Proteome and Interactome Linked to Metabolism, Genetic Information Processing, and Abiotic Stress in Gametophytes of Two Woodferns
Abstract
:1. Introduction
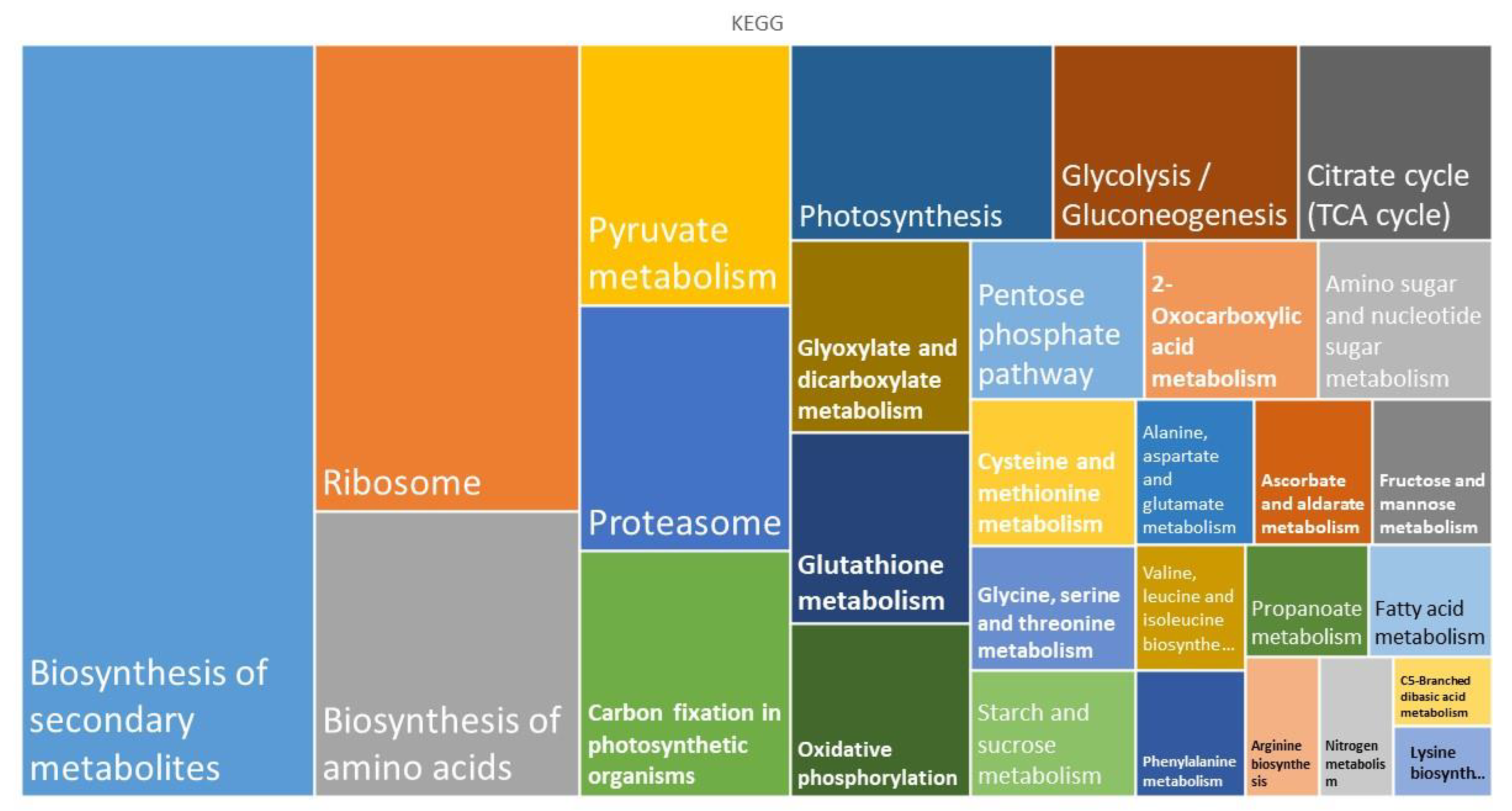
2. Results
3. Discussion
3.1. Metabolism
- Carbohydrates
- Tricarboxylic acid cycle and pentose phosphate pathway
- Metabolism of lipids
- Biosynthesis of amino acids and nucleotides
- Metabolism of energy
- Sulfur and nitrogen metabolism
- Metabolism of secondary compounds
3.2. Genetic Information Processing
- Transcription and translation
- Protein folding and sorting
- Protein degradation
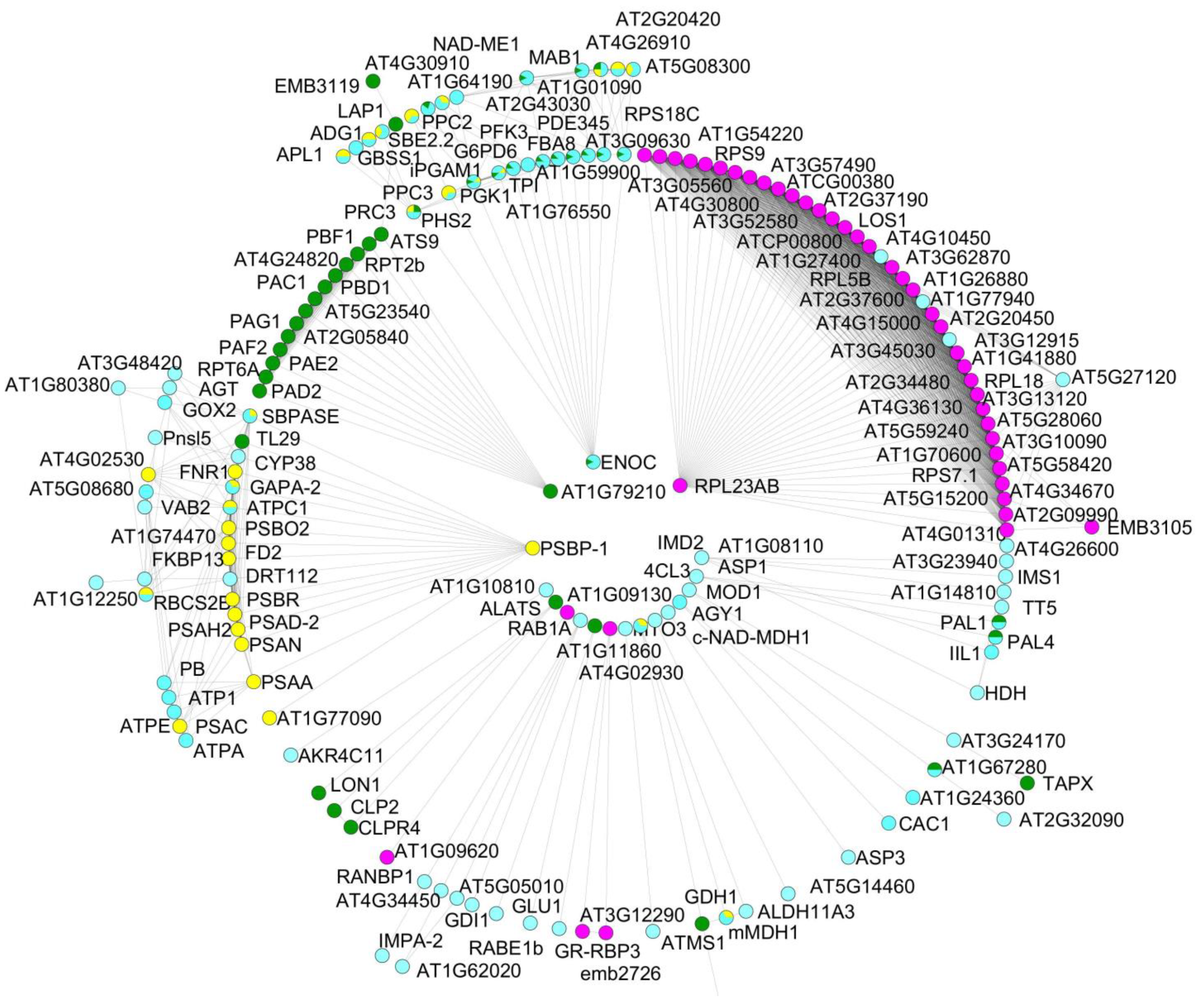
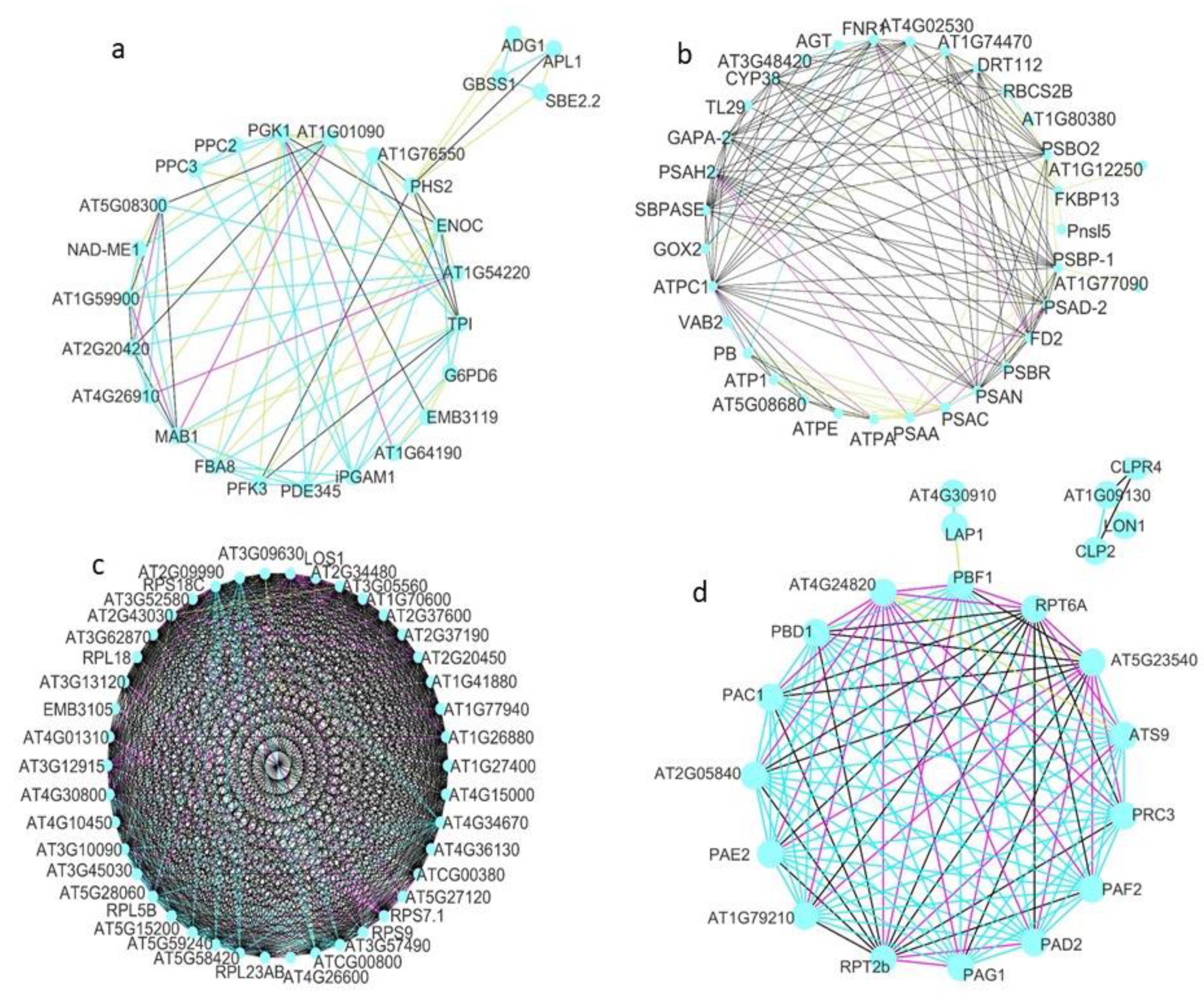
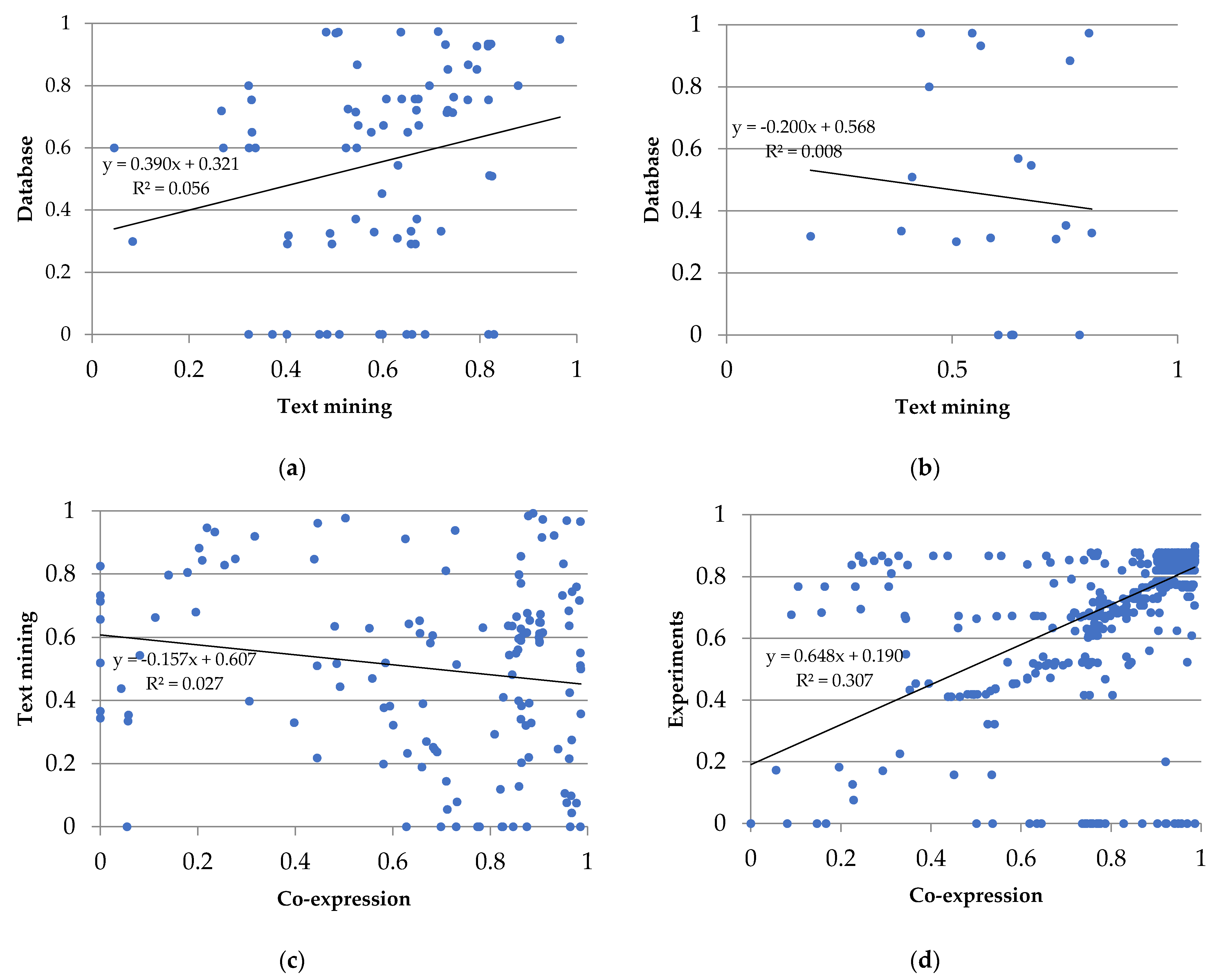
3.3. Protein–Protein Interactions
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Protein Extraction, Separation, and In-Gel Digestion
4.3. Protein Separation and In-Gel Digestion
4.4. Protein Identification, Verification, and Bioinformatic Downstream Analyses
4.5. Protein Analysis Using the STRING Platform
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wada, M. The fern as a model system to study photomorphogenesis. J. Plant Res. 2007, 120, 3–16. [Google Scholar] [CrossRef]
- Salmi, M.L.; Bushart, T.; Stout, S.; Roux, S. Profile and analysis of gene expression changes during early development in germinating spores of Ceratopteris richardii. Plant Physiol. 2005, 138, 1734–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmi, M.L.; Morris, K.E.; Roux, S.J.; Porterfield, D.M. Nitric oxide and CGMP signaling in calcium-dependent development of cell polarity in Ceratopteris richardii. Plant Physiol. 2007, 144, 94–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suo, J.; Zhao, Q.; Zhang, Z.; Chen, S.; Cao, J.; Liu, G.; Wei, X.; Wang, T.; Yang, C.; Dai, S. Cytological and proteomic analyses of Osmunda cinnamomea germinating spores reveal characteristics of fern spore germination and rhizoid tip growth. Mol. Cell. Proteom. 2015, 14, 2510–2534. [Google Scholar] [CrossRef] [Green Version]
- Salmi, M.L.; Bushart, T.J. Cellular, molecular, and genetic changes during the development of Ceratopteris richardii gametophytes. In Working with Ferns: Issues and Applications; Fernández, H., Kumar, A., Revilla, M.A., Eds.; Springer International Publishing: New York, NY, USA, 2010; pp. 11–24. [Google Scholar]
- Eeckhout, S.; Leroux, O.; Willats, W.G.; Popper, Z.A.; Viane, R.L. Comparative glycan profiling of Ceratopteris richardii ‘C-fern’ gametophytes and sporophytes links cell-wall composition to functional specialization. Ann. Bot. 2014, 114, 1295–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, H.; Revilla, M.A. In vitro culture of ornamental ferns. Plant Cell. Tissue Organ Cult. 2003, 73, 1–13. [Google Scholar] [CrossRef]
- Rivera, A.; Cañal, M.J.; Grossniklaus, U.; Fernández, H. The gametophyte of fern: Born to reproduce. In Current Advances in Fern Research; Fernández, H., Ed.; Springer International Publishing: New York, NY, USA, 2018; pp. 3–19. [Google Scholar]
- Chen, C.-Y.; Chiu, F.-Y.; Lin, Y.; Huang, W.-J.; Hsieh, P.-S.; Hsu, F.-L. Chemical constituents analysis and antidiabetic activity validation of four fern species from Taiwan. Int. J. Mol. Sci 2015, 16, 2497–2516. [Google Scholar] [CrossRef] [Green Version]
- Femi-Adepoju, A.G.; Dada, A.O.; Otun, K.O.; Adepoju, A.O.; Fatoba, O.P. Green synthesis of silver nanoparticles using terrestrial fern (Gleichenia pectinata (Willd.) C. Presl.): Characterization and antimicrobial studies. Heliyon 2019, 5, e01543. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, S.; Zhang, H.; Shi, L.; Cao, F.; Guo, L.; Xie, Y.; Wang, T.; Yan, X.; Dai, S. Desiccation tolerance mechanism in resurrection fern-ally Selaginella tamariscina revealed by physiological and proteomic analysis. J. Proteome Res. 2010, 9, 6561–6577. [Google Scholar] [CrossRef]
- Rathinasabapathi, B. Ferns represent an untapped biodiversity for improving crops for environmental stress tolerance. New Phytol. 2006, 172, 385–390. [Google Scholar] [CrossRef]
- Dhir, B. Role of ferns in environmental cleanup. In Current Advances in Fern Research; Fernández, H., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 517–531. [Google Scholar]
- Barker, M.S.; Wolf, P.G. Unfurling fern biology in the genomics age. Bioscience 2010, 60, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Der, J.P.; Barker, M.S.; Wickett, N.J.; de Pamphilis, C.W.; Wolf, P.G. De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genom. 2011, 12, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bona, E.; Marsano, F.; Massa, N.; Cattaneo, C. Proteomic analysis as a tool for investigating arsenic stress in Pteris vittata roots colonized or not by arbuscular mycorrhizal symbiosis. J. Proteom. 2011, 74, 1338–1350. [Google Scholar] [CrossRef]
- Valledor, L.; Menéndez, V.; Canal, M.J.; Revilla, A.; Fernández, H. Proteomic approaches to sexual development mediated by antheridiogen in the fern Blechnum spicant L. Proteomics 2014, 14, 2061–2071. [Google Scholar] [CrossRef] [PubMed]
- Aya, K.; Kobayashi, M.; Tanaka, J.; Ohyanagi, H.; Suzuki, T.; Yano, K.; Takano, T.; Yano, K.; Matsuoka, M. De novo transcriptome assembly of a fern, Lygodium japonicum, and a web resource database Ljtrans DB. Plant Cell Physiol. 2015, 56, e5. [Google Scholar] [CrossRef] [Green Version]
- Domżalska, L.; Kędracka-Krok, S.; Jankowska, U.; Grzyb, M.; Sobczak, M.; Rybczyński, J.J.; Mikuła, A. Proteomic analysis of stipe explants reveals differentially expressed proteins involved in early direct somatic embryogenesis of the tree fern Cyathea delgadii Sternb. Plant Sci. 2017, 258, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Sigel, E.M.; Schuettpelz, E.; Pryer, K.M.; Der, J.P. Overlapping patterns of gene expression between gametophyte and sporophyte phases in the fern Polypodium amorphum (Polypodiales). Front. Plant Sci. 2018, 9, 1450. [Google Scholar] [CrossRef]
- Sareen, B.; Thapa, P.; Joshi, R.; Bhattacharya, A. Proteome analysis of the gametophytes of a western Himalayan fern Diplazium maximum reveals their adaptive responses to changes in their micro-environment. Front. Plant Sci. 2019, 10, 1623. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Z.; Huang, W.; Fu, H.; Wang, Q.; Wang, Y.; Cao, J. Proteomic analysis of gametophytic sex expression in the fern Ceratopteris thalictroides. PLoS ONE 2019, 14, e0221470. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Chen, L. Comparative transcriptome analysis of two reproductive modes in Adiantum reniforme var. sinense targeted to explore possible mechanism of apogamy. BMC Genet. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Cordle, A.; Irish, E.; Cheng, C.L. Gene expression associated with apogamy commitment in Ceratopteris richardii. Sex. Plant Reprod. 2012, 25, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Atallah, N.M.; Vitek, O.; Gaiti, F.; Tanurdzic, M.; Banks, J.A. Sex determination in Ceratopteris richardii is accompanied by transcriptome changes that drive epigenetic reprogramming of the young gametophyte. G3 2018, 8, 2205–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youngstrom, C.E.; Geadelmann, L.F.; Irish, E.E.; Cheng, C.-L. A fern WUSCHEL-RELATED HOMEOBOX gene functions in both gametophyte and sporophyte generations. BMC Plant Biol. 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aragón-Raygoza, A.; Herrera-Estrella, L.; Cruz-Ramirez, A. Transcriptional analysis of Ceratopteris richardii young sporophyte reveals conservation of stem cell factors in the root apical meristem. Front. Plant Sci. 2022, 13, 924660. [Google Scholar] [CrossRef]
- Hong, Y.; Wang, Z.; Li, M.; Su, Y.; Wang, T. First multi-organ full-length transcriptome of tree fern Alsophila spinulosa highlights the stress-resistant and light-adapted genes. Front. Plant Sci. 2022, 12, 784546. [Google Scholar] [CrossRef]
- Huang, X.; Wang, W.; Gong, T.; Wickell, D.; Kuo, L.-Y.; Zhang, X.; Wen, J.; Kim, H.; Lu, F.; Zhao, H.; et al. The flying spider-monkey tree fern genome provides insights into fern evolution and arborescence. Nat. Plants 2022, 8, 500–512. [Google Scholar] [CrossRef]
- Xia, Z.; Liu, L.; Wei, Z.; Wang, F.; Shen, H.; Yan, Y. Analysis of comparative transcriptome and positively selected genes reveal adaptive evolution in leaf-less and root-less whisk ferns. Plants 2022, 11, 1198. [Google Scholar] [CrossRef]
- Grossmann, J.; Fernández, H.; Chaubey, P.M.; Valdés, A.E.; Gagliardini, V.; Cañal, M.J.; Russo, G.; Grossniklaus, U. Proteogenomic analysis greatly expands the identification of proteins related to reproduction in the apogamous fern Dryopteris affinis ssp. affinis. Front. Plant Sci. 2017, 8, 336. [Google Scholar] [CrossRef] [Green Version]
- Wyder, S.; Rivera, A.; Valdés, A.E.; Cañal, M.J.; Gagliardini, V.; Fernández, H.; Grossniklaus, U. Differential gene expression profiling of one- and two-dimensional apogamous gametophytes of the fern Dryopteris affinis ssp. affinis. Plant Physiol. Biochem. 2020, 148, 302–311. [Google Scholar] [CrossRef]
- Fernández, H.; Grossmann, J.; Gagliardini, V.; Feito, I.; Rivera, A.; Rodríguez, L.; Quintanilla, L.G.; Quesada, V.; Cañal, M.J.; Grossniklaus, U. Sexual and apogamous species of woodferns show different protein and phytohormone profiles. Front. Plant Sci. 2021, 12, 718932. [Google Scholar] [CrossRef]
- Ojosnegros, S.; Alvarez, J.M.; Grossmann, J.; Gagliardini, V.; Quintanilla, L.G.; Grossniklaus, U.; Fernández, H. The shared proteome of the apomictic fern Dryopteris affinis ssp. affinis and its sexual relative Dryopteris oreades. Int. J. Mol. Sci. 2022, 23, 14027. [Google Scholar] [CrossRef]
- Tronconi, M.A.; Fahnenstich, H.; Gerrard Weehler, M.C.; Andreo, C.S.; Flügge, U.I.; Drincovich, M.F.; Maurino, V.G. Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol. 2008, 146, 1540. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Assmann, S.M. The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 5179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomaz, T.; Bagard, M.; Pracharoenwattana, I.; Lindén, P.; Lee, C.P.; Carroll, A.J.; Ströher, E.; Smith, S.M.; Gardeström, P.; Millar, A.H. Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 2010, 154, 1143–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howles, P.A.; Birch, R.J.; Collings, D.A.; Gebbie, L.K.; Hurley, U.A.; Hocart, C.H.; Arioli, T.; Williamson, R.E. A mutation in an Arabidopsis ribose 5-phosphate isomerase reduces cellulose synthesis and is rescued by exogenous uridine. Plant J. 2006, 48, 606–618. [Google Scholar] [CrossRef]
- Wakao, S.; Benning, C. Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J. 2005, 41, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; He, Y.; Dai, Y.; Liu, X.; Li, J. Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 2000, 12, 405–417. [Google Scholar] [CrossRef] [Green Version]
- Fatland, B.L.; Ke, J.; Anderson, M.D.; Mentzen, W.I.; Wei Cui, L.; Christy Allred, C.; Johnston, J.L.; Nikolau, B.J.; Syrkin Wurtele, E.; Biology LWC, M. Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol. 2002, 130, 740–756. [Google Scholar] [CrossRef] [Green Version]
- Fatland, B.L.; Nikolau, B.J.; Wurtele, E.S. Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. Plant Cell 2005, 17, 182–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pracharoenwattana, I.; Cornah, J.E.; Smith, S.M. Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 2005, 17, 2037–2048. [Google Scholar] [CrossRef] [Green Version]
- Knill, T.; Reichelt, M.; Paetz, C.; Gershenzon, J.; Binder, S. Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. Plant Mol. Biol. 2009, 71, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Ishizaki, T.; Ohsumi, C.; Totsuka, K.; Igarashi, D. Analysis of glutamate homeostasis by overexpression of Fd-GOGAT gene in Arabidopsis thaliana. Amin. Acids 2009, 38, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, K.; Sandoval, F.J.; Santiago, K.; Roje, S. One-carbon metabolism in plants: Characterization of a plastid serine hydroxymethyltransferase. Biochem. J. 2010, 430, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, L.N.; Marineo, S.; Mandalà, S.; Davids, F.; Sewell, B.T.; Ingle, R.A. The missing link in plant histidine biosynthesis: Arabidopsis MYOINOSITOL MONOPHOSPHATASE-LIKE2 encodes a functional histidinol-phosphate phosphatase. Plant Physiol. 2010, 152, 1186–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B.; Li, C.; Tarczynski, M.C. High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-ADENOSYL-L-METHIONINE SYNTHETASE 3 gene. Plant J. 2002, 29, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Lundin, B.; Hansson, M.; Schoefs, B.; Vener, A.V.; Spetea, C. The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. Plant J. 2007, 49, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Last, R.L. A chloroplast thylakoid lumen protein is required for proper photosynthetic acclimation of plants under fluctuating light environments. Proc. Natl. Acad. Sci. USA 2017, 114, E8110–E8117. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, B.; Xu, Y.; Li, H.; Li, S.; Zhang, D.; Mao, Z.; Guo, S.; Yang, C.; Weng, Y.; et al. The cyclophilin CYP20-2 modulates the conformation of BRASSINAZOLE-RESISTANT1, which binds the promoter of FLOWERING LOCUS D to regulate flowering in Arabidopsis. Plant Cell 2013, 25, 2504–2521. [Google Scholar] [CrossRef] [Green Version]
- Bracher, A.; Sharma, A.; Starling-Windhof, A.; Hartl, F.U.; Hayer-Hartl, M. Degradation of potent RUBISCO inhibitor by selective sugar phosphatase. Nat. Plants 2014, 112015, 1–7. [Google Scholar] [CrossRef]
- Boldt, R.; Edner, C.; Kolukisaoglu, Ü.; Hagemann, M.; Weckwerth, W.; Wienkoop, S.; Morgenthal, K.; Bauwe, H. D-glycerate 3-kinase, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 2005, 17, 2413–2420. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Yang, L.; Han, X.; Zhao, Y.; Zhao, L.; Xiang, B.; Zhu, Y.; Bai, Y.; Wang, Y. Overexpression of AtAGT1 promoted root growth and development during seedling establishment. Plant Cell Rep. 2019, 38, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Sanda, S.; Leustek, T.; Theisen, M.J.; Garavito, R.M.; Benning, C. Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J. Biol. Chem. 2001, 276, 3941–3946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, H.; Abdel-Ghany, S.E.; Anderson, T.D.; Pilon-Smits, E.A.; Pilon, M. CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J. Biol. Chem. 2006, 281, 8958–8969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrario-Méry, S.; Meyer, C.; Hodges, M. Chloroplast nitrite uptake is enhanced in Arabidopsis PII mutants. FEBS Lett. 2008, 582, 1061–1066. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, M.; Sasaki, Y.; Ida, S.; Morikawa, H. Nitrite reductase gene enrichment improves assimilation of NO2 in Arabidopsis. Plant Physiol. 2001, 126, 731–741. [Google Scholar] [CrossRef] [Green Version]
- Shirley, B.W.; Hanley, S.; Goodman, H.M. Effects of ionizing radiation on a plant genome: Analysis of two Arabidopsis transparent testa mutations. Plant Cell 1992, 4, 333–347. [Google Scholar] [CrossRef]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate: Coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef]
- Klee, H.J.; Muskopf, Y.M.; Gasser, C.S. Cloning of an Arabidopsis thaliana gene encoding 5-enolpyruvyl shikimate-3-phosphate synthase: Sequence analysis and manipulation to obtain glyphosate-tolerant plants. Mol. Gen. Genet. 1987, 210, 437–442. [Google Scholar] [CrossRef]
- Dixon, D.P.; Edwards, R. Roles for stress-inducible lambda glutathione transferases in flavonoid metabolism in plants as identified by ligand fishing. J. Biol. Chem. 2010, 285, 36322–36329. [Google Scholar] [CrossRef] [Green Version]
- Rosenquist, M.; Alsterfjord, M.; Larsson, C.; Sommarin, M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes: Expression is demonstrated for two out of five novel genes. Plant Physiol. 2001, 127, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Hanson, M.R.; Bentolila, S. Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Res. 2015, 43, 3814–3825. [Google Scholar] [CrossRef]
- Lambermon, M.H.L.; Fu, Y.; Kirk, D.A.W.; Dupasquier, M.; Filipowicz, W.; Lorković, Z.J. UBA1 and UBA2, two proteins that interact with UBP1, a multifunctional effector of pre-mRNA maturation in plants. Mol. Cell. Biol. 2002, 22, 4346–4357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Xiong, L.; Ishitani, M.; Zhu, J.K. An Arabidopsis mutation in TRANSLATION ELONGATION FACTOR 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl. Acad. Sci. USA 2002, 99, 7786–7791. [Google Scholar] [CrossRef]
- Skalitzky, C.A.; Martin, J.R.; Harwood, J.H.; Beirne, J.J.; Adamczyk, B.J.; Heck, G.R.; Cline, K.; Fernandez, D.E. Plastids contain a second Sec translocase system with essential functions. Plant Physiol. 2011, 155, 354–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharjee, S.; Lee, L.Y.; Oltmanns, H.; Cao, H.; Veena; Cuperus, J.; Gelvin, S.B. IMPa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell 2008, 20, 2661–2680. [Google Scholar] [CrossRef] [Green Version]
- Haferkamp, I.; Hackstein, J.H.P.; Voncken, F.G.J.; Schmit, G.; Tjaden, J. Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur. J. Biochem. 2002, 269, 3172–3181. [Google Scholar] [CrossRef]
- Küchler, M.; Decker, S.; Hörmann, F.; Soll, J.; Heins, L. Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 2002, 21, 6136–6145. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Chung, G.C.; Jang, J.Y.; Ahn, S.J.; Zwiazek, J.J. Overexpression of PIP2;5 aquaporin alleviates effects of low root temperature on cell hydraulic conductivity and growth in Arabidopsis. Plant Physiol. 2012, 159, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Stanislas, T.; Hüser, A.; Barbosa, I.C.R.; Kiefer, C.S.; Brackmann, K.; Pietra, S.; Gustavsson, A.; Zourelidou, M.; Schwechheimer, C.; Grebe, M. Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nat. Plants 2015, 1, 15162. [Google Scholar] [CrossRef] [PubMed]
- Scranton, M.A.; Yee, A.; Park, S.Y.; Walling, L.L. Plant leucine aminopeptidases moonlight as molecular chaperones to alleviate stress-induced damage. J. Biol. Chem. 2012, 287, 18408–18417. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.-F.; O’Toole, N.; Taylor, N.L.; Millar, A.H. Divalent metal ions in plant mitochondria and their role in interactions with proteins and oxidative stress-induced damage to respiratory function. Plant Physiol. 2010, 152, 747–761. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Swart, C.; Alseekh, S.; Scossa, F.; Jiang, L.; Obata, T.; Graf, A.; Fernie, A.R. The extra-pathway interactome of the TCA cycle: Expected and unexpected metabolic interactions. Plant Physiol. 2018, 177, 966–979. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Liu, Q.; Zang, X.; Yuan, S.; Bat-Erdene, U.; Nguyen, C.; Gan, J.; Zhou, J.; Jacobsen, S.E.; Tang, Y. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature 2018, 559, 415–418. [Google Scholar] [CrossRef]
- de Kraker, J.W.; Luck, K.; Textor, S.; Tokuhisa, J.G.; Gershenzon, J. Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol. 2007, 143, 970–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Chen, L.; Zhou, Y.; Mawhinney, T.P.; Chen, B.; Kang, B.H.; Hauser, B.A.; Chen, S. Functional characterization of Arabidopsis thaliana isopropylmalate dehydrogenases reveals their important roles in gametophyte development. New Phytol. 2011, 189, 160–175. [Google Scholar] [CrossRef]
- Varotto, C.; Pesaresi, P.; Meurer, J.; Oelmüller, R.; Steiner-Lange, S.; Salamini, F.; Leister, D. Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 2000, 22, 115–124. [Google Scholar] [CrossRef]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef]
- Gargano, D.; Maple-Groedem, J.; Moeller, S.G. In vivo phosphorylation of FtsZ2 in Arabidopsis thaliana. Biochem. J. 2012, 446, 517–521. [Google Scholar] [CrossRef]
- Takagi, D.; Amako, K.; Hashiguchi, M.; Fukaki, H.; Ishizaki, K.; Goh, T.; Fukao, Y.; Sano, R.; Kurata, T.; Demura, T.; et al. Chloroplastic ATP synthase builds up a proton motive force preventing production of reactive oxygen species in photosystem I. Plant J. 2017, 91, 306–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, A.J.; Heazlewood, J.L.; Ito, J.; Millar, A.H. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell Proteom. 2008, 7, 347–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 1962, 15, 473–497. [Google Scholar] [CrossRef]

| Category | Accession Number | UniProtKB/ Swiss-Prot | Gene Name | Protein Name | MW (kDa) | % Coverage | Exclusive Unique Peptides | Total Spectrum | E-Value |
|---|---|---|---|---|---|---|---|---|---|
| Ribogenesis | 1-12177_3_ORF5 | P56799 | RPS4 | SMALL RIBOSOMAL SUBUNIT PROTEIN US4C | 25.6 | 0 | 0 | 3 | 7.63 × 10−70 |
| Ribogenesis | 16646-605_4_ORF2 | Q9SX68 | RPL18 | LARGE RIBOSOMAL SUBUNIT PROTEIN UL18C | 20.9 | 7 | 1 | 3 | 7.35 × 10−57 |
| Ribogenesis | 24557-506_3_ORF2 | Q9SKX4 | RPL3A | LARGE RIBOSOMAL SUBUNIT PROTEIN UL3C | 29.3 | 14 | 2 | 9 | 5.37 × 10−121 |
| Ribogenesis | 139931-207_5_ORF1 | O04603 | RPL5 | LARGE RIBOSOMAL SUBUNIT PROTEIN UL5C | 32.4 | 8 | 2 | 14 | 2.41 × 10−92 |
| Ribogenesis | 26987-486_4_ORF2 | Q9M3C3 | RPL23AB | LARGE RIBOSOMAL SUBUNIT PROTEIN UL23Y | 20.2 | 31 | 1 | 39 | 3.98 × 10−60 |
| Ribogenesis | 6809-878_4_ORF2 | P49227 | RPL5B | LARGE RIBOSOMAL SUBUNIT PROTEIN UL18Y | 34.7 | 13 | 4 | 12 | 1.09 × 10−53 |
| Ribogenesis | 87113-269_4_ORF1 | Q9M9W1 | RPL22B | LARGE RIBOSOMAL SUBUNIT PROTEIN EL22Z | 16.1 | 14 | 1 | 3 | 1.35 × 10−52 |
| Ribogenesis | 21899-533_1_ORF2 | P51419 | RPL27C | LARGE RIBOSOMAL SUBUNIT PROTEIN EL27X | 18.2 | 19 | 3 | 22 | 7.19 × 10−65 |
| Ribogenesis | 24282-509_2_ORF1 | P50883 | RPL12A | LARGE RIBOSOMAL SUBUNIT PROTEIN UL11Z | 19.1 | 13 | 0 | 6 | 1.24 × 10−104 |
| Ribogenesis | 36225-421_1_ORF2 | Q9SIM4 | RPL14A | LARGE RIBOSOMAL SUBUNIT PROTEIN EL14Z | 17.9 | 19 | 2 | 11 | 2.62 × 10−68 |
| Ribogenesis | 10983-723_3_ORF2 | P51418 | RPL18AB | LARGE RIBOSOMAL SUBUNIT PROTEIN EL20Y | 22.9 | 10 | 1 | 9 | 1.99 × 10−119 |
| Ribogenesis | 34727-430_6_ORF2 | P49637 | RPL27AC | LARGE RIBOSOMAL SUBUNIT PROTEIN UL15X | 16.3 | 7 | 1 | 4 | 8.73 × 10−78 |
| Ribogenesis | 73912-293_6_ORF2 | Q42064 | RPL8C | LARGE RIBOSOMAL SUBUNIT PROTEIN UL2X | 28.7 | 25 | 4 | 42 | 2.24 × 10−167 |
| Ribogenesis | 37807-413_3_ORF1 | Q93VI3 | RPL17A | LARGE RIBOSOMAL SUBUNIT PROTEIN UL22Z | 23.5 | 4 | 1 | 7 | 5.22 × 10−107 |
| Ribogenesis | 176389-160_3_ORF2 | Q42351 | RPL34A | LARGE RIBOSOMAL SUBUNIT PROTEIN EL34Z | 18.2 | 12 | 1 | 15 | 1.23 × 10−65 |
| Ribogenesis | 8788-797_5_ORF2 | Q9FZH0 | RPL35AB | LARGE RIBOSOMAL SUBUNIT PROTEIN EL33Z | 13.6 | 18 | 0 | 17 | 6.54 × 10−60 |
| Ribogenesis | 27503-481_2_ORF2 | O80929 | RPL36A | LARGE RIBOSOMAL SUBUNIT PROTEIN EL36Z | 13 | 16 | 1 | 28 | 8.82 × 10−52 |
| Ribogenesis | 75664-290_4_ORF2 | Q9SF40 | RPL4A | LARGE RIBOSOMAL SUBUNIT PROTEIN UL4Z | 46.3 | 12 | 3 | 20 | 0 |
| Ribogenesis | 45247-378_5_ORF2 | Q9SZX9 | RPL9D | LARGE RIBOSOMAL SUBUNIT PROTEIN UL6X | 25.1 | 7 | 1 | 11 | 1.63 × 10−105 |
| Ribogenesis | 86488-270_3_ORF2 | Q9LZH9 | RPL7AB | LARGE RIBOSOMAL SUBUNIT PROTEIN EL8Y | 32.5 | 26 | 8 | 33 | 7.63 × 10−154 |
| Ribogenesis | 163051-176_2_ORF2 | Q8VZ19 | RPL30B | LARGE RIBOSOMAL SUBUNIT PROTEIN EL30Y | 16.7 | 13 | 2 | 9 | 1.91 × 10−61 |
| Ribogenesis | 69050-304_1_ORF1 | P49200 | RPS20A | SMALL RIBOSOMAL SUBUNIT PROTEIN US10Z | 21.1 | 12 | 2 | 14 | 3.32 × 10−72 |
| Ribogenesis | 5816-941_6_ORF2 | P42036 | RPS14C | SMALL RIBOSOMAL SUBUNIT PROTEIN US11X | 18.7 | 22 | 3 | 7 | 2.52 × 10−87 |
| Ribogenesis | 39126-407_1_ORF2 | Q8LC83 | RPS24B | SMALL RIBOSOMAL SUBUNIT PROTEIN ES24Y | 20.7 | 7 | 1 | 2 | 8.38 × 10−74 |
| Ribogenesis | 108940-238_6_ORF2 | Q42262 | RPS3AB | SMALL RIBOSOMAL SUBUNIT PROTEIN ES1Y | 32.7 | 22 | 5 | 21 | 8.87 × 10−155 |
| Ribogenesis | 85164-272_6_ORF2 | Q8VYK6 | RPS4D | SMALL RIBOSOMAL SUBUNIT PROTEIN ES4X | 30 | 7 | 2 | 18 | 9.28 × 10−165 |
| Ribogenesis | 45770-376_1_ORF2 | Q9LXG1 | RPS9B | SMALL RIBOSOMAL SUBUNIT PROTEIN US4Z | 24.7 | 4 | 0 | 9 | 1.32 × 10−121 |
| Ribogenesis | 140134-206_2_ORF2 | F4JB06 | MGH6.2 | RIBOSOMAL PROTEIN S5/ELONGATION FACTOR G/III/V FAMILY PROTEIN | 16.9 | 7 | 0 | 1 | 2 × 10−50 |
| Ribogenesis | 1627-1498_1_ORF1 | P61841 | RPS7-A | SMALL RIBOSOMAL SUBUNIT PROTEIN US7CZ | 18.8 | 13 | 3 | 11 | 3.24 × 10−77 |
| Ribogenesis | 31704-450_4_ORF1 | Q9FIF3 | ES8Y | RIBOSOMAL PROTEIN ES8Y | 16.8 | 28 | 1 | 19 | 1.93 × 10−73 |
| Ribogenesis | 11320-714_2_ORF2 | Q9XJ27 | RPS9 | SMALL RIBOSOMAL SUBUNIT PROTEIN US9C | 25.9 | 3 | 1 | 3 | 1.03 × 10−76 |
| Ribogenesis | 21394-539_3_ORF2 | P42791 | RPL18B | LARGE RIBOSOMAL SUBUNIT PROTEIN EL18Y | 23.4 | 25 | 3 | 21 | 1.03 × 10−105 |
| Ribogenesis | 40578-399_1_ORF2 | P56798 | RPS3 | SMALL RIBOSOMAL SUBUNIT PROTEIN US3C | 25.3 | 6 | 1 | 6 | 6.73 × 10−82 |
| Ribogenesis | 10791-728_3_ORF2 | O65569 | RPS11B | SMALL RIBOSOMAL SUBUNIT PROTEIN US17Y | 20.5 | 12 | 2 | 36 | 5.17 × 10−84 |
| Ribogenesis | 66444-310_2_ORF2 | Q1PEP5 | NUCL2 | NUCLEOLIN 2 | 61.5 | 12 | 6 | 11 | 6.83 × 10−49 |
| Ribogenesis | 260531-93_3_ORF2 | O04658 | NOP5-1 | PROBABLE NUCLEOLAR PROTEIN 5-1 | 62.5 | 11 | 5 | 10 | 0 |
| Proteasome | 93086-259_4_ORF1 | Q9LT08 | RPN11 | 26S PROTEASOME NON-ATPASE REGULATORY SUBUNIT 14 HOMOLOG | 34.9 | 3 | 1 | 5 | 0 |
| Proteasome | 78122-285_4_ORF2 | Q93Y35 | RPN7 | 26S PROTEASOME NON-ATPASE REGULATORY SUBUNIT 6 HOMOLOG | 44.8 | 4 | 1 | 9 | 0 |
| Proteasome | 353924-42_4_ORF1 | O23712 | PAF2 | PROTEASOME SUBUNIT ALPHA TYPE-1-B | 13,9 | 8 | 1 | 2 | 2.8 × 10−136 |
| Proteasome | 7073-864_3_ORF1 | O23708 | PAB1 | PROTEASOME SUBUNIT ALPHA TYPE-2-A | 26.4 | 9 | 2 | 10 | 4.87 × 10−158 |
| Proteasome | 326729-54_1_ORF2 | O81148 | PAC1 | PROTEASOME SUBUNIT ALPHA TYPE-4-A | 30.6 | 13 | 3 | 9 | 3.69 × 10−154 |
| Proteasome | 68626-305_4_ORF2 | Q42134 | PAE2 | PROTEASOME SUBUNIT ALPHA TYPE-5-B | 28.7 | 28 | 3 | 13 | 4.68 × 10−158 |
| Proteasome | 23928-513_3_ORF1 | O81147 | PAA2 | PROTEASOME SUBUNIT ALPHA TYPE-6-B | 30.8 | 7 | 3 | 12 | 9.67 × 10−153 |
| Proteasome | 340935-47_4_ORF1 | O24616 | PAD2 | PROTEASOME SUBUNIT ALPHA TYPE-7-B | 27.4 | 18 | 4 | 11 | 3.86 × 10−148 |
| Proteasome | 14462-637_1_ORF2 | O23714 | PBD1 | PROTEASOME SUBUNIT BETA TYPE-2-A | 24.3 | 4 | 1 | 7 | 1.43 × 10−103 |
| Biological Function | Protein | Nº of Interactions |
|---|---|---|
| Metabolism of carbohydrates | PHOSPHOGLYCERATE KINASE 1 | 11 |
| Biosynthesis of amino acids | 3-ISOPROPYLMALATE DEHYDROGENASE | 5 |
| DIHYDROXY-ACID DEHYDRATASE | 5 | |
| ASPARTATE-SEMIALDEHYDE DEHYDROGENASE | 5 | |
| Metabolism of energy | ATP SYNTHASE GAMMA CHAIN 1 | 18 |
| Secondary metabolism | 4-COUMARATE-COA LIGASE 3 | 3 |
| Transcription & Translation | LARGE RIBOSOMAL SUBUNIT PROTEIN UL4Z | 44 |
| SMALL RIBOSOMAL SUBUNIT PROTEIN US11X | 44 | |
| SMALL RIBOSOMAL SUBUNIT PROTEIN US17Y | 44 | |
| Transport | PROTEIN TRANSLOCASE SUBUNIT SECA1 | 2 |
| COATOMER SUBUNIT GAMMA | 2 |
| Category | Accession Number | UniProtKB/ Swiss-Prot | Gene Name | Protein Name | MW (kDa) | % Coverage | Exclusive Unique Peptides | Total Spectra | E-Value |
|---|---|---|---|---|---|---|---|---|---|
| Carbohydrates | 58787-330_2_ORF2 | Q94AA4 | PFK3 | ATP-DEPENDENT 6-PHOSPHOFRUCTOKINASE 3 | 64.5 | 2 | 1 | 1 | 0 |
| Carbohydrates | 135690-210_1_ORF2 | Q9ZU52 | FBA3 | FRUCTOSE-BISPHOSPHATE ALDOLASE 3 | 42.7 | 32 | 7 | 38 | 0 |
| Carbohydrates | 38153-411_5_ORF2 | Q38799 | PDH2 | PYRUVATE DEHYDROGENASE E1 COMPONENT SUBUNIT BETA-1 | 40.3 | 14 | 4 | 6 | 0 |
| Carbohydrates | 83096-276_3_ORF2 | Q5GM68 | PPC2 | PHOSPHOENOLPYRUVATE CARBOXYLASE 2 | 112.8 | 6 | 5 | 5 | 0 |
| Carbohydrates | 54280-344_1_ORF1 | Q84VW9 | PPC3 | PHOSPHOENOLPYRUVATE CARBOXYLASE 3 | 111.8 | 1 | 0 | 1 | 0 |
| Carbohydrates | 113756-233_2_ORF1 | Q9SIU0 | NAD-ME1 | NAD-DEPENDENT MALIC ENZYME 1 | 71.4 | 3 | 2 | 3 | 0 |
| Carbohydrates | 102811-246_6_ORF2 | O04499 | PGM1 | 2,3-BIPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE 1 | 63.2 | 4 | 1 | 2 | 0 |
| Carbohydrates | 64100-316_1_ORF1 | O82662 | AT2G20420 | SUCCINATE-COA LIGASE [ADP-FORMING] SUBUNIT BETA | 50 | 9 | 0 | 4 | 0 |
| Carbohydrates | 8279-816_3_ORF2 | P68209 | AT5G08300 | SUCCINATE-COA LIGASE [ADP-FORMING] SUBUNIT ALPHA-1 | 34.6 | 9 | 2 | 3 | 0 |
| Carbohydrates | 222487-119_2_ORF2 | P93819 | MDH1 | MALATE DEHYDROGENASE 1 | 38.4 | 20 | 2 | 16 | 0 |
| Carbohydrates | 156827-185_4_ORF1 | Q9SH69 | PGD1 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING 1 | 58.8 | 16 | 2 | 9 | 1.37 × 10−112 |
| Carbohydrates | 12493-682_6_ORF2 | Q9FJI5 | G6PD6 | GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE 6 | 65.1 | 6 | 3 | 3 | 0 |
| Carbohydrates | 20760-547_4_ORF1 | Q9LD57 | PGK1 | PHOSPHOGLYCERATE KINASE 1 | 19.5 | 14 | 0 | 3 | 5.63 × 10−43 |
| Carbohydrates | 69882-302_6_ORF2 | Q9LZS3 | SBE2.2 | 1,4-ALPHA-GLUCAN-BRANCHING ENZYME 2-2 | 98.6 | 5 | 4 | 6 | 0 |
| Carbohydrates | 96049-255_6_ORF1 | Q9MAQ0 | GBSS1 | GRANULE BOUND STARCH SYNTHASE 1 | 70.16 | 1 | 1 | 1 | 0 |
| Lipids | 20213-554_2_ORF1 | Q9SLA8 | MOD1 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] | 41.8 | 7 | 1 | 6 | 4.48 × 10−180 |
| Lipids | 387953-27_4_ORF1 | Q9SGY2 | ACLA-1 | ATP-CITRATE SYNTHASE ALPHA CHAIN PROTEIN 1 | 46.8 | 10 | 2 | 4 | 0 |
| Lipids | 211149-128_1_ORF1 | Q9LXS6 | CSY2 | CITRATE SYNTHASE 2 | 57.9 | 2 | 0 | 1 | 0 |
| Amino acids | 47558-369_4_ORF2 | P46643 | ASP1 | ASPARTATE AMINOTRANSFERASE | 51.5 | 5 | 0 | 5 | 0 |
| Amino acids | 72506-296_4_ORF1 | Q94AR8 | IIL1 | 3-ISOPROPYLMALATE DEHYDRATASE LARGE SUBUNIT | 57.6 | 6 | 3 | 4 | 0 |
| Amino acids | 125905-219_3_ORF2 | Q9ZNZ7 | GLU1 | FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1 | 181.3 | 6 | 5 | 13 | 0 |
| Amino acids | 14065-645_3_ORF2 | Q9C5U8 | HISN8 | HISTIDINOL DEHYDROGENASE | 55.2 | 1 | 2 | 1 | 0 |
| Amino acids | 294436-71_4_ORF2 | Q9LUT2 | METK4 | S-ADENOSYLMETHIONINE SYNTHASE 4 | 42.7 | 5 | 0 | 2 | 0 |
| Nucleotides | 2121-1366_3_ORF2 | Q9SF85 | ADK1 | ADENOSINE KINASE 1 | 39.2 | 14 | 3 | 4 | 0 |
| Nucleotides | 59309-329_5_ORF1 | Q96529 | PURA | ADENYLOSUCCINATE SYNTHETASE | 57.9 | 2 | 1 | 1 | 0 |
| Nucleotides | 152024-193_3_ORF2 | Q9S726 | RPI3 | PROBABLE RIBOSE-5-PHOSPHATE ISOMERASE 3 | 36.1 | 2 | 0 | 2 | 5.33 × 10−120 |
| Nucleotides | 27769-479_3_ORF1 | P55228 | ADG1 | GLUCOSE-1-PHOSPHATE ADENYLYLTRANSFERASE SMALL SUBUNIT | 12.5 | 7 | 0 | 1 | 4.43 × 10−63 |
| Nucleotides | 181563-155_3_ORF2 | P55229 | ADG2 | GLUCOSE-1-PHOSPHATE ADENYLYLTRANSFERASE LARGE SUBUNIT 1 | 57.4 | 6 | 2 | 4 | 0 |
| Energy | 154679-189_1_ORF2 | Q9S841 | PSBO2 | OXYGEN-EVOLVING ENHANCER PROTEIN 1-2 | 35.3 | 35 | 7 | 24 | 6.62 × 10−141 |
| Energy | 218625-122_1_ORF2 | O22773 | AT4G02530 | THYLAKOID LUMENAL 16.5 kDa PROTEIN | 24.7 | 5 | 1 | 1 | 8.55 × 10−47 |
| Category | Accession Number | UniProtKB/ Swiss-Prot | Gene Name | Protein Name | MW (kDa) | % Coverage | Exclusive Unique Peptides | Total Spectra | E-Value |
| Energy | 6036-926_2_ORF1 | Q9ASS6 | PNSL5 | PHOTOSYNTHETIC NDH SUBUNIT OF LUMENAL LOCATION 5 | 32.2 | 15 | 4 | 10 | 2.45 × 10−93 |
| Energy | 250817-99_2_ORF2 | Q94K71 | CBBY | CBBY-LIKE PROTEIN | 34.9 | 7 | 2 | 3 | 6.24 × 10−131 |
| Energy | 235330-110_2_ORF1 | Q944I4 | GLYK | D-GLYCERATE 3-KINASE | 43.9 | 3 | 0 | 1 | 2.82 × 10−153 |
| Energy | 297118-70_2_ORF2 | Q56YA5 | AGT1 | SERINE-GLYOXYLATE AMINOTRANSFERASE | 47.8 | 2 | 0 | 2 | 0 |
| S&N metabolism | 33137-439_6_ORF2 | O48917 | SQD1 | UDP-SULFOQUINOVOSE SYNTHASE | 54.9 | 10 | 3 | 4 | 0 |
| S&N metabolism | 227095-115_1_ORF2 | Q84W65 | SUFE1 | SUFE-LIKE PROTEIN 1 | 40.7 | 2 | 1 | 1 | 3.22 × 10−106 |
| S&N metabolim | 311596-62_2_ORF2 | Q9ZST4 | GLB1 | NITROGEN REGULATORY PROTEIN P-II HOMOLOG | 23.4 | 15 | 1 | 1 | 3.28 × 10−62 |
| S&N metabolim | 318906-58_1_ORF1 | Q39161 | NIR1 | FERREDOXIN-NITRITE REDUCTASE | 69.6 | 7 | 4 | 8 | 0 |
| Secondary compounds | 156331-186_3_ORF2 | P41088 | CHI1 | CHALCONE-FLAVANONE ISOMERASE 1 | 26.2 | 6 | 0 | 2 | 6.89 × 10−56 |
| Secondary compounds | 230420-113_2_ORF2 | P34802 | GGPPS1 | HETERODIMERIC GERANYLGERANYL PYROPHOSPHATE SYNTHASE LARGE SUBUNIT 1 | 41 | 5 | 1 | 1 | 1.48 × 10−147 |
| Secondary compounds | 85783-271_1_ORF2 | Q9T030 | PCBER1 | PHENYLCOUMARAN BENZYLIC ETHER REDUCTASE 1 | 34.9 | 33 | 9 | 23 | 4.03 × 10−115 |
| Secondary compounds | 156554-185_2_ORF1 | Q9S777 | 4CL3 | 4-COUMARATE-COA LIGASE 3 | 51.9 | 2 | 1 | 2 | 0 |
| Secondary compounds | 223603-118_1_ORF1 | P05466 | AT2G45300 | 3-PHOSPHOSHIKIMATE 1-CARBOXYVINYLTRANSFERASE | 44.3 | 2 | 1 | 1 | 0 |
| Oxido -reduction | 133847-212_2_ORF2 | Q9SID3 | AT2G31350 | HYDROXYACYLGLUTATHIONE HYDROLASE 2 | 33.1 | 6 | 1 | 1 | 6.51 × 10−131 |
| Oxido -reduction | 34437-432_2_ORF1 | Q9M2W2 | GSTL2 | GLUTATHIONE S-TRANSFERASE L2 | 16.1 | 15 | 2 | 5 | 3.24 × 10−29 |
| Oxido -reduction | 115571-230_4_ORF1 | Q9LZ06 | GSTL3 | GLUTATHIONE S-TRANSFERASE L3 | 35.3 | 1 | 2 | 1 | 6.13 × 10−75 |
| Transcription | 181200-155_2_ORF2 | Q96300 | GRF7 | 14-3-3-LIKE PROTEIN GF14 NU | 32.8 | 12 | 1 | 9 | 1.11 × 10−161 |
| Transcription | 287872-75_1_ORF1 | Q9C5W6 | GRF12 | 14-3-3-LIKE PROTEIN GF14 IOTA | 32.8 | 12 | 1 | 9 | 3.83 × 10−151 |
| Translation | 209284-130_2_ORF2 | Q9FNR1 | RBG3 | GLYCINE-RICH RNA-BINDING PROTEIN 3 | 19.8 | 15 | 2 | 4 | 1.45 × 10−31 |
| Translation | 293356-72_1_ORF1 | Q9LR72 | PCMP-E3 | PUTATIVE PENTATRICOPEPTIDE REPEAT-CONTAINING PROTEIN AT1G03510 (POLIPASA) | 26.1 | 5 | 1 | 1 | 0.4 |
| Translation | 26795-487_6_ORF2 | Q0WW84 | RBP47B | POLYADENYLATE-BINDING PROTEIN RBP47B | 45.9 | 2 | 0 | 1 | 3.27 × 10−136 |
| Translation | 174433-162_1_ORF1 | Q9ASR1 | LOS1 | ELONGATION FACTOR 2 | 73.8 | 9 | 1 | 12 | 0 |
| Folding | 26640-489_1_ORF2 | Q9M1C2 | CPN10-1 | 10 kDa CHAPERONIN 1 | 19.4 | 15 | 2 | 6 | 9.48 × 10−39 |
| Folding | 189606-147_1_ORF2 | Q9SR70 | FKBP16-4 | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FKBP16-4 | 26.4 | 13 | 3 | 6 | 1.35 × 10−89 |
| Folding | 2524-1285_6_ORF2 | Q9SKQ0 | CYP19-2 | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE CYP19-2 | 21.6 | 27 | 5 | 24 | 2.24 × 10−90 |
| Sorting | 19573-562_5_ORF2 | Q9SYI0 | SECA1 | PROTEIN TRANSLOCASE SUBUNIT SECA1 | 115.7 | 2 | 1 | 2 | 0 |
| Sorting | 146969-201_2_ORF1 | F4JL11 | IMPA2 | IMPORTIN SUBUNIT ALPHA-2 | 59.1 | 5 | 0 | 2 | 6 × 10−61 |
| Sorting | 151836-193_1_ORF2 | P40941 | AAC2 | ADP, ATP CARRIER PROTEIN 2 | 42.3 | 7 | 1 | 5 | 0 |
| Sorting | 161087-178_2_ORF2 | Q8H0U5 | TIC62 | PROTEIN TIC 62 | 73.4 | 6 | 4 | 6 | 5.77 × 10−115 |
| Sorting | 82340-277_1_ORF2 | Q39196 | PIP1.4 | PROBABLE AQUAPORIN PIP1-4 | 33 | 6 | 2 | 3 | 3.75 × 10−170 |
| Sorting | 272341-85_2_ORF2 | Q94A40 | AT1G62020 | COATOMER SUBUNIT ALPHA-1 | 137 | 1 | 0 | 1 | 0 |
| Sorting | 29489-466_3_ORF1 | Q0WW26 | AT4G34450 | COATOMER SUBUNIT GAMMA | 103 | 2 | 1 | 1 | 0 |
| Sorting | 43675-385_1_ORF2 | Q67YI9 | EPSIN2 | CLATHRIN INTERACTOR EPSIN 2 | 85.2 | 1 | 1 | 1 | 1.51 × 10−117 |
| Sorting | 68824-304_5_ORF2 | Q9LQ55 | DRP2B | DYNAMIN-2B | 105.3 | 1 | 1 | 1 | 0 |
| Degradation | 141778-205_4_ORF2 | Q8L770 | CLPR3 | ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT-RELATED PROTEIN 3 | 38.9 | 2 | 1 | 1 | 1.73 × 10−136 |
| Degradation | 172993-163_5_ORF1 | Q9XJ36 | CLPR2 | ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT-RELATED PROTEIN 2 | 32.7 | 4 | 1 | 1 | 4.49 × 10−126 |
| Degradation | 170504-166_2_ORF2 | P30184 | LAP1 | LEUCINE AMINOPEPTIDASE 1 | 62.5 | 4 | 1 | 4 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ojosnegros, S.; Alvarez, J.M.; Grossmann, J.; Gagliardini, V.; Quintanilla, L.G.; Grossniklaus, U.; Fernández, H. Proteome and Interactome Linked to Metabolism, Genetic Information Processing, and Abiotic Stress in Gametophytes of Two Woodferns. Int. J. Mol. Sci. 2023, 24, 12429. https://doi.org/10.3390/ijms241512429
Ojosnegros S, Alvarez JM, Grossmann J, Gagliardini V, Quintanilla LG, Grossniklaus U, Fernández H. Proteome and Interactome Linked to Metabolism, Genetic Information Processing, and Abiotic Stress in Gametophytes of Two Woodferns. International Journal of Molecular Sciences. 2023; 24(15):12429. https://doi.org/10.3390/ijms241512429
Chicago/Turabian StyleOjosnegros, Sara, José Manuel Alvarez, Jonas Grossmann, Valeria Gagliardini, Luis G. Quintanilla, Ueli Grossniklaus, and Helena Fernández. 2023. "Proteome and Interactome Linked to Metabolism, Genetic Information Processing, and Abiotic Stress in Gametophytes of Two Woodferns" International Journal of Molecular Sciences 24, no. 15: 12429. https://doi.org/10.3390/ijms241512429
APA StyleOjosnegros, S., Alvarez, J. M., Grossmann, J., Gagliardini, V., Quintanilla, L. G., Grossniklaus, U., & Fernández, H. (2023). Proteome and Interactome Linked to Metabolism, Genetic Information Processing, and Abiotic Stress in Gametophytes of Two Woodferns. International Journal of Molecular Sciences, 24(15), 12429. https://doi.org/10.3390/ijms241512429