Metabolic Pathway Reconstruction Indicates the Presence of Important Medicinal Compounds in Coffea Such as L-DOPA
Abstract
:1. Introduction
2. Results
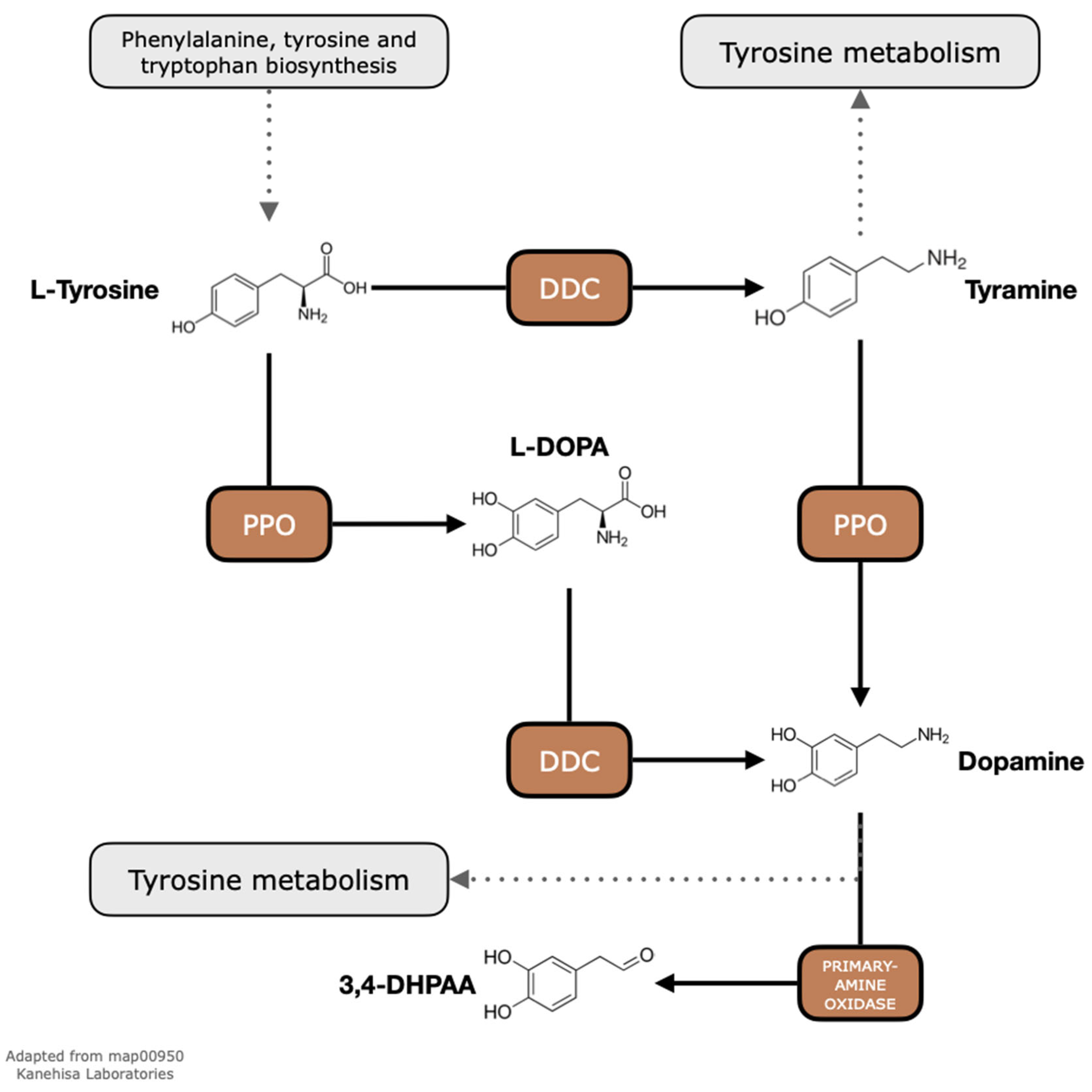
2.1. Exploratory Analyses of Metabolic Pathways Show That PPOs and DDCs Are Present in the Genomes of C. arabica and C. canephora
2.2. Chromatographic Analyses
3. Discussion
3.1. Multiple PPO Copies Are Present in C. arabica and C. canephora Genomes
3.2. Multiple PPO Copies Are Expressed in C. arabica Leaves
3.3. Multiple DDC Copies Are Present in C. arabica and C. canephora Genomes but Only One Copy Is Expressed in C. arabica Fully Expanded Leaves
3.4. Chromatography Analyses Confirmed the Presence of L-DOPA in Coffee Leaves and Fruits
4. Material and Methods
4.1. Identification of Enzyme-Coding Genes in Coffee Genome and Inference of Metabolic Pathways
4.2. Characterization of PPO- and DDC-Coding Genes in Coffea
4.3. Phylogenetic Analysis
4.4. Expression Evaluation of PPOs and DDCs
4.5. Extraction of L-DOPA from C. arabica Leaves
4.6. Liquid Chromatographic Analysis and Validation Parameters
4.7. LC–MS/MS for Qualitative Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- International Coffee Organization—Trade Statistics Tables. Available online: https://www.ico.org/trade_statistics.asp (accessed on 22 August 2022).
- Pham, Y.; Reardon-Smith, K.; Mushtaq, S.; Cockfield, G. The Impact of Climate Change and Variability on Coffee Production: A Systematic Review. Clim. Chang. 2019, 156, 609–630. [Google Scholar] [CrossRef]
- Lashermes, P.; Combes, M.-C.; Robert, J.; Trouslot, P.; D’Hont, A.; Anthony, F.; Charrier, A. Molecular Characterisation and Origin of the Coffea arabica L. Genome. Mol. Gen. Genet. MGG 1999, 261, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, B.; Bardil, A.; Baraille, H.; Dussert, S.; Doulbeau, S.; Dubois, E.; Severac, D.; Dereeper, A.; Etienne, H. The Greater Phenotypic Homeostasis of the Allopolyploid Coffea arabica Improved the Transcriptional Homeostasis over That of Both Diploid Parents. Plant Cell Physiol. 2015, 56, 2035–2051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, R.O.; Mondego, J.M.C.; Pot, D.; Ambrósio, A.B.; Andrade, A.C.; Pereira, L.F.P.; Colombo, C.A.; Vieira, L.G.E.; Carazzolle, M.F.; Pereira, G.A.G. A High-Throughput Data Mining of Single Nucleotide Polymorphisms in Coffea Species Expressed Sequence Tags Suggests Differential Homeologous Gene Expression in the Allotetraploid Coffea arabica. Plant Physiol. 2010, 154, 1053–1066. [Google Scholar] [CrossRef] [Green Version]
- Silva, F.L.F.; Nascimento, G.O.; Lopes, G.S.; Matos, W.O.; Cunha, R.L.; Malta, M.R.; Liska, G.R.; Owen, R.W.; Trevisan, M.T.S. The Concentration of Polyphenolic Compounds and Trace Elements in the Coffea arabica Leaves: Potential Chemometric Pattern Recognition of Coffee Leaf Rust Resistance. Food Res. Int. 2020, 134, 109221. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, Â.; Colomban, S.; Azinheira, H.G.; Guerra-Guimarães, L.; Do Céu Silva, M.; Navarini, L.; Resmini, M. Dietary Antioxidants in Coffee Leaves: Impact of Botanical Origin and Maturity on Chlorogenic Acids and Xanthones. Antioxidants 2019, 9, 6. [Google Scholar] [CrossRef] [Green Version]
- Ashihara, H.; Monteiro, A.M.; Gillies, F.M.; Crozier, A. Biosynthesis of Caffeine in Leaves of Coffee. Plant Physiol. 1996, 111, 747–753. [Google Scholar] [CrossRef] [Green Version]
- Campa, C.; Mondolot, L.; Rakotondravao, A.; Bidel, L.P.R.; Gargadennec, A.; Couturon, E.; La Fisca, P.; Rakotomalala, J.-J.; Jay-Allemand, C.; Davis, A.P. A Survey of Mangiferin and Hydroxycinnamic Acid Ester Accumulation in Coffee (Coffea) Leaves: Biological Implications and Uses. Ann. Bot. 2012, 110, 595–613. [Google Scholar] [CrossRef] [Green Version]
- de Almeida, R.F.; Trevisan, M.T.S.; Thomaziello, R.A.; Breuer, A.; Klika, K.D.; Ulrich, C.M.; Owen, R.W. Nutraceutical Compounds: Echinoids, Flavonoids, Xanthones and Caffeine Identified and Quantitated in the Leaves of Coffea arabica Trees from Three Regions of Brazil. Food Res. Int. 2019, 115, 493–503. [Google Scholar] [CrossRef]
- Molitor, C.; Mauracher, S.G.; Pargan, S.; Mayer, R.L.; Halbwirth, H.; Rompel, A. Latent and Active Aurone Synthase from Petals of C. Grandiflora: A Polyphenol Oxidase with Unique Characteristics. Planta 2015, 242, 519–537. [Google Scholar] [CrossRef] [Green Version]
- Mayer, A.M.; Harel, E. Polyphenol Oxidases in Plants. Phytochemistry 1979, 18, 193–215. [Google Scholar] [CrossRef]
- Goldfeder, M.; Kanteev, M.; Isaschar-Ovdat, S.; Adir, N.; Fishman, A. Determination of Tyrosinase Substrate-Binding Modes Reveals Mechanistic Differences between Type-3 Copper Proteins. Nat. Commun. 2014, 5, 4505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauracher, S.G.; Molitor, C.; Michael, C.; Kragl, M.; Rizzi, A.; Rompel, A. High Level Protein-Purification Allows the Unambiguous Polypeptide Determination of Latent Isoform PPO4 of Mushroom Tyrosinase. Phytochemistry 2014, 99, 14–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Yeon, Y.J.; Choi, Y.R.; Song, W.; Pack, S.P.; Choi, Y.S. A Cold-Adapted Tyrosinase with an Abnormally High Monophenolase/Diphenolase Activity Ratio Originating from the Marine Archaeon Candidatus Nitrosopumilus Koreensis. Biotechnol. Lett. 2016, 38, 1535–1542. [Google Scholar] [CrossRef]
- Mayer, A.M. Polyphenol Oxidases in Plants and Fungi: Going Places? A Review. Phytochemistry 2006, 67, 2318–2331. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.T.; Taylor, J.S.; Constabel, C.P. The Polyphenol Oxidase Gene Family in Land Plants: Lineage-Specific Duplication and Expansion. BMC Genom. 2012, 13, 395. [Google Scholar] [CrossRef] [Green Version]
- Lai, X.; Soler-Lopez, M.; Wichers, H.J.; Dijkstra, B.W. Large-scale recombinant expression and purification of human tyrosinase suitable for structural studies. PLoS ONE 2016, 11, e0161697. [Google Scholar] [CrossRef] [PubMed]
- Fernández, E.; Sanchez-Amat, A.; Solano, F. Location and Catalytic Characteristics of a Multipotent Bacterial Polyphenol Oxidase. Pigment Cell Res. 1999, 12, 331–339. [Google Scholar] [CrossRef]
- Solano, F. Melanins: Skin Pigments and Much More—Types, Structural Models, Biological Functions, and Formation Routes. New J. Sci. 2014, 2014, e498276. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Ferrer, Á.; Neptuno Rodríguez-López, J.; García-Cánovas, F.; García-Carmona, F. Tyrosinase: A Comprehensive Review of Its Mechanism. Biochim. Biophys. Acta BBA—Protein Struct. Mol. Enzymol. 1995, 1247, 1–11. [Google Scholar] [CrossRef]
- Körner, A.; Pawelek, J. Mammalian Tyrosinase Catalyzes Three Reactions in the Biosynthesis of Melanin. Science 1982, 217, 1163–1165. [Google Scholar] [CrossRef] [PubMed]
- Thipyapong, P.; Hunt, M.D.; Steffens, J.C. Antisense Downregulation of Polyphenol Oxidase Results in Enhanced Disease Susceptibility. Planta 2004, 220, 105–117. [Google Scholar] [CrossRef]
- Queiroz, C.; Mendes Lopes, M.L.; Fialho, E.; Valente-Mesquita, V.L. Polyphenol Oxidase: Characteristics and Mechanisms of Browning Control. Food Rev. Int. 2008, 24, 361–375. [Google Scholar] [CrossRef]
- Mayer, A.; Harel, E. Phenoloxidases and Their Significance in Fruit and Vegetables. Food Enzymol. 1991, 1, 373–398. [Google Scholar]
- Martinez, M.V.; Whitaker, J.R. The Biochemistry and Control of Enzymatic Browning. Trends Food Sci. Technol. 1995, 6, 195–200. [Google Scholar] [CrossRef]
- Li, L.; Steffens, J.C. Overexpression of Polyphenol Oxidase in Transgenic Tomato Plants Results in Enhanced Bacterial Disease Resistance. Planta 2002, 215, 239–247. [Google Scholar] [CrossRef]
- Li, Y.; Zafar, A.; Kilmartin, P.A.; Reynisson, J.; Leung, I.K.H. Development and Application of an NMR-Based Assay for Polyphenol Oxidases. ChemistrySelect 2017, 2, 10435–10441. [Google Scholar] [CrossRef]
- Kampatsikas, I.; Bijelic, A.; Rompel, A. Biochemical and Structural Characterization of Tomato Polyphenol Oxidases Provide Novel Insights into Their Substrate Specificity. Sci. Rep. 2019, 9, 4022. [Google Scholar] [CrossRef] [Green Version]
- Gasparetti, C.; Faccio, G.; Arvas, M.; Buchert, J.; Saloheimo, M.; Kruus, K. Discovery of a New Tyrosinase-like Enzyme Family Lacking a C-Terminally Processed Domain: Production and Characterization of an Aspergillus Oryzae Catechol Oxidase. Appl. Microbiol. Biotechnol. 2010, 86, 213–226. [Google Scholar] [CrossRef]
- Hakulinen, N.; Gasparetti, C.; Kaljunen, H.; Kruus, K.; Rouvinen, J. The Crystal Structure of an Extracellular Catechol Oxidase from the Ascomycete Fungus Aspergillus Oryzae. JBIC J. Biol. Inorg. Chem. 2013, 18, 917–929. [Google Scholar] [CrossRef]
- McLarin, M.-A.; Leung, I.K.H. Substrate Specificity of Polyphenol Oxidase. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 274–308. [Google Scholar] [CrossRef]
- Mazzafera, P.; Robinson, S.P. Characterization of Polyphenol Oxidase in Coffee. Phytochemistry 2000, 55, 285–296. [Google Scholar] [CrossRef]
- Mondolot, L.; La Fisca, P.; Buatois, B.; Talansier, E.; De Kochko, A.; Campa, C. Evolution in Caffeoylquinic Acid Content and Histolocalization During Coffea Canephora Leaf Development. Ann. Bot. 2006, 98, 33–40. [Google Scholar] [CrossRef]
- Ovallath, S.; Sulthana, B. Levodopa: History and Therapeutic Applications. Ann. Indian Acad. Neurol. 2017, 20, 185–189. [Google Scholar] [CrossRef]
- Guigoni, C.; Li, Q.; Aubert, I.; Dovero, S.; Bioulac, B.H.; Bloch, B.; Crossman, A.R.; Gross, C.E.; Bezard, E. Involvement of Sensorimotor, Limbic, and Associative Basal Ganglia Domains in L-3,4-Dihydroxyphenylalanine-Induced Dyskinesia. J. Neurosci. 2005, 25, 2102–2107. [Google Scholar] [CrossRef] [PubMed]
- Höglinger, G.U.; Rizk, P.; Muriel, M.P.; Duyckaerts, C.; Oertel, W.H.; Caille, I.; Hirsch, E.C. Dopamine Depletion Impairs Precursor Cell Proliferation in Parkinson Disease. Nat. Neurosci. 2004, 7, 726–735. [Google Scholar] [CrossRef]
- Hardebo, J.E.; Owman, C. Barrier Mechanisms for Neurotransmitter Monoamines and Their Precursors at the Blood-Brain Interface. Ann. Neurol. 1980, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, T.; Zhang, Z.; Yuan, X.; Chen, Q.; Zheng, J.; Chen, S.; Ma, F.; Li, C. Overexpression of the Tyrosine Decarboxylase Gene MdTyDC Confers Salt Tolerance in Apple. Environ. Exp. Bot. 2020, 180, 104244. [Google Scholar] [CrossRef]
- Li, C.; Sun, X.; Chang, C.; Jia, D.; Wei, Z.; Li, C.; Ma, F. Dopamine Alleviates Salt-Induced Stress in Malus Hupehensis. Physiol. Plant. 2015, 153, 584–602. [Google Scholar] [CrossRef]
- Liang, B.; Li, C.; Ma, C.; Wei, Z.; Wang, Q.; Huang, D.; Chen, Q.; Li, C.; Ma, F. Dopamine Alleviates Nutrient Deficiency-Induced Stress in Malus Hupehensis. Plant Physiol. Biochem. 2017, 119, 346–359. [Google Scholar] [CrossRef]
- Lan, G.; Jiao, C.; Wang, G.; Sun, Y.; Sun, Y. Effects of Dopamine on Growth, Carbon Metabolism, and Nitrogen Metabolism in Cucumber under Nitrate Stress. Sci. Hortic. 2020, 260, 108790. [Google Scholar] [CrossRef]
- Monente, C.; Ludwig, I.A.; Irigoyen, A.; De Peña, M.-P.; Cid, C. Assessment of Total (Free and Bound) Phenolic Compounds in Spent Coffee Extracts. J. Agric. Food Chem. 2015, 63, 4327–4334. [Google Scholar] [CrossRef]
- Farah, A.; Donangelo, C.M. Phenolic Compounds in Coffee. Braz. J. Plant Physiol. 2006, 18, 23–36. [Google Scholar] [CrossRef]
- Aoki, K.F.; Kanehisa, M. Using the KEGG Database Resource. Curr. Protoc. Bioinform. 2005, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Stevens, P.F. Angiosperm Phylogeny Website, Version 13; Angiosperm Phylogeny Website Version 13 2016. Available online: http://www.mobot.org/MOBOT/research/APweb/welcome.html (accessed on 4 September 2022).
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Snyder, L.R.; Kirkland, J.J.; Dolan, J.W. Introduction to Modern Liquid Chromatography; John Wiley & Sons: Hoboken, NJ, USA, 2011; ISBN 1-118-21039-5. [Google Scholar]
- Harris, D.C. Análise Química Quantitativa, 7th ed.; LTC: Rio Jan, Brazil, 2008. [Google Scholar]
- Elbarbry, F.; Nguyen, V.; Mirka, A.; Zwickey, H.; Rosenbaum, R. A New Validated HPLC Method for the Determination of Levodopa: Application to Study the Impact of Ketogenic Diet on the Pharmacokinetics of Levodopa in Parkinson’s Participants. Biomed. Chromatogr. 2019, 33, e4382. [Google Scholar] [CrossRef] [Green Version]
- César, I.C.; Byrro, R.M.D.; de Santana e Silva Cardoso, F.F.; Mundim, I.M.; de Souza Teixeira, L.; Gomes, S.A.; Bonfim, R.R.; Pianetti, G.A. Development and Validation of a High-Performance Liquid Chromatography–Electrospray Ionization–MS/MS Method for the Simultaneous Quantitation of Levodopa and Carbidopa in Human Plasma. J. Mass Spectrom. 2011, 46, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Chi, J.; Ling, Y.; Jenkins, R.; Li, F. Quantitation of Levodopa and Carbidopa in Rat Plasma by LC–MS/MS: The Key Role of Ion-Pairing Reversed-Phase Chromatography. J. Chromatogr. B 2017, 1054, 1–9. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, F.; Deng, L.; Chen, C.; Cheng, Z.; Huang, J.; Liu, J.; Jiang, H. Development and Validation of an LC-MS/MS Method for Simultaneous Quantification of Levodopa and MD01 in Rat Plasma and Its Application to a Pharmacokinetic Study of Mucuna pruriens Extract. Biomed. Chromatogr. 2016, 30, 1506–1514. [Google Scholar] [CrossRef]
- Patay, É.B.; Bencsik, T.; Papp, N. Phytochemical Overview and Medicinal Importance of Coffea Species from the Past until Now. Asian Pac. J. Trop. Med. 2016, 9, 1127–1135. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-M.; Ma, Z.; Kitts, D.D. Effects of Processing Method and Age of Leaves on Phytochemical Profiles and Bioactivity of Coffee Leaves. Food Chem. 2018, 249, 143–153. [Google Scholar] [CrossRef]
- Kampatsikas, I.; Bijelic, A.; Pretzler, M.; Rompel, A. A Peptide-Induced Self-Cleavage Reaction Initiates the Activation of Tyrosinase. Angew. Chem. Int. Ed. 2019, 58, 7475–7479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, L.T.; Constabel, C.P. The Polyphenol Oxidase Gene Family in Poplar: Phylogeny, Differential Expression and Identification of a Novel, Vacuolar Isoform. Planta 2011, 234, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Ono, E.; Hatayama, M.; Isono, Y.; Sato, T.; Watanabe, R.; Yonekura-Sakakibara, K.; Fukuchi-Mizutani, M.; Tanaka, Y.; Kusumi, T.; Nishino, T. Localization of a Flavonoid Biosynthetic Polyphenol Oxidase in Vacuoles. Plant J. 2006, 45, 133–143. [Google Scholar] [CrossRef]
- Virador, V.M.; Reyes Grajeda, J.P.; Blanco-Labra, A.; Mendiola-Olaya, E.; Smith, G.M.; Moreno, A.; Whitaker, J.R. Cloning, Sequencing, Purification, and Crystal Structure of Grenache (Vitis Vinifera) Polyphenol Oxidase. J. Agric. Food Chem. 2010, 58, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Kanteev, M.; Goldfeder, M.; Fishman, A. Structure-Function Correlations in Tyrosinases. Protein Sci. Publ. Protein Soc. 2015, 24, 1360–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dereeper, A.; Bocs, S.; Rouard, M.; Guignon, V.; Ravel, S.; Tranchant-Dubreuil, C.; Poncet, V.; Garsmeur, O.; Lashermes, P.; Droc, G. The Coffee Genome Hub: A Resource for Coffee Genomes. Nucleic Acids Res. 2015, 43, D1028–D1035. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Leebens-Mack, J.; Ayyampalayam, S.; Bowers, J.E.; McKain, M.R.; McNeal, J.; Rolf, M.; Ruzicka, D.R.; Wafula, E.; Wickett, N.J. A Genome Triplication Associated with Early Diversification of the Core Eudicots. Genome Biol. 2012, 13, R3. [Google Scholar] [CrossRef] [Green Version]
- Freeling, M.; Thomas, B.C. Gene-Balanced Duplications, like Tetraploidy, Provide Predictable Drive to Increase Morphological Complexity. Genome Res. 2006, 16, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Pavón-Pérez, J.; Oviedo, C.A.; Elso-Freudenberg, M.; Henríquez-Aedo, K.; Aranda, M. Lc-ms/ms method for l-dopa quantification in different tissues of vicia faba. J. Chil. Chem. Soc. 2019, 64, 4651–4653. [Google Scholar] [CrossRef]
- Ribani, M.; Bottoli, C.B.G.; Collins, C.H.; Jardim, I.C.S.F.; Melo, L.F.C. Validação Em Métodos Cromatográficos e Eletroforéticos. Quím. Nova 2004, 27, 771–780. [Google Scholar] [CrossRef] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Johns Hopkins University Coffea arabica V. Caturra Genome 2018. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_003713225.1 (accessed on 2 June 2022).
- Hoff, K.J.; Stanke, M. Predicting Genes in Single Genomes with AUGUSTUS. Curr. Protoc. Bioinform. 2019, 65, e57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Klabunde, T.; Eicken, C.; Sacchettini, J.C.; Krebs, B. Crystal Structure of a Plant Catechol Oxidase Containing a Dicopper Center. Nat. Struct. Biol. 1998, 5, 1084–1090. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Recent Developments in the MAFFT Multiple Sequence Alignment Program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package), Version 3.5 c. 1993. Available online: http://www.dbbm.fiocruz.br/molbiol/main.html (accessed on 2 June 2022).
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Sperschneider, J.; Catanzariti, A.-M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular Localization Prediction of Both Plant and Effector Proteins in the Plant Cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardon, C.H.; de Oliveira, R.R.; Lesy, V.; Ribeiro, T.H.C.; Fust, C.; Pereira, L.P.; Colasanti, J.; Chalfun-Junior, A. Expression of Coffee Florigen CaFT1 Reveals a Sustained Floral Induction Window Associated with Asynchronous Flowering in Tropical Perennials. Plant Sci. 2022, 325, 111479. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; McCarthy, D.; Ritchie, M.; Robinson, M.; Smyth, G.; Hall, E. EdgeR: Differential Analysis of Sequence Read Count Data User’s Guide; R Package; 2020; pp. 1–121. Available online: http://52.71.54.154/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf (accessed on 8 March 2023).
- Warnes, M.G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A. “Package ‘gplots’”. Various R Programming Tools for Plotting Data: 2016. Available online: https://www.researchgate.net/profile/Arni-Magnusson-2/publication/303186599_gplots_Various_R_programming_tools_for_plotting_data/data/5a8fd3b80f7e9ba4296a11a1/gplots.pdf (accessed on 5 December 2022).
- Polanowska, K.; Łukasik, R.; Kuligowski, M.; Nowak, J. Development of a Sustainable, Simple, and Robust Method for Efficient l-DOPA Extraction. Molecules 2019, 24, 2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Parameter | L-DOPA |
|---|---|
| B (linear coefficient) | 1694.5 |
| A (angular coefficient) | 177.4 |
| R2 | 0.99998 |
| DL (µg·mL−1) | 0.81 |
| QL (µg·mL−1) | 2.73 |
| Recovery (%) | 81 to 104 |
| CV (%) | 0.38 to 1.11 |
| Species | Tissue | Sample ID | Counts | Signal Above Noise Level |
|---|---|---|---|---|
| Coffea arabica | Leaves | 1 | 48 | Yes |
| 2 | 48 | Yes | ||
| 3 | 48 | Yes | ||
| Flowers | 4 | 47 | No | |
| 5 | 47 | No | ||
| 6 | 47 | No | ||
| Fruits | 7 | 48.5 | Yes | |
| 8 | 49 | Yes | ||
| 9 | 49.5 | Yes | ||
| Coffea canephora | Leaves | 10 | 48.5 | Yes |
| 11 | 48 | Yes | ||
| 12 | 49 | Yes | ||
| Flowers | 13 | 48 | No | |
| 14 | 48 | No | ||
| 15 | 48 | No | ||
| Fruits | 16 | 47 | No | |
| 17 | 48 | No | ||
| 18 | 48 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cherubino Ribeiro, T.H.; de Oliveira, R.R.; das Neves, T.T.; Santiago, W.D.; Mansur, B.L.; Saczk, A.A.; Vilela de Resende, M.L.; Chalfun-Junior, A. Metabolic Pathway Reconstruction Indicates the Presence of Important Medicinal Compounds in Coffea Such as L-DOPA. Int. J. Mol. Sci. 2023, 24, 12466. https://doi.org/10.3390/ijms241512466
Cherubino Ribeiro TH, de Oliveira RR, das Neves TT, Santiago WD, Mansur BL, Saczk AA, Vilela de Resende ML, Chalfun-Junior A. Metabolic Pathway Reconstruction Indicates the Presence of Important Medicinal Compounds in Coffea Such as L-DOPA. International Journal of Molecular Sciences. 2023; 24(15):12466. https://doi.org/10.3390/ijms241512466
Chicago/Turabian StyleCherubino Ribeiro, Thales Henrique, Raphael Ricon de Oliveira, Taís Teixeira das Neves, Wilder Douglas Santiago, Bethania Leite Mansur, Adelir Aparecida Saczk, Mario Lucio Vilela de Resende, and Antonio Chalfun-Junior. 2023. "Metabolic Pathway Reconstruction Indicates the Presence of Important Medicinal Compounds in Coffea Such as L-DOPA" International Journal of Molecular Sciences 24, no. 15: 12466. https://doi.org/10.3390/ijms241512466
APA StyleCherubino Ribeiro, T. H., de Oliveira, R. R., das Neves, T. T., Santiago, W. D., Mansur, B. L., Saczk, A. A., Vilela de Resende, M. L., & Chalfun-Junior, A. (2023). Metabolic Pathway Reconstruction Indicates the Presence of Important Medicinal Compounds in Coffea Such as L-DOPA. International Journal of Molecular Sciences, 24(15), 12466. https://doi.org/10.3390/ijms241512466







