11, 662–670. [Google Scholar] [CrossRef]
- Beck, S.; Le Good, J.A.; Guzman, M.; Ben Haim, N.; Roy, K.; Beermann, F.; Constam, D.B. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat. Cell Biol. 2002, 4, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, N.; Lu, C.; Guzman-Ayala, M.; Pescatore, L.; Mesnard, D.; Bischofberger, M.; Naef, F.; Robertson, E.J.; Constam, D.B. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell 2006, 11, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Mesnard, D.; Donnison, M.; Fuerer, C.; Pfeffer, P.L.; Constam, D.B. The microenvironment patterns the pluripotent mouse epiblast through paracrine Furin and Pace4 proteolytic activities. Genes Dev. 2011, 25, 1871–1880. [Google Scholar] [CrossRef]
- Sozen, B.; Cornwall-Scoones, J.; Zernicka-Goetz, M. The dynamics of morphogenesis in stem cell-based embryology: Novel insights for symmetry breaking. Dev. Biol. 2021, 474, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Francou, A.; Saint-Michel, E.; Mesbah, K.; Theveniau-Ruissy, M.; Rana, M.S.; Christoffels, V.M.; Kelly, R.G. Second heart field cardiac progenitor cells in the early mouse embryo. Biochim. Biophys. Acta 2013, 1833, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, M.; Kannan, S.; Anderson, M.J.; Liu, X.; Suh, D.; Htet, M.; Li, B.; Kakani, T.; Murphy, S.; Tampakakis, E.; et al. Cardiac progenitors instruct second heart field fate through Wnts. Proc. Natl. Acad. Sci. USA 2023, 120, e2217687120. [Google Scholar] [CrossRef]
- Devine, W.P.; Wythe, J.D.; George, M.; Koshiba-Takeuchi, K.; Bruneau, B.G. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife 2014, 3, e03848. [Google Scholar] [CrossRef]
- Hemberger, M.; Dean, W. The placenta: Epigenetic insights into trophoblast developmental models of a generation-bridging organ with long-lasting impact on lifelong health. Physiol. Rev. 2023, 103, 2523–2560. [Google Scholar] [CrossRef]
- Amadei, G.; Handford, C.E.; Qiu, C.; De Jonghe, J.; Greenfeld, H.; Tran, M.; Martin, B.K.; Chen, D.Y.; Aguilera-Castrejon, A.; Hanna, J.H.; et al. Embryo model completes gastrulation to neurulation and organogenesis. Nature 2022, 610, 143–153. [Google Scholar] [CrossRef]
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef]
- Nichols, J.; Silva, J.; Roode, M.; Smith, A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 2009, 136, 3215–3222. [Google Scholar] [CrossRef] [PubMed]
- Ficz, G.; Hore, T.A.; Santos, F.; Lee, H.J.; Dean, W.; Arand, J.; Krueger, F.; Oxley, D.; Paul, Y.L.; Walter, J.; et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 2013, 13, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Juhasz, O.; Li, J.; Tarasova, Y.S.; Boheler, K.R. Embryonic stem cells and cardiomyocyte differentiation: Phenotypic and molecular analyses. J. Cell. Mol. Med. 2005, 9, 804–817. [Google Scholar] [CrossRef] [PubMed]
- Jeyarajah, M.; Patterson, V.; Bhatttad, G.J.; Zhao, L.; Whitehead, S.; Renaud, S.J. Placental extracellular vesicles promote cardiomyocyte maturation and fetal heart development. Res. Square 2023. [Google Scholar] [CrossRef]
- Langford, M.B.; Outhwaite, J.E.; Hughes, M.; Natale, D.R.C.; Simmons, D.G. Deletion of the Syncytin A receptor Ly6e impairs syncytiotrophoblast fusion and placental morphogenesis causing embryonic lethality in mice. Sci. Rep. 2018, 8, 3961. [Google Scholar] [CrossRef] [PubMed]
- Torregrosa-Carrion, R.; Pineiro-Sabaris, R.; Siguero-Alvarez, M.; Grego-Bessa, J.; Luna-Zurita, L.; Fernandes, V.S.; MacGrogan, D.; Stainier, D.Y.R.; de la Pompa, J.L. Adhesion G protein-coupled receptor Gpr126/Adgrg6 is essential for placental development. Sci. Adv. 2021, 7, eabj5445. [Google Scholar] [CrossRef]
- Raffel, G.D.; Chu, G.C.; Jesneck, J.L.; Cullen, D.E.; Bronson, R.T.; Bernard, O.A.; Gilliland, D.G. Ott1 (Rbm15) is essential for placental vascular branching morphogenesis and embryonic development of the heart and spleen. Mol. Cell. Biol. 2009, 29, 333–341. [Google Scholar] [CrossRef]
- Shalom-Barak, T.; Zhang, X.; Chu, T.; Timothy Schaiff, W.; Reddy, J.K.; Xu, J.; Sadovsky, Y.; Barak, Y. Placental PPARgamma regulates spatiotemporally diverse genes and a unique metabolic network. Dev. Biol. 2012, 372, 143–155. [Google Scholar] [CrossRef]
- Nagy, A.; Rossant, J.; Nagy, R.; Abramow-Newerly, W.; Roder, J.C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 1993, 90, 8424–8428. [Google Scholar] [CrossRef]
- Riley, P.; Anson-Cartwright, L.; Cross, J.C. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 1998, 18, 271–275. [Google Scholar] [CrossRef]
- Riley, P.R.; Gertsenstein, M.; Dawson, K.; Cross, J.C. Early exclusion of hand1-deficient cells from distinct regions of the left ventricular myocardium in chimeric mouse embryos. Dev. Biol. 2000, 227, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Risebro, C.A.; Smart, N.; Dupays, L.; Breckenridge, R.; Mohun, T.J.; Riley, P.R. Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development 2006, 133, 4595–4606. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.; Sienerth, A.R.; Hemberger, M. Plet1 is an epigenetically regulated cell surface protein that provides essential cues to direct trophoblast stem cell differentiation. Sci. Rep. 2016, 6, 25112. [Google Scholar] [CrossRef] [PubMed]
- Toms, D.; Deardon, R.; Ungrin, M. Climbing the mountain: Experimental design for the efficient optimization of stem cell bioprocessing. J. Biol. Eng. 2017, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Okishio, K.; Ui-Tei, K.; Saigo, K.; Kinoshita-Toyoda, A.; Toyoda, H.; Nishimura, T.; Suda, Y.; Hayasaka, M.; Hanaoka, K.; et al. Heparan sulfate regulates self-renewal and pluripotency of embryonic stem cells. J. Biol. Chem. 2008, 283, 3594–3606. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Radford, B.N.; Ungrin, M.; Dean, W.; Hemberger, M. The Trophoblast Compartment Helps Maintain Embryonic Pluripotency and Delays Differentiation towards Cardiomyocytes. Int. J. Mol. Sci. 2023, 24, 12423. https://doi.org/10.3390/ijms241512423
Zhao X, Radford BN, Ungrin M, Dean W, Hemberger M. The Trophoblast Compartment Helps Maintain Embryonic Pluripotency and Delays Differentiation towards Cardiomyocytes. International Journal of Molecular Sciences. 2023; 24(15):12423. https://doi.org/10.3390/ijms241512423
Chicago/Turabian StyleZhao, Xiang, Bethany N. Radford, Mark Ungrin, Wendy Dean, and Myriam Hemberger. 2023. "The Trophoblast Compartment Helps Maintain Embryonic Pluripotency and Delays Differentiation towards Cardiomyocytes" International Journal of Molecular Sciences 24, no. 15: 12423. https://doi.org/10.3390/ijms241512423
APA StyleZhao, X., Radford, B. N., Ungrin, M., Dean, W., & Hemberger, M. (2023). The Trophoblast Compartment Helps Maintain Embryonic Pluripotency and Delays Differentiation towards Cardiomyocytes. International Journal of Molecular Sciences, 24(15), 12423. https://doi.org/10.3390/ijms241512423




