Multi-Omics Analysis Reveals Mechanisms of Strong Phosphorus Adaptation in Tea Plant Roots
Abstract
:1. Introduction
2. Results
2.1. 250P Inhibited Tea Plant Root Growth
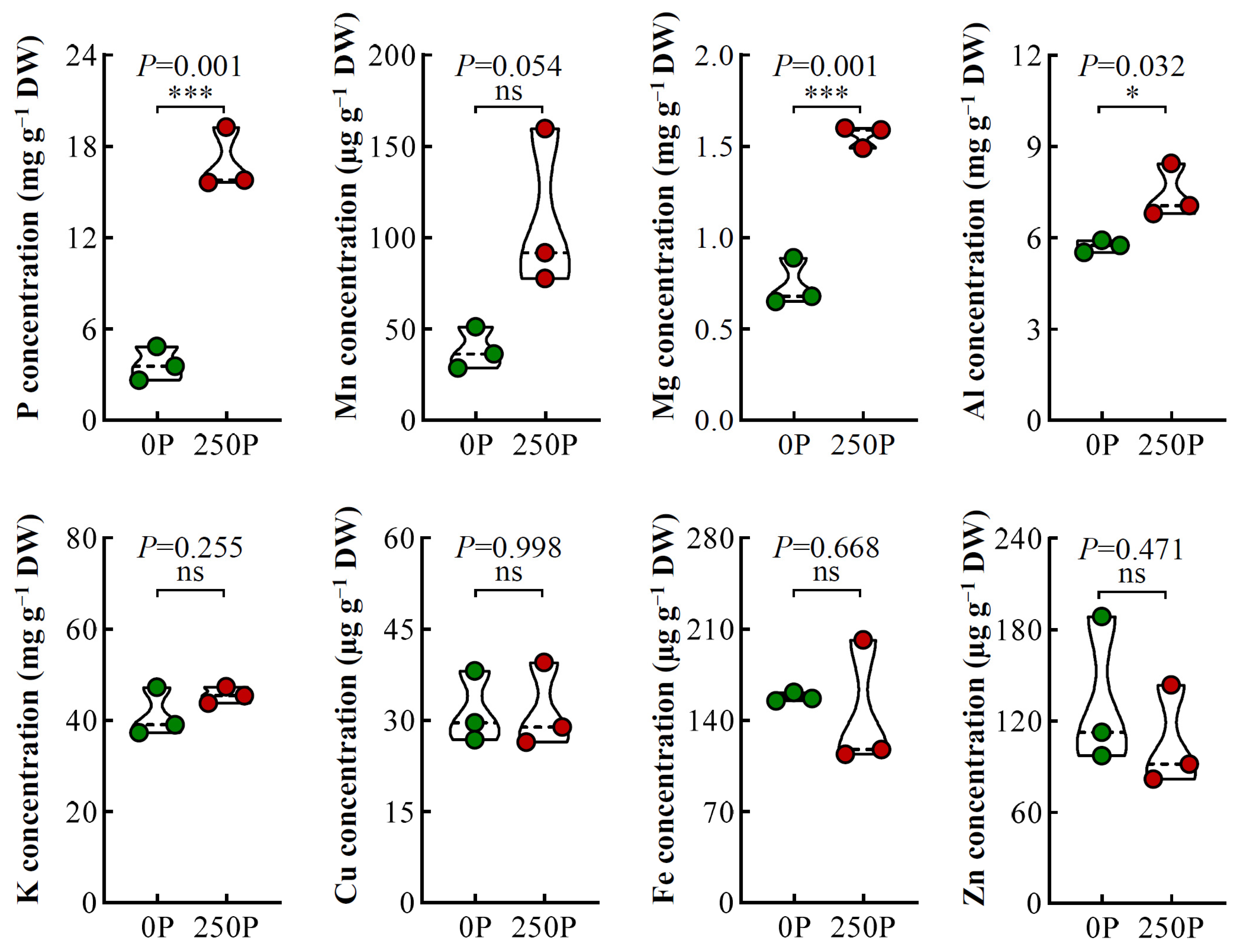
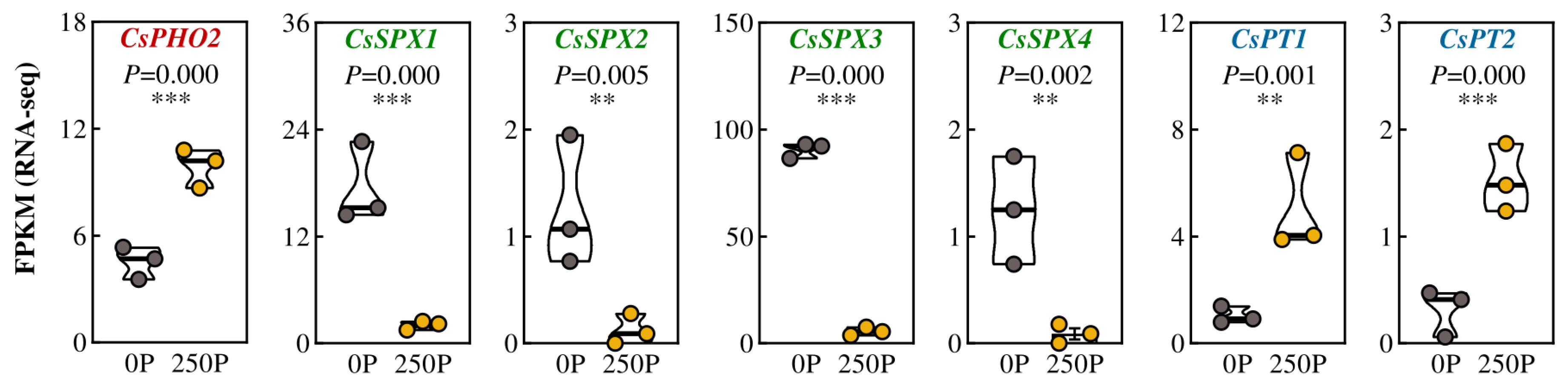
2.2. 250P-Increased Expression of PT and Over-Accumulation of P in Tea Plant Roots
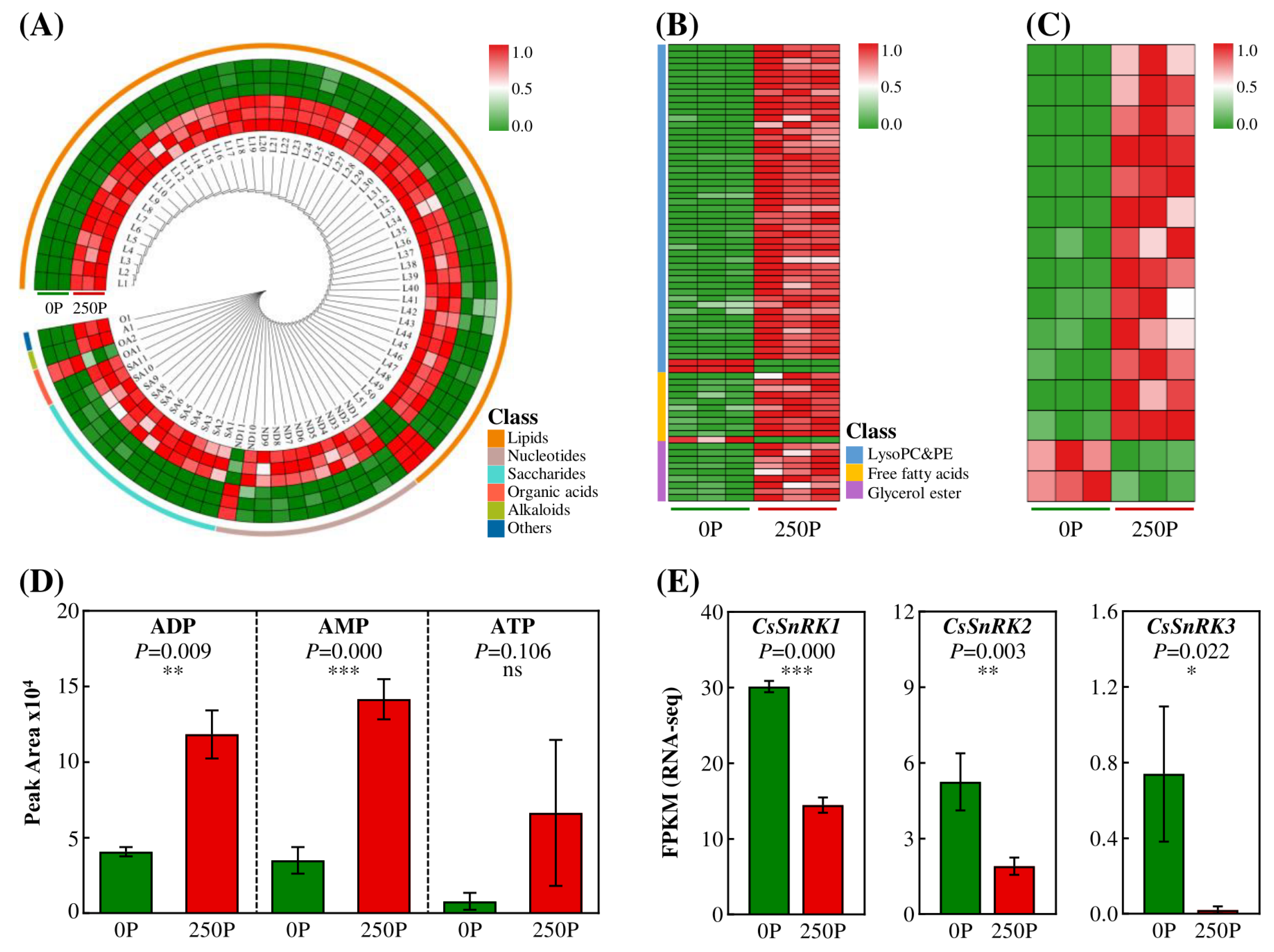
2.3. 250P Reduced Expression of SCPLs and Synthesis of Esterified Catechins
2.4. 250P-Decreased Synthesis of AMP/ATP Ratio and Expression of SnRKs
3. Discussion
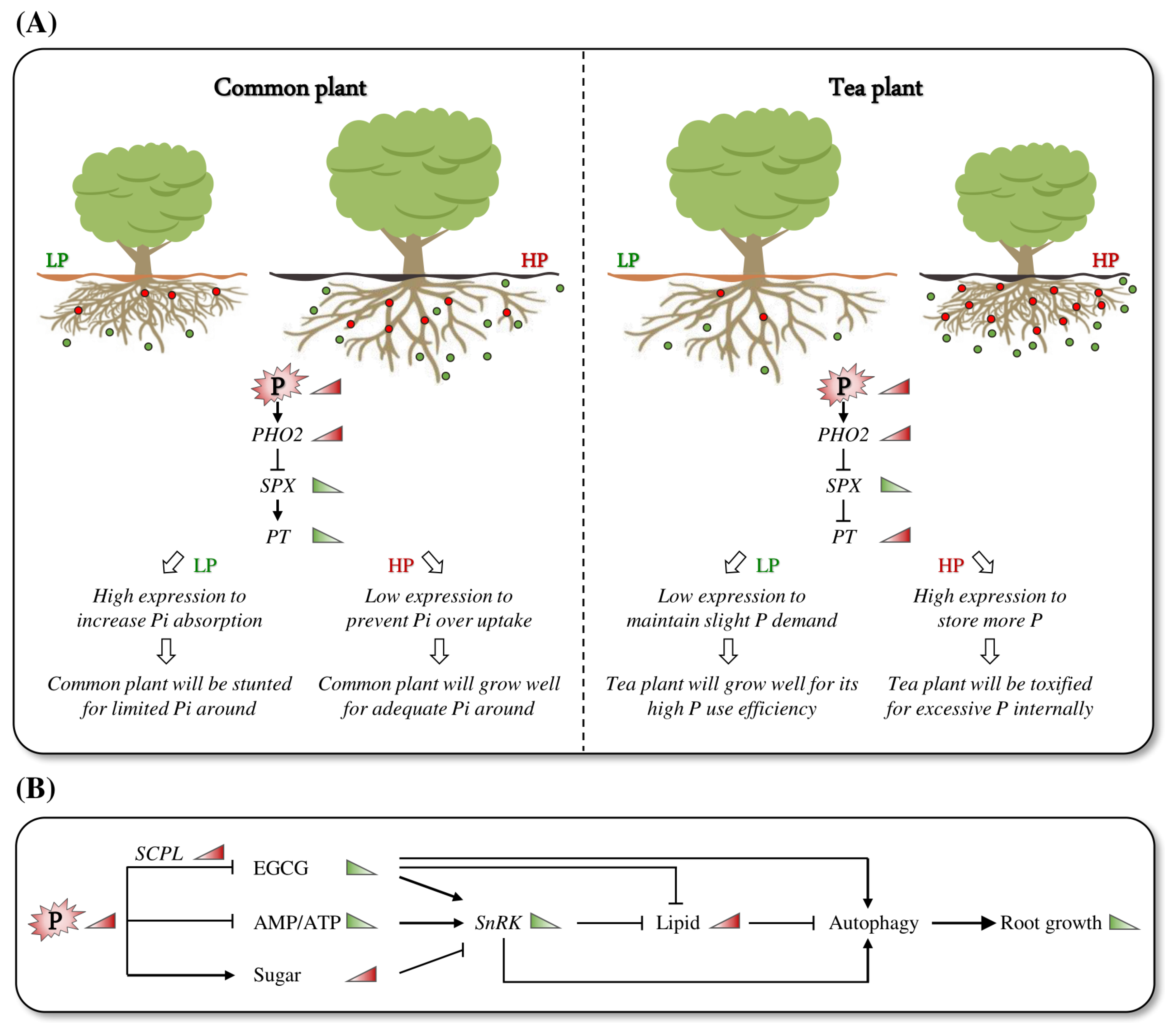
3.1. Tea Plant Roots Exhibit Unique Adaptation to Low-P Growing Environments
3.2. Adaptation of Tea Plants to Low P May Be Primarily Regulated by Energy Metabolism
4. Materials and Methods
4.1. Plant Growth
4.2. Phenotypic Analysis
4.3. Ionomic Analysis
4.4. Metabolomic Analysis
4.5. Transcriptomic Analysis
4.6. Gene Expression Analysis
4.7. Evolutionary Tree Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lambers, H. Phosphorus acquisition and utilization in plants. Annu. Rev. Plant Biol. 2022, 73, 17–42. [Google Scholar] [CrossRef]
- Wang, X.R.; Shen, J.B.; Liao, H. Acquisition or utilization, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Sci. 2010, 179, 302–306. [Google Scholar] [CrossRef]
- Kochian, L.V.; Hoekenga, O.A.; Piñeros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Puga, M.I.; Rojas-Triana, M.; Martinez-Hevia, I.; Diaz, S.; Poza-Carrion, C.; Minambres, M.; Leyva, A. Plant adaptation to low phosphorus availability: Core signaling, crosstalks, and applied implications. Mol. Plant. 2022, 15, 104–124. [Google Scholar] [CrossRef]
- Jia, X.Q.; Wang, L.; Zeng, H.Q.; Yi, K.K. Insights of intracellular/intercellular phosphate transport and signaling in unicellular green algae and multicellular land plants. New Phytol. 2021, 232, 1566–1571. [Google Scholar] [CrossRef]
- Duan, K.; Yi, K.K.; Dang, L.; Huang, H.J.; Wu, W.; Wu, P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef]
- Yao, Z.F.; Liang, C.Y.; Zhang, Q.; Chen, Z.J.; Xiao, B.X.; Tian, J.; Liao, H. SPX1 is an important component in the phosphorus signalling network of common bean regulating root growth and phosphorus homeostasis. J. Exp. Bot. 2014, 65, 3299–3310. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Yan, Y.; Zhang, F.; Wang, H.D.; Gu, M.; Wu, X.X.; Sun, S.B.; Xu, G.H. Fine characterization of OsPHO2 knockout mutants reveals its key role in Pi utilization in rice. J. Plant Physiol. 2014, 171, 340–348. [Google Scholar] [CrossRef]
- Wang, C.; Ying, S.; Huang, J.; Li, K.; Wu, P.; Shou, H.X. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 2009, 57, 895–904. [Google Scholar] [CrossRef]
- Chien, P.S.; Chao, Y.T.; Chou, C.H.; Hsu, Y.Y.; Chiang, S.F.; Tung, C.W.; Chiou, T.J. Phosphate transporter PHT1;1 is a key determinant of phosphorus acquisition in Arabidopsis natural accessions. Plant Physiol. 2022, 190, 682–697. [Google Scholar] [CrossRef]
- Sun, S.B.; Gu, M.; Cao, Y.; Huang, X.P.; Zhang, X.; Ai, P.H.; Zhao, J.N.; Fan, X.R.; Xu, G.H. A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 2012, 159, 1571–1581. [Google Scholar] [CrossRef] [Green Version]
- Nussaume, L.; Kanno, S.; Javot, H.; Marin, E.; Pochon, H.; Ayadi, A.; Nakanishi, T.M.; Thibaud, M.C. Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2011, 2, 83. [Google Scholar] [CrossRef] [Green Version]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef]
- Misson, J.; Thibaud, M.C.; Bechtold, N.; Raghothama, K.; Nussaume, L. Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 2004, 55, 727–741. [Google Scholar] [CrossRef]
- Sun, L.L.; Zhang, M.S.; Liu, X.M.; Mao, Q.Z.; Shi, C.; Kochian, L.V.; Liao, H. Aluminium is essential for root growth and development of tea plants (Camellia sinensis). J. Integr. Plant Biol. 2020, 62, 984–997. [Google Scholar] [CrossRef]
- Salehi, S.Y.; Hajiboland, R. A high internal phosphorus use efficiency in tea (Camellia sinensis L.) plants. Asian J. Plant Sci. 2008, 7, 30–36. [Google Scholar] [CrossRef]
- Lin, Z.H.; Chen, L.S.; Chen, R.B.; Zhang, F.Z.; Jiang, H.X.; Tang, N.; Smith, B.R. Root release and metabolism of organic acids in tea plants in response to phosphorus supply. J. Plant Physiol. 2011, 168, 644–652. [Google Scholar] [CrossRef]
- Lin, Z.H.; Chen, L.S.; Chen, R.B.; Zhang, F.Z.; Jiang, H.X.; Tan, N. CO2 assimilation, ribulose-1,5-bisphosphate carboxylase/oxygenase, carbohydrates and photosynthetic electron transport probed by the JIP-test, of tea leaves in response to phosphorus supply. BMC Plant Biol. 2009, 21, 9–43. [Google Scholar]
- Lin, Z.H.; Chen, L.S.; Chen, R.B.; Zhang, F.Z. Antioxidant system of tea (Camellia sinensis) leaves in response to phosphorus supply. Acta Physiol. Plant. 2012, 34, 2443–2448. [Google Scholar] [CrossRef]
- Zhu, G.T.; Wang, S.C.; Huang, Z.J.; Zhang, S.B.; Liao, Q.G.; Zhang, C.Z.; Lin, T.; Qin, M.; Peng, M.; Yang, C.K.; et al. Rewiring of the fruit metabolome in tomato breeding. Cell 2018, 172, 249–261. [Google Scholar] [CrossRef]
- Ding, Z.T.; Jia, S.S.; Wang, Y.; Xiao, J.; Zhang, Y.F. Phosphate stresses affect ionome and metabolome in tea plants. Plant Physiol. Biochem. 2017, 120, 30–39. [Google Scholar] [CrossRef]
- KC, S.; Liu, M.Y.; Zhang, Q.F.; Fan, K.; Shi, Y.Z.; Ruan, J.Y. Metabolic changes of amino acids and flavonoids in tea plants in response to inorganic phosphate limitation. Int. J. Mol. Sci. 2018, 19, 3683. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Shou, H.X.; Xu, G.H.; Lian, X.M. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 2013, 16, 205–212. [Google Scholar] [CrossRef] [Green Version]
- Raghothama, K.G. Phosphate transport and signaling. Curr. Opin. Plant Biol. 2000, 3, 182–187. [Google Scholar] [CrossRef]
- Almatroodi, S.A.; Almatroudi, A.; Khan, A.A.; Alhumaydhi, F.A.; Alsahli, M.A.; Rahma, A.H. Potential therapeutic targets of epigallocatechin gallate (EGCG), the most abundant catechin in green tea, and its role in the therapy of various types of cancer. Molecules 2020, 25, 3146. [Google Scholar] [CrossRef]
- Yao, S.B.; Liu, Y.J.; Zhuang, J.H.; Zhao, Y.; Dai, X.L.; Jiang, C.J.; Wang, Z.H.; Jiang, X.L.; Zhang, S.X.; Qian, Y.M.; et al. Insights into acylation mechanisms: Co-expression of serine carboxypeptidase-like acyltransferases and their non-catalytic companion paralogs. Plant J. 2022, 111, 117–133. [Google Scholar] [CrossRef]
- Robaglia, C.; Thomas, M.; Meyer, C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 2012, 15, 301–307. [Google Scholar] [CrossRef]
- Lynch, J.P. Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [Green Version]
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef]
- Crombez, H.; Motte, H.; Beeckman, T. Tackling plant phosphate starvation by the roots. Dev. Cell 2019, 48, 599–615. [Google Scholar] [CrossRef] [Green Version]
- Yao, Z.F.; Tian, J.; Liao, H. Comparative characterization of GmSPX members reveals that GmSPX3 is involved in phosphate homeostasis in soybean. Ann. Bot. 2014, 114, 477–488. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.R.; Wang, B.M.; Zhao, Y.J.; Zhang, J.R.; Li, Z.X. Auxin and GA signaling play important roles in the maize response to phosphate deficiency. Plant Sci. 2019, 283, 177–188. [Google Scholar] [CrossRef]
- Liu, D. Root developmental responses to phosphorus nutrition. J. Integr. Plant Biol. 2021, 63, 1065–1090. [Google Scholar] [CrossRef]
- Lin, S.C.; Hardie, D.G. AMPK: Sensing glucose as well as cellular energy status. Cell Metab. 2018, 27, 299–313. [Google Scholar] [CrossRef] [Green Version]
- Rutter, G.A.; Xavier, G.D.S.; Leclerc, I. Roles of 5′-AMP-activated protein kinase (AMPK) in mammalian glucose homoeostasis. Biochem. J. 2003, 375, 1–16. [Google Scholar] [CrossRef]
- Sugden, C.; Crawford, R.M.; Halford, N.G.; Hardie, D.G. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J. 1999, 19, 433–439. [Google Scholar] [CrossRef] [Green Version]
- Wilson, W.A.; Hawley, S.A.; Hardie, D.G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 1996, 6, 1426–1634. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Yan, Y.; Novick, S.J.; Kovach, A.; Goswami, D.; Ke, J.Y.; Tan, M.H.E.; Wang, L.L.; Li, X.D.; Waal, P.W.D.; et al. Deconvoluting AMP-dependent kinase (AMPK) adenine nucleotide binding and sensing. J. Biol. Chem. 2017, 292, 12653–12666. [Google Scholar] [CrossRef] [Green Version]
- Leene, J.V.; Eeckhout, D.; Gadeyne, A.; Matthijs, C.; Han, C.; Winne, N.D.; Persiau, G.; Slijke, E.V.D.; Persyn, F.; Mertens, T.; et al. Mapping of the plant SnRK1 kinase signalling network reveals a key regulatory role for the class II T6P synthase-like proteins. Nat. Plants 2022, 8, 1245–1261. [Google Scholar] [CrossRef]
- Murase, T.; Misawa, K.; Haramizu, S.; Hase, T. Catechin-induced activation of the LKB1/AMP-activated protein kinase pathway. Biochem. Pharmacol. 2009, 78, 78–84. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Violle, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 2007, 3, 257–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papinski, D.; Schuschnig, M.; Reiter, W.; Wilhelm, L.; Barnes, C.A.; Maiolica, A.; Hansmann, I.; Pfaffenwimmer, T.; Kijanska, M.; Stoffel, I.; et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell 2014, 53, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Su, Z.Z.; Huang, L.; Xia, F.N.; Qi, H.; Xie, L.J.; Xiao, S.; Chen, Q.F. The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci. 2017, 8, 1201. [Google Scholar] [CrossRef] [Green Version]
- Woods, A.; Munday, M.R.; Scott, J.; Yang, X.L.; Carlson, M.; Carling, D. Yeast SNFl is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 1994, 269, 19509–19515. [Google Scholar] [CrossRef]
- Nosheen, S.; Naz, T.; Yang, J.H.; Hussain, S.A.; Fazili, A.B.A.; Nazir, Y.; Li, S.Q.; Mohamed, H.; Yang, W.; Mustafa, K.; et al. Role of Snf-β in lipid accumulation in the high lipid-producing fungus Mucor circinelloides WJ11. Microb. Cell Fact. 2021, 20, 52. [Google Scholar] [CrossRef]
- Guan, B.; Jiang, Y.T.; Lin, D.L.; Lin, W.H.; Xue, H.W. Phosphatidic acid suppresses autophagy through competitive inhibition by binding GAPC (glyceraldehyde-3-phosphate dehydrogenase) and PGK (phosphoglycerate kinase) proteins. Autophagy 2022, 18, 2656–2670. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, J.; Shu, W.; Gao, B.; Xiong, S. Epigallocatechin-3-gallate opposes HBV-induced incomplete autophagy by enhancing lysosomal acidification, which is unfavorable for HBV replication. Cell Death Dis. 2015, 6, e1770. [Google Scholar] [CrossRef] [Green Version]
- Muoki, R.C.; Paul, A.; Kumari, A.; Singh, K.; Kumar, S. An improved protocol for the isolation of RNA from roots of tea (Camellia sinensis (L.) O. Kuntze). Mol. Biotechnol. 2012, 52, 82–88. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Tian, J.; Liu, G.; Sun, L. Multi-Omics Analysis Reveals Mechanisms of Strong Phosphorus Adaptation in Tea Plant Roots. Int. J. Mol. Sci. 2023, 24, 12431. https://doi.org/10.3390/ijms241512431
Liu X, Tian J, Liu G, Sun L. Multi-Omics Analysis Reveals Mechanisms of Strong Phosphorus Adaptation in Tea Plant Roots. International Journal of Molecular Sciences. 2023; 24(15):12431. https://doi.org/10.3390/ijms241512431
Chicago/Turabian StyleLiu, Xiaomei, Jing Tian, Guodao Liu, and Lili Sun. 2023. "Multi-Omics Analysis Reveals Mechanisms of Strong Phosphorus Adaptation in Tea Plant Roots" International Journal of Molecular Sciences 24, no. 15: 12431. https://doi.org/10.3390/ijms241512431
APA StyleLiu, X., Tian, J., Liu, G., & Sun, L. (2023). Multi-Omics Analysis Reveals Mechanisms of Strong Phosphorus Adaptation in Tea Plant Roots. International Journal of Molecular Sciences, 24(15), 12431. https://doi.org/10.3390/ijms241512431





