Role of microRNAs in Chronic Lymphocytic Leukemia
Abstract
1. Introduction
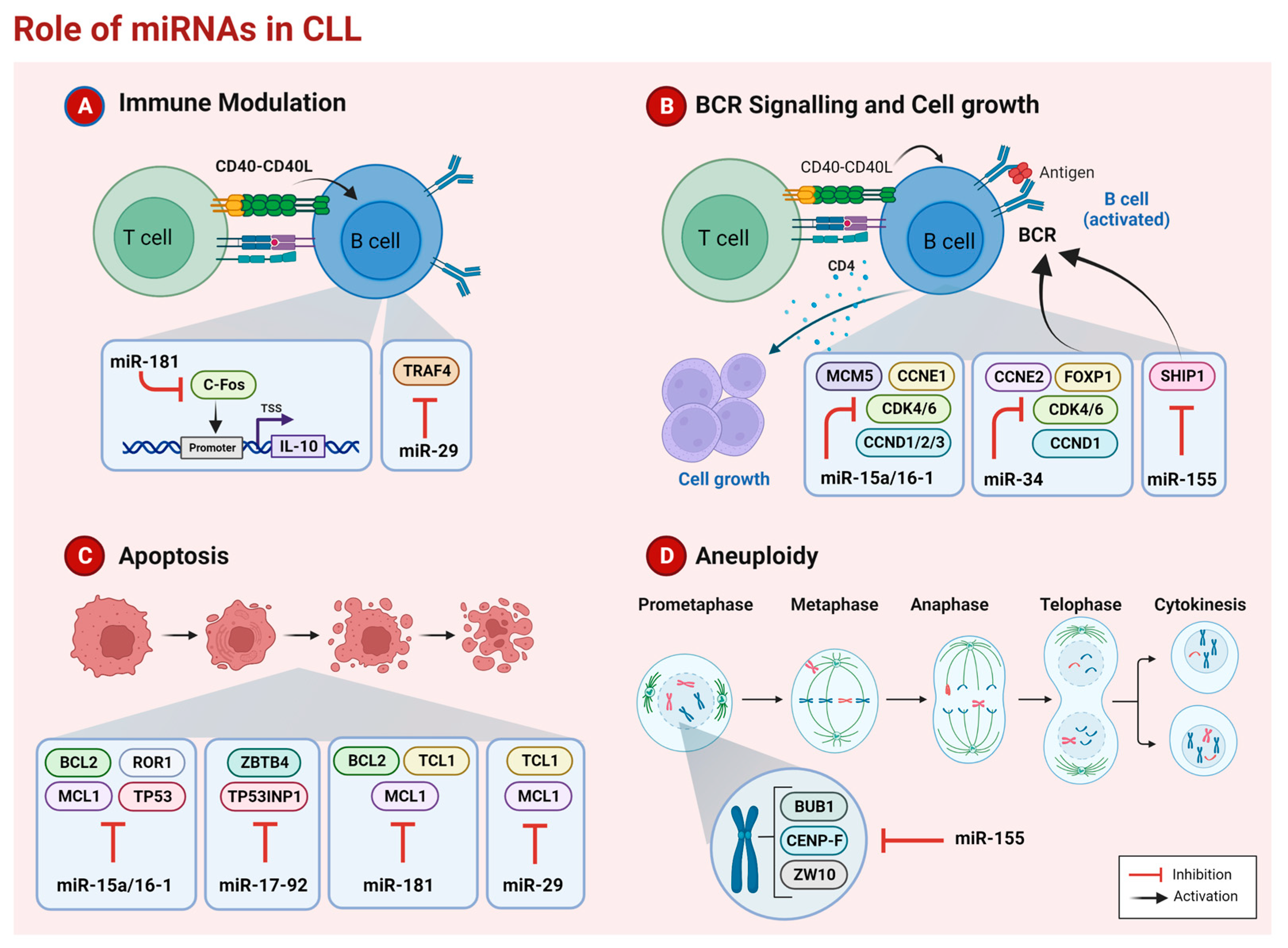
2. microRNAs in CLL
2.1. miR-15a/16-1
Therapeutic Strategies
2.2. miR-29 Family
Therapeutic Strategies
2.3. miR-34 Family
Therapeutic Strategies
2.4. miR-17-92 Cluster
Therapeutic Strategies
2.5. miR-155
Therapeutic Strategies
2.6. miR-181 Family
Therapeutic Strategies
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghia, P.; Ferreri, A.M.; Galigaris-Cappio, F. Chronic Lymphocytic Leukemia. Crit. Rev. Oncol. Hematol. 2007, 64, 234–246. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.J.; Montserrat, E.; Rai, K.R.; et al. Guidelines for the Diagnosis and Treatment of Chronic Lymphocytic Leukemia: A Report from the International Workshop on Chronic Lymphocytic Leukemia Updating the National Cancer Institute-Working Group 1996 Guidelines. Blood 2008, 111, 5446–5456. [Google Scholar] [CrossRef]
- Zenz, T.; Mertens, D.; Küppers, R.; Döhner, H.; Stilgenbauer, S. From Pathogenesis to Treatment of Chronic Lymphocytic Leukaemia. Nat. Rev. Cancer 2010, 10, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Eichhorst, B.; Dreyling, M.; Robak, T.; Montserrat, E.; Hallek, M. Chronic Lymphocytic Leukemia: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2011, 22, vi50–vi54. [Google Scholar] [CrossRef] [PubMed]
- Till, K.J.; Pettitt, A.R.; Slupsky, J.R. Expression of Functional Sphingosine-1 Phosphate Receptor-1 Is Reduced by B Cell Receptor Signaling and Increased by Inhibition of PI3 Kinase δ but Not SYK or BTK in Chronic Lymphocytic Leukemia Cells. J. Immunol. 2015, 194, 2439–2446. [Google Scholar] [CrossRef] [PubMed]
- Herman, S.E.M.; Mustafa, R.Z.; Gyamfi, J.A.; Pittaluga, S.; Chang, S.; Chang, B.; Farooqui, M.; Wiestner, A. Ibrutinib Inhibits BCR and NF-ΚB Signaling and Reduces Tumor Proliferation in Tissue-Resident Cells of Patients with CLL. Blood 2014, 123, 3286–3295. [Google Scholar] [CrossRef]
- Byrd, J.C.; Brown, J.R.; O’Brien, S.; Barrientos, J.C.; Kay, N.E.; Reddy, N.M.; Coutre, S.; Tam, C.S.; Mulligan, S.P.; Jaeger, U.; et al. Ibrutinib versus Ofatumumab in Previously Treated Chronic Lymphoid Leukemia. N. Engl. J. Med. 2014, 371. [Google Scholar] [CrossRef]
- O′Brien, S.; Furman, R.R.; Coutre, S.E.; Sharman, J.P.; Burger, J.A.; Blum, K.A.; Grant, B.; Richards, D.A.; Coleman, M.; Wierda, W.G.; et al. Ibrutinib as Initial Therapy for Elderly Patients with Chronic Lymphocytic Leukaemia or Small Lymphocytic Lymphoma: An Open-Label, Multicentre, Phase 1b/2 Trial. Lancet Oncol. 2014, 15, 48–58. [Google Scholar] [CrossRef]
- Byrd, J.C.; Jones, J.J.; Woyach, J.A.; Johnson, A.J.; Flynn, J.M. Entering the Era of Targeted Therapy for Chronic Lymphocytic Leukemia: Impact on the Practicing Clinician. J. Clin. Oncol. 2014, 32, 3039–3047. [Google Scholar] [CrossRef][Green Version]
- Furman, R.R.; Sharman, J.P.; Coutre, S.E.; Cheson, B.D.; Pagel, J.M.; Hillmen, P.; Barrientos, J.C.; Zelenetz, A.D.; Kipps, T.J.; Flinn, I.; et al. Idelalisib and Rituximab in Relapsed Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2014, 370. [Google Scholar] [CrossRef]
- Brown, J.R.; Byrd, J.C.; Coutre, S.E.; Benson, D.M.; Flinn, I.W.; Wagner-Johnston, N.D.; Spurgeon, S.E.; Kahl, B.S.; Bello, C.; Webb, H.K.; et al. Idelalisib, an Inhibitor of Phosphatidylinositol 3-Kinase P110δ, for Relapsed/Refractory Chronic Lymphocytic Leukemia. Blood 2014, 123, 3390–3397. [Google Scholar] [CrossRef]
- Thompson, P.A.; Burger, J.A. Bruton’s Tyrosine Kinase Inhibitors: First and Second Generation Agents for Patients with Chronic Lymphocytic Leukemia (CLL). Expert Opin. Investig. Drugs 2018, 27, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Al-Sawaf, O.; Bahlo, J.; Fink, A.-M.; Tandon, M.; Dixon, M.; Robrecht, S.; Warburton, S.; Humphrey, K.; Samoylova, O.; et al. Venetoclax and Obinutuzumab in Patients with CLL and Coexisting Conditions. N. Engl. J. Med. 2019, 380, 2225–2236. [Google Scholar] [CrossRef] [PubMed]
- Kater, A.P.; Wu, J.Q.; Kipps, T.; Eichhorst, B.; Hillmen, P.; D’Rozario, J.; Assouline, S.; Owen, C.; Robak, T.; de la Serna, J.; et al. Venetoclax plus Rituximab in Relapsed Chronic Lymphocytic Leukemia: 4-Year Results and Evaluation of Impact of Genomic Complexity and Gene Mutations from the MURANO Phase III Study. J. Clin. Oncol. 2020, 38, 4042. [Google Scholar] [CrossRef]
- Byrd, J.C.; Hillmen, P.; Ghia, P.; Kater, A.P.; Chanan-Khan, A.; Furman, R.R.; O’Brien, S.; Yenerel, M.N.; Illés, A.; Kay, N.; et al. Acalabrutinib Versus Ibrutinib in Previously Treated Chronic Lymphocytic Leukemia: Results of the First Randomized Phase III Trial. J. Clin. Oncol. 2021, 39, 3441–3452. [Google Scholar] [CrossRef]
- Byrd, J.C.; Woyach, J.A.; Furman, R.R.; Martin, P.; O’Brien, S.; Brown, J.R.; Stephens, D.M.; Barrientos, J.C.; Devereux, S.; Hillmen, P.; et al. Acalabrutinib in Treatment-Naive Chronic Lymphocytic Leukemia. Blood 2021, 137, 3327–3338. [Google Scholar] [CrossRef] [PubMed]
- Byrd, J.C.; Furman, R.R.; Coutre, S.E.; Flinn, I.W.; Burger, J.A.; Blum, K.A.; Grant, B.; Sharman, J.P.; Coleman, M.; Wierda, W.G.; et al. Targeting BTK with Ibrutinib in Relapsed Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2013, 369, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Byrd, J.C.; O’Brien, S.; James, D.F. Ibrutinib in Relapsed Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2013, 369, 1277–1279. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent Deletions and Down-Regulation of Micro-RNA Genes MiR15 and MiR16 at 13q14 in Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Balatti, V.; Pekarky, Y.; Rizzotto, L.; Croce, C.M. MiR Deregulation in CLL. Adv. Exp. Med. Biol. 2013, 792, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Zagatti, B.; Rizzotto, L.; Cavazzini, F.; Veronese, A.; Ciccone, M.; Saccenti, E.; Lupini, L.; Grilli, A.; De Angeli, C.; et al. MicroRNAs Involvement in Fludarabine Refractory Chronic Lymphocytic Leukemia. Mol. Cancer 2010, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.G.; Sevignani, C.; Ferracin, M.; Felli, N.; Dumitru, C.D.; Shimizu, M.; Cimmino, A.; Zupo, S.; Dono, M.; et al. MicroRNA Profiling Reveals Distinct Signatures in B Cell Chronic Lymphocytic Leukemias. Proc. Natl. Acad. Sci. USA 2004, 101, 11755–11760. [Google Scholar] [CrossRef]
- Döhner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Kröber, A.; Bullinger, L.; Döhner, K.; Bentz, M.; Lichter, P. Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef] [PubMed]
- Ouillette, P.; Erba, H.; Kujawski, L.; Kaminski, M.; Shedden, K.; Malek, S.N. Integrated Genomic Profiling of Chronic Lymphocytic Leukemia Identifies Subtypes of Deletion 13q14. Cancer Res. 2008, 68, 1012–1021. [Google Scholar] [CrossRef]
- Pepe, F.; Rassenti, L.Z.; Pekarsky, Y.; Labanowska, J.; Nakamura, T.; Nigita, G. A Large Fraction of Trisomy 12, 17p2,and 11q2 CLL Cases Carry Unidenti Fi Ed Microdeletions of MiR-15a/16-1. Proc. Natl. Acad. Sci. USA 2022, 119, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Raveche, E.S.; Salerno, E.; Scaglione, B.J.; Manohar, V.; Abbasi, F.; Lin, Y.C.; Fredrickson, T.; Landgraf, P.; Ramachandra, S.; Huppi, K.; et al. Abnormal MicroRNA-16 Locus with Synteny to Human 13q14 Linked to CLL in NZB Mice. Blood 2007, 109, 5079–5086. [Google Scholar] [CrossRef]
- Kasar, S.; Underbayev, C.; Hassan, M.; Ilev, I.; Degheidy, H.; Bauer, S.; Marti, G.; Lutz, C.; Raveche, E.; Batish, M. Alterations in the MiR-15a/16-1 Loci Impairs Its Processing and Augments B-1 Expansion in De Novo Mouse Model of Chronic Lymphocytic Leukemia (CLL). PLoS ONE 2016, 11, e0149331. [Google Scholar] [CrossRef]
- Veronese, A.; Pepe, F.; Chiacchia, J.; Pagotto, S.; Lanuti, P.; Veschi, S.; Di Marco, M.; D’Argenio, A.; Innocenti, I.; Vannata, B.; et al. Allele-Specific Loss and Transcription of the MiR-15a/16-1 Cluster in Chronic Lymphocytic Leukemia. Leukemia 2015, 29, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Lerner, M.; Harada, M.; Lovén, J.; Castro, J.; Davis, Z.; Oscier, D.; Henriksson, M.; Sangfelt, O.; Grandér, D.; Corcoran, M.M. DLEU2, Frequently Deleted in Malignancy, Functions as a Critical Host Gene of the Cell Cycle Inhibitory MicroRNAs MiR-15a and MiR-16-1. Exp. Cell Res. 2009, 315, 2941–2952. [Google Scholar] [CrossRef] [PubMed]
- Kasar, S.; Underbayev, C.; Yuan, Y.; Hanlon, M.; Aly, S.; Khan, H.; Chang, V.; Batish, M.; Gavrilova, T.; Badiane, F.; et al. Therapeutic Implications of Activation of the Host Gene (Dleu2) Promoter for MiR-15a/16-1 in Chronic Lymphocytic Leukemia. Oncogene 2014, 33, 3307–3315. [Google Scholar] [CrossRef] [PubMed]
- Sampath, D.; Liu, C.; Vasan, K.; Sulda, M.; Puduvalli, V.K.; Wierda, W.G.; Keating, M.J. Histone Deacetylases Mediate the Silencing of MiR-15a, MiR-16, and MiR-29b in Chronic Lymphocytic Leukemia. Blood 2012, 119, 1162–1172. [Google Scholar] [CrossRef] [PubMed]
- Allegra, D.; Bilan, V.; Garding, A.; Döhner, H.; Stilgenbauer, S.; Kuchenbauer, F.; Mertens, D. Defective DROSHA Processing Contributes to Downregulation of MiR-15/-16 in Chronic Lymphocytic Leukemia. Leukemia 2014, 28, 98–107. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. MiR-15 and MiR-16 Induce Apoptosis by Targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Calin, G.A.; Cimmino, A.; Fabbri, M.; Ferracin, M.; Wojcik, S.E.; Shimizu, M.; Taccioli, C.; Zanesi, N.; Garzon, R.; Aqeilan, R.I.; et al. MiR-15a and MiR-16-1 Cluster Functions in Human Leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 5166–5171. [Google Scholar] [CrossRef]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G.; et al. The DLEU2/MiR-15a/16-1 Cluster Controls B Cell Proliferation and Its Deletion Leads to Chronic Lymphocytic Leukemia. Cancer Cell 2010, 17, 28–40. [Google Scholar] [CrossRef]
- Rassenti, L.Z.; Balatti, V.; Ghia, E.M.; Palamarchuk, A.; Tomasello, L.; Fadda, P.; Pekarsky, Y.; Widhopf, G.F.; Kipps, T.J.; Croce, C.M. MicroRNA Dysregulation to Identify Therapeutic Target Combinations for Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. USA 2017, 114, 10731–10736. [Google Scholar] [CrossRef]
- Lin, K.; Farahani, M.; Yang, Y.; Johnson, G.G.; Oates, M.; Atherton, M.; Douglas, A.; Kalakonda, N.; Pettitt, A.R. Loss of MIR15A and MIR16-1 at 13q14 Is Associated with Increased TP53 MRNA De-repression of BCL2 and adverse outcome in chronic lymphocytic leukaemia. Br. J. Haematol. 2014, 167, 346–355. [Google Scholar] [CrossRef]
- Fabbri, M.; Bottoni, A.; Ph, D.; Shimizu, M.; Ph, D.; Nicoloso, M.S.; Rossi, S.; Ph, D.; Barbarotto, E.; Ph, D.; et al. Association of a MicroRNA/TP53 Feedback Circuitry With Pathogenesis and Outcome of B-Cell Chronic Lymphocytic Leukemia. JAMA 2011, 305, 59–67. [Google Scholar] [CrossRef]
- Cutrona, G.; Matis, S.; Colombo, M.; Massucco, C.; Baio, G.; Valdora, F.; Emionite, L.; Fabris, S.; Recchia, A.G.; Gentile, M.; et al. Effects of MiRNA-15 and MiRNA-16 Expression Replacement in Chronic Lymphocytic Leukemia: Implication for Therapy. Leukemia 2017, 31, 1894–1904. [Google Scholar] [CrossRef] [PubMed]
- Kasar, S.; Salerno, E.; Yuan, Y.; Underbayev, C.; Vollenweider, D.; Laurindo, M.F.; Fernandes, H.; Bonci, D.; Addario, A.; Mazzella, F.; et al. Systemic in Vivo Lentiviral Delivery of MiR-15a/16 Reduces Malignancy in the NZB de Novo Mouse Model of Chronic Lymphocytic Leukemia. Genes Immun. 2012, 13, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Kollinerova, S.; Vassanelli, S.; Modriansky, M. The Role of miR-29 Family Members in Malignant Hematopoiesis. Biomed. Pap. 2014, 158, 489–501. [Google Scholar] [CrossRef]
- Horita, M.; Farquharson, C.; Stephen, L.A. The Role of miR-29 Family in Disease. J. Cell. Biochem. 2021, 122, 696–715. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread MicroRNA Repression by Myc Contributes to Tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Santanam, U.; Cimmino, A.; Palamarchuk, A.; Efanov, A.; Maximov, V.; Volinia, S.; Alder, H.; Liu, C.G.; Rassenti, L.; et al. Tcl1 Expression in Chronic Lymphocytic Leukemia Is Regulated by miR-29 and miR-181. Cancer Res. 2006, 66, 11590–11593. [Google Scholar] [CrossRef] [PubMed]
- Herling, M.; Patel, K.A.; Khalili, J.; Schlette, E.; Kobayashi, R.; Medeiros, L.J.; Jones, D. TCL1 Shows a Regulated Expression Pattern in Chronic Lymphocytic Leukemia That Correlates with Molecular Subtypes and Proliferative State. Leukemia 2006, 20, 280–285. [Google Scholar] [CrossRef]
- Santanam, U.; Zanesi, N.; Efanov, A.; Costinean, S.; Palamarchuk, A.; Hagan, J.P.; Volinia, S.; Alder, H.; Rassenti, L.; Kipps, T.; et al. Chronic Lymphocytic Leukemia Modeled in Mouse by Targeted miR-29 Expression. Proc. Natl. Acad. Sci. USA 2010, 107, 12210–12215. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Croce, C.M. Is miR-29 an Oncogene or Tumor Suppressor in CLL? Oncotarget 2010, 1, 224–227. [Google Scholar] [CrossRef]
- Mott, J.L.; Kobayashi, S.; Bronk, S.F.; Gores, G.J. MiR-29 Regulates Mcl-1 Protein Expression and Apoptosis. Oncogene 2007, 26, 6133–6140. [Google Scholar] [CrossRef]
- Sharma, S.; Pavlasova, G.M.; Seda, V.; Cerna, K.A.; Vojackova, E.; Filip, D.; Ondrisova, L.; Sandova, V.; Kostalova, L.; Zeni, P.F.; et al. MiR-29 Modulates CD40 Signaling in Chronic Lymphocytic Leukemia by Targeting TRAF4: An Axis Affected by BCR Inhibitors. Blood 2021, 137, 2481–2494. [Google Scholar] [CrossRef] [PubMed]
- Pascutti, M.F.; Jak, M.; Tromp, J.M.; Derks, I.A.M.; Remmerswaal, E.B.M.; Thijssen, R.; Van Attekum, M.H.A.; Van Bochove, G.G.; Luijks, D.M.; Pals, S.T.; et al. IL-21 and CD40L Signals from Autologous T Cells Can Induce Antigen-Independent Proliferation of CLL Cells. Blood 2013, 122, 3010–3019. [Google Scholar] [CrossRef] [PubMed]
- Baskar, S.; Kwong, K.Y.; Hofer, T.; Levy, J.M.; Kennedy, M.G.; Lee, E.; Staudt, L.M.; Wilson, W.H.; Wiestner, A.; Rader, C. Unique Cell Surface Expression of Receptor Tyrosine Kinase ROR1 in Human B-Cell Chronic Lymphocytic Leukemia. Clin. Cancer Res. 2008, 14, 396–404. [Google Scholar] [CrossRef]
- Chiang, C.L.; Goswami, S.; Frissora, F.W.; Xie, Z.; Yan, P.S.; Bundschuh, R.; Walker, L.A.; Huang, X.; Mani, R.; Mo, X.M.; et al. ROR1-Targeted Delivery of MiR-29b Induces Cell Cycle Arrest and Therapeutic Benefit in Vivo in a CLL Mouse Model. Blood 2019, 134, 432–444. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human MicroRNA Genes Are Frequently Located at Fragile Sites and Genomic Regions Involved in Cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. Chronic Lymphocytic Leukemia: Interplay between Noncoding RNAs and Protein-Coding Genes. Blood 2009, 114, 4761–4770. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lim, L.P.; De Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A MicroRNA Component of the P53 Tumour Suppressor Network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef]
- Malek, S.; Ouillette, P. Invariant Deletion of One miR-34b/c Locus in CLL with Del11q. Blood 2008, 112, 2074. [Google Scholar] [CrossRef]
- Asslaber, D.; Piñón, J.D.; Seyfried, I.; Desch, P.; Stöcher, M.; Tinhofer, I.; Egle, A.; Merkel, O.; Greil, R. MicroRNA-34a Expression Correlates with MDM2 SNP309 Polymorphism and Treatment-Free Survival in Chronic Lymphocytic Leukemia. Blood 2010, 115, 4191–4197. [Google Scholar] [CrossRef]
- Zenz, T.; Mohr, J.; Eldering, E.; Kater, A.P.; Bühler, A.; Kienle, D.; Winkler, D.; Dürig, J.; Van Oers, M.H.J.; Mertens, D.; et al. MiR-34a as Part of the Resistance Network in Chronic Lymphocytic Leukemia. Blood 2009, 113, 3801–3808. [Google Scholar] [CrossRef]
- Zenz, T.; Häbe, S.; Denzel, T.; Mohr, J.; Winkler, D.; Bühler, A.; Sarno, A.; Groner, S.; Mertens, D.; Busch, R.; et al. Detailed Analysis of P53 Pathway Defects in Fludarabine-Refractory Chronic Lymphocytic Leukemia (CLL): Dissecting the Contribution of 17p Deletion, TP53 Mutation, P53-P21 Dysfunction, and MiR34a in a Prospective Clinical Trial. Blood 2009, 114, 2589–2597. [Google Scholar] [CrossRef]
- Cerna, K.; Oppelt, J.; Chochola, V.; Musilova, K.; Seda, V.; Pavlasova, G.; Radova, L.; Arigoni, M.; Calogero, R.A.; Benes, V.; et al. MicroRNA miR-34a Downregulates FOXP1 during DNA Damage Response to Limit BCR Signalling in Chronic Lymphocytic Leukaemia B Cells. Leukemia 2019, 33, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Balatti, V.; Tomasello, L.; Rassenti, L.Z.; Veneziano, D.; Nigita, G.; Wang, H.Y.; Thorson, J.A.; Kipps, T.J.; Pekarsky, Y.; Croce, C.M. MiR-125a and miR-34a Expression Predicts Richter Syndrome in Chronic Lymphocytic Leukemia Patients. Blood 2018, 132, 2179–2182. [Google Scholar] [CrossRef]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the P53 Network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lowe, S.W.; Hannon, G.J. MicroRNAs Join the P53 Network—Another Piece in the Tumour-Suppression Puzzle. Nat. Rev. Cancer 2007, 7, 819–822. [Google Scholar] [CrossRef]
- Zenz, T.; Eichhorst, B.; Busch, R.; Denzel, T.; Häbe, S.; Winkler, D.; Bühler, A.; Edelmann, J.; Bergmann, M.; Hopfinger, G.; et al. TP53 Mutation and Survival in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2010, 28, 4473–4479. [Google Scholar] [CrossRef]
- Campo, E.; Cymbalista, F.; Ghia, P.; Jäger, U.; Pospisilova, S.; Rosenquist, R.; Schuh, A.; Stilgenbauer, S. TP53 Aberrations in Chronic Lymphocytic Leukemia: An Overview of the Clinical Implications of Improved Diagnostics. Haematologica 2018, 103. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, M.K.; van Lom, K.; Tielemans, D.; Elstrodt, F.; Langerak, A.W.; Veer, M.B.v.; Jongen-Lavrencic, M. 17p13/TP53 Deletion in B-CLL Patients Is Associated with MicroRNA-34a Downregulation. Leukemia 2009, 23, 1956–1968. [Google Scholar] [CrossRef]
- Mraz, M.; Malinova, K.; Kotaskova, J.; Pavlova, S.; Tichy, B.; Malcikova, J.; Kozubik, K.S.; Smardova, J.; Brychtova, Y.; Doubek, M.; et al. MiR-34a, MiR-29c and MiR-17-5p Are Downregulated in CLL Patients with TP53 Abnormalities. Leukemia 2009, 23, 1159–1163. [Google Scholar] [CrossRef]
- Nag, S.; Qin, J.; Srivenugopal, K.S.; Wang, M.; Zhang, R. The MDM2-P53 Pathway Revisited. J. Biomed. Res. 2013, 27, 254. [Google Scholar]
- Bond, G.L.; Hu, W.; Bond, E.E.; Robins, H.; Lutzker, S.G.; Arva, N.C.; Bargonetti, J.; Bartel, F.; Taubert, H.; Wuerl, P.; et al. A Single Nucleotide Polymorphism in the MDM2 Promoter Attenuates the P53 Tumor Suppressor Pathway and Accelerates Tumor Formation in Humans. Cell 2004, 119, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Gryshchenko, I.; Hofbauer, S.; Stoecher, M.; Daniel, P.T.; Steurer, M.; Gaiger, A.; Eigenberger, K.; Greil, R.; Tinhofer, I. MDM2 SNP309 Is Associated with Poor Outcome in B-Cell Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2008, 26, 2257. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Q.; Kwong, Y.L.; Wong, K.F.; Kho, C.S.B.; Jin, D.Y.; Tse, E.; Rosèn, A.; Chim, C.S. Epigenetic Inactivation of miR-34b/c in Addition to miR-34a and DAPK1 in Chronic Lymphocytic Leukemia. J. Transl. Med. 2014, 12, 52. [Google Scholar] [CrossRef]
- Deneberg, S.; Kanduri, M.; Ali, D.; Bengtzen, S.; Karimi, M.; Qu, Y.; Kimby, E.; Mansouri, L.; Rosenquist, R.; Lennartsson, A.; et al. MicroRNA-34b/c on Chromosome 11q23 Is Aberrantly Methylated in Chronic Lymphocytic Leukemia. Epigenetics 2014, 9, 910–917. [Google Scholar] [CrossRef]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gullà, A.; Tagliaferri, P.; Tassone, P.; et al. MiR-34: A New Weapon against Cancer? Mol. Ther. Nucleic Acids 2014, 3, E195. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 Study of MRX34, a Liposomal miR-34a Mimic, in Patients with Advanced Solid Tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I Study of MRX34, a Liposomal miR-34a Mimic, Administered Twice Weekly in Patients with Advanced Solid Tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Diener, C.; Keller, A.; Meese, E. Emerging Concepts of MiRNA Therapeutics: From Cells to Clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Ota, A.; Tagawa, H.; Karnan, S.; Tsuzuki, S.; Karpas, A.; Kira, S.; Yoshida, Y.; Seto, M. Identification and Characterization of a Novel Gene, C13orf25, as a Target for 13q31-Q32 Amplification in Malignant Lymphoma. Cancer Res. 2004, 64, 3087–3095. [Google Scholar] [CrossRef]
- Chocholska, S.; Zarobkiewicz, M.; Szymańska, A.; Lehman, N.; Woś, J.; Bojarska-Junak, A. Prognostic Value of the MiR-17-92 Cluster in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2023, 24, 1705. [Google Scholar] [CrossRef]
- Selven, H.; Andersen, S.; Pedersen, M.I.; Lombardi, A.P.G.; Busund, L.T.R.; Kilvær, T.K. High Expression of MiR-17-5p and MiR-20a-5p Predicts Favorable Disease-Specific Survival in Stage I-III Colon Cancer. Sci. Rep. 2022, 12, 7080. [Google Scholar] [CrossRef] [PubMed]
- Kuo, G.; Wu, C.Y.; Yang, H.Y. MiR-17-92 Cluster and Immunity. J. Formos. Med. Assoc. 2019, 118, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Vecchiarelli-Federico, L.M.; Li, Y.J.; Egan, S.E.; Spaner, D.; Hough, M.R.; Ben-David, Y. The miR-17-92 Cluster Expands Multipotent Hematopoietic Progenitors Whereas Imbalanced Expression of Its Individual Oncogenic MiRNAs Promotes Leukemia in Mice. Blood 2012, 119, 4486–4498. [Google Scholar] [CrossRef] [PubMed]
- Bomben, R.; Gobessi, S.; Dal Bo, M.; Volinia, S.; Marconi, D.; Tissino, E.; Benedetti, D.; Zucchetto, A.; Rossi, D.; Gaidano, G.; et al. The MiR-17-92 Family Regulates the Response to Toll-like Receptor 9 Triggering of CLL Cells with Unmutated IGHV Genes. Leukemia 2012, 26, 1584–1593. [Google Scholar] [CrossRef]
- Weber, A.; Marquardt, J.; Elzi, D.; Forster, N.; Starke, S.; Glaum, A.; Yamada, D.; Defossez, P.A.; Delrow, J.; Eisenman, R.N.; et al. Zbtb4 Represses Transcription of P21CIP1 and Controls the Cellular Response to P53 Activation. EMBO J. 2008, 27, 1563–1574. [Google Scholar] [CrossRef]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of MicroRNA Function by AntimiR Oligonucleotides. Silence 2012, 3, 1. [Google Scholar] [CrossRef]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of MicroRNAs in Vivo with ‘Antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Dereani, S.; Macor, P.; D’Agaro, T.; Mezzaroba, N.; Dal-Bo, M.; Capolla, S.; Zucchetto, A.; Tissino, E.; Del Poeta, G.; Zorzet, S.; et al. Potential Therapeutic Role of AntagomiR17 for the Treatment of Chronic Lymphocytic Leukemia. J. Hematol. Oncol. 2014, 7, 79. [Google Scholar] [CrossRef]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; Van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A.; et al. Requirement of Bic/MicroRNA-155 for Normal Immune Function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 Is Induced during the Macrophage Inflammatory Response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef]
- Yee, D.; Shah, K.M.; Coles, M.C.; Sharp, T.V.; Lagos, D. MicroRNA-155 Induction via TNF-α and IFN-γ Suppresses Expression of Programmed Death Ligand-1 (PD-L1) in Human Primary Cells. J. Biol. Chem. 2017, 292, 20683–20693. [Google Scholar] [CrossRef]
- Masaki, S.; Ohtsuka, R.; Abe, Y.; Muta, K.; Umemura, T. Expression Patterns of MicroRNAs 155 and 451 during Normal Human Erythropoiesis. Biochem. Biophys. Res. Commun. 2007, 364, 509–514. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a Role in Cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Soneji, S.; Marafioti, T.; Cooper, C.D.O.; Palazzo, S.; Paterson, J.C.; Cattan, H.; Enver, T.; Mager, R.; Boultwood, J.; et al. MicroRNA Expression Distinguishes between Germinal Center B Cell-like and Activated B Cell-like Subtypes of Diffuse Large B Cell Lymphoma. Int. J. Cancer 2007, 121, 1156–1161. [Google Scholar] [CrossRef]
- Van den Berg, A.; Kroesen, B.J.; Kooistra, K.; De Jong, D.; Briggs, J.; Blokzijl, T.; Jacobs, S.; Kluiver, J.; Diepstra, A.; Maggio, E.; et al. High Expression of B-Cell Receptor Inducible Gene BIC in All Subtypes of Hodgkin Lymphoma. Genes Chromosom. Cancer 2003, 37, 20–28. [Google Scholar] [CrossRef]
- Kluiver, J.; Poppema, S.; de Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Kroesen, B.-J.; van den Berg, A. BIC and miR-155 Are Highly Expressed in Hodgkin, Primary Mediastinal and Diffuse Large B Cell Lymphomas. J. Pathol. 2005, 207, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Faraoni, I.; Laterza, S.; Ardiri, D.; Ciardi, C.; Fazi, F.; Lo-Coco, F. MiR-424 and miR-155 Deregulated Expression in Cytogenetically Normal Acute Myeloid Leukaemia: Correlation with NPM1 and FLT3 Mutation Status. J. Hematol. Oncol. 2012, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Fulci, V.; Chiaretti, S.; Goldoni, M.; Azzalin, G.; Carucci, N.; Tavolaro, S.; Castellano, L.; Magrelli, A.; Citarella, F.; Messina, M.; et al. Quantitative Technologies Establish a Novel MicroRNA Profile of Chronic Lymphocytic Leukemia. Blood 2007, 109, 4944–4951. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A MicroRNA Expression Signature of Human Solid Tumors Defines Cancer Gene Targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Gusev, Y.; Jiang, J.; Nuovo, G.J.; Lerner, M.R.; Frankel, W.L.; Morgan, D.L.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. Expression Profiling Identifies MicroRNA Signature in Pancreatic Cancer. Int. J. Cancer 2007, 120, 1046–1054. [Google Scholar] [CrossRef]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. MiR-155 Gene: A Typical Multifunctional MicroRNA. Biochim. Biophys. Acta Mol. Basis Dis. 2009, 1792, 497–505. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D.; Diorio, D.; Nikiforov, Y.E. MicroRNA Expression Profiling of Thyroid Tumors: Biological Significance and Diagnostic Utility. J. Clin. Endocrinol. Metab. 2008, 93, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Jay, C.; Nemunaitis, J.; Chen, P.; Fulgham, P.; Tong, A.W. MiRNA Profiling for Diagnosis and Prognosis of Human Cancer. DNA Cell Biol. 2007, 26, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.; Ben-Yehuda, D.; Hayward, W.S. Bic, a Novel Gene Activated by Proviral Insertions in Avian Leukosis Virus-Induced Lymphomas, Is Likely to Function through Its Noncoding RNA. Mol. Cell. Biol. 1997, 17, 1490–1502. [Google Scholar] [CrossRef]
- Tam, W. Identification and Characterization of Human BIC, a Gene on Chromosome 21 That Encodes a Noncoding RNA. Gene 2001, 274, 157–167. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Zhou, H.; Huang, X.; Cui, H.; Luo, X.; Tang, Y.; Chen, S.; Wu, L.; Shen, N. MiR-155 and Its Star-Form Partner MiR-155* Cooperatively Regulate Type I Interferon Production by Human Plasmacytoid Dendritic Cells. Blood 2010, 116, 5885–5894. [Google Scholar] [CrossRef] [PubMed]
- Yim, R.L.; Wong, K.Y.; Kwong, Y.L.; Loong, F.; Leung, C.Y.; Chu, R.; Lam, W.W.L.; Hui, P.K.; Lai, R.; Chim, C.S. Methylation of MiR-155-3p in Mantle Cell Lymphoma and Other Non-Hodgkin’s Lymphomas. Oncotarget 2014, 5, 9770–9782. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, G.; Zhong, L.; Luo, H.; Wang, S. MicroRNA-155-3p Promotes Breast Cancer Progression through down-Regulating CADM1. Onco. Targets. Ther. 2019, 12, 7993–8002. [Google Scholar] [CrossRef]
- Dawson, O.; Piccinini, A.M. MiR-155-3p: Processing by-Product or Rising Star in Immunity and Cancer? Open Biol. 2022, 12, 220070. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, J.; Shao, H.; Liu, J.; Jin, M.; Chen, J.; Huang, Y. Transforming Growth Factor Β1/Smad4 Signaling Affects Osteoclast Differentiation via Regulation of miR-155 Expression. Mol. Cells 2017, 40, 211–221. [Google Scholar] [CrossRef]
- Wang, L.; Toomey, N.L.; Diaz, L.A.; Walker, G.; Ramos, J.C.; Barber, G.N.; Ning, S. Oncogenic IRFs Provide a Survival Advantage for Epstein-Barr Virus- or Human T-Cell Leukemia Virus Type 1-Transformed Cells through Induction of BIC Expression. J. Virol. 2011, 85, 8328–8337. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Wang, X.; McBride, J.; Fewell, C.; Flemington, E. B-Cell Receptor Activation Induces BIC/MiR-155 Expression through a Conserved AP-1 Element. J. Biol. Chem. 2008, 283, 2654–2662. [Google Scholar] [CrossRef] [PubMed]
- Quinn, S.R.; Mangan, N.E.; Caffrey, B.E.; Gantier, M.P.; Williams, B.R.G.; Hertzog, P.J.; McCoy, C.E.; O’Neill, L.A.J. The Role of Ets2 Transcription Factor in the Induction of Microrna-155 (MiR-155) by Lipopolysaccharide and Its Targeting by Interleukin-10. J. Biol. Chem. 2014, 289, 4316–4325. [Google Scholar] [CrossRef] [PubMed]
- Bruning, U.; Cerone, L.; Neufeld, Z.; Fitzpatrick, S.F.; Cheong, A.; Scholz, C.C.; Simpson, D.A.; Leonard, M.O.; Tambuwala, M.M.; Cummins, E.P.; et al. MicroRNA-155 Promotes Resolution of Hypoxia-Inducible Factor 1α Activity during Prolonged Hypoxia. Mol. Cell. Biol. 2011, 31, 4087–4096. [Google Scholar] [CrossRef] [PubMed]
- Gatto, G.; Rossi, A.; Rossi, D.; Kroening, S.; Bonatti, S.; Mallardo, M. Epstein-Barr Virus Latent Membrane Protein 1 Trans-Activates miR-155 Transcription through the NF-ΚB Pathway. Nucleic Acids Res. 2008, 36, 6608–6619. [Google Scholar] [CrossRef]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in Human B Cell Lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef]
- Ferrajoli, A.; Shanafelt, T.D.; Ivan, C.; Shimizu, M.; Rabe, K.G.; Nouraee, N.; Ikuo, M.; Ghosh, A.K.; Lerner, S.; Rassenti, L.Z.; et al. Prognostic Value of miR-155 in Individuals with Monoclonal B-Cell Lymphocytosis and Patients with B Chronic Lymphocytic Leukemia. Blood 2013, 122, 1891–1899. [Google Scholar] [CrossRef]
- Costinean, S.; Zanesi, N.; Pekarsky, Y.; Tili, E.; Volinia, S.; Heerema, N.; Croce, C.M. Pre-B Cell Proliferation and Lymphoblastic Leukemia/High-Grade Lymphoma in Eμ-MiR155 Transgenic Mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7024–7029. [Google Scholar] [CrossRef]
- Vargova, K.; Curik, N.; Burda, P.; Basova, P.; Kulvait, V.; Pospisil, V.; Savvulidi, F.; Kokavec, J.; Necas, E.; Berkova, A.; et al. MYB Transcriptionally Regulates the miR-155 Host Gene in Chronic Lymphocytic Leukemia. Blood 2011, 117, 3816–3825. [Google Scholar] [CrossRef]
- Cui, B.; Chen, L.; Zhang, S.; Mraz, M.; Fecteau, J.F.; Yu, J.; Ghia, E.M.; Zhang, L.; Bao, L.; Rassenti, L.Z.; et al. Micro RNA-155 Influences B-Cell Receptor Signaling and Associates with Aggressive Disease in Chronic Lymphocytic Leukemia. Blood 2014, 124, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Pagotto, S.; Veronese, A.; Soranno, A.; Lanuti, P.; Marco, M.D.; Russo, M.V.; Ramassone, A.; Marchisio, M.; Simeone, P.; Guanciali-Franchi, P.E.; et al. Hsa-MiR-155-5p Drives Aneuploidy at Early Stages of Cellular Transformation. Oncotarget 2018, 9, 13036–13047. [Google Scholar] [CrossRef] [PubMed]
- Pagotto, S.; Veronese, A.; Soranno, A.; Balatti, V.; Ramassone, A.; Guanciali-Franchi, P.E.; Palka, G.; Innocenti, I.; Autore, F.; Rassenti, L.Z.; et al. Hnrnpl Restrains miR-155 Targeting of BUB1 to Stabilize Aberrant Karyotypes of Transformed Cells in Chronic Lymphocytic Leukemia. Cancers 2019, 11, 575. [Google Scholar] [CrossRef] [PubMed]
- Querfeld, C.; Pacheco, T.; Foss, F.M.; Halwani, A.S.; Porcu, P.; Seto, A.G.; Ruckman, J.; Landry, M.L.; Jackson, A.L.; Pestano, L.A.; et al. Preliminary Results of a Phase 1 Trial Evaluating MRG-106, a Synthetic MicroRNA Antagonist (LNA AntimiR) of MicroRNA-155, in Patients with CTCL. Blood 2016, 128, 1829. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an Oligonucleotide Inhibitor of MiR-155, Slows DLBCL Tumor Cell Growth in Vitro and in Vivo. Clin. Cancer Res. 2021, 27, 1139–1149. [Google Scholar] [CrossRef]
- Sun, X.; Sit, A.; Feinberg, M.W. Role of MiR-181 Family in Regulating Vascular Inflammation and Immunity. Trends Cardiovasc. Med. 2014, 24, 105–112. [Google Scholar] [CrossRef]
- Visone, R.; Veronese, A.; Rassenti, L.Z.; Balatti, V.; Pearl, D.K.; Acunzo, M.; Volinia, S.; Taccioli, C.; Kipps, T.J.; Croce, C.M. MiR-181b Is a Biomarker of Disease Progression in Chronic Lymphocytic Leukemia. Blood 2011, 118, 3072–3079. [Google Scholar] [CrossRef]
- Visone, R.; Veronese, A.; Balatti, V.; Croce, C.M. MiR-181b: New Perspective to Evaluate Disease Progression in Chronic Lymphocytic Leukemia. Oncotarget 2012, 3, 195–202. [Google Scholar] [CrossRef]
- Marton, S.; Garcia, M.R.; Robello, C.; Persson, H.; Trajtenberg, F.; Pritsch, O.; Rovira, C.; Naya, H.; Dighiero, G.; Cayota, A. Small RNAs Analysis in CLL Reveals a Deregulation of MiRNA Expression and Novel MiRNA Candidates of Putative Relevance in CLL Pathogenesis. Leukemia 2008, 22, 330–338. [Google Scholar] [CrossRef]
- Li, S.; Moffett, H.F.; Lu, J.; Werner, L.; Zhang, H.; Ritz, J.; Neuberg, D.; Wucherpfennig, K.W.; Brown, J.R.; Novina, C.D. Microrna Expression Profiling Identifies Activated B Cell Status in Chronic Lymphocytic Leukemia Cells. PLoS ONE 2011, 6, e16956. [Google Scholar] [CrossRef]
- Bichi, R.; Shinton, S.A.; Martin, E.S.; Koval, A.; Calin, G.A.; Cesari, R.; Russo, G.; Hardy, R.R.; Croce, C.M. Human Chronic Lymphocytic Leukemia Modeled in Mouse by Targeted TCL1 Expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6955–6960. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.X.; Zhu, W.; Fang, C.; Fan, L.; Zou, Z.J.; Wang, Y.H.; Liu, P.; Hong, M.; Miao, K.R.; Liu, P.; et al. MiR-181a/b Significantly Enhances Drug Sensitivity in Chronic Lymphocytic Leukemia Cells via Targeting Multiple Anti-Apoptosis Genes. Carcinogenesis 2012, 33, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Pekarsky, Y.; Croce, C.M. The Role of MicroRNA and Other Non-Coding RNA in the Pathogenesis of Chronic Lymphocytic Leukemia. Best Pract. Res. Clin. Haematol. 2007, 20, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Mraz, M.; Kipps, T.J. MicroRNAs and B Cell Receptor Signaling in Chronic Lymphocytic Leukemia. Leuk. Lymphoma 2013, 54, 1836–1839. [Google Scholar] [CrossRef] [PubMed]
- Mraz, M.; Pospisilova, S.; Malinova, K.; Slapak, I.; Mayer, J. MicroRNAs in Chronic Lymphocytic Leukemia Pathogenesis and Disease Subtypes. Leuk. Lymphoma 2009, 50, 506–509. [Google Scholar] [CrossRef]
- Ramkissoon, S.H.; Mainwaring, L.A.; Ogasawara, Y.; Keyvanfar, K.; Philip McCoy, J.; Sloand, E.M.; Kajigaya, S.; Young, N.S. Hematopoietic-Specific MicroRNA Expression in Human Cells. Leuk. Res. 2006, 30, 643–647. [Google Scholar] [CrossRef]
- Zhu, W.; Shan, X.; Wang, T.; Shu, Y.; Liu, P. MiR-181b Modulates Multidrug Resistance by Targeting BCL2 in Human Cancer Cell Lines. Int. J. Cancer 2010, 127, 2520–2529. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Palamarchuk, A.; Maximov, V.; Efanov, A.; Nazaryan, N.; Santanam, U.; Rassenti, L.; Kipps, T.; Croce, C.M. Tcl1 Functions as a Transcriptional Regulator and Is Directly Involved in the Pathogenesis of CLL. Proc. Natl. Acad. Sci. USA 2008, 105, 19643–19648. [Google Scholar] [CrossRef]
- Vogler, M.; Butterworth, M.; Majid, A.; Walewska, R.J.; Sun, X.M.; Dyer, M.J.S.; Cohen, G.M. Concurrent Up-Regulation of BCL-XL and BCL2A1 Induces Approximately 1000-Fold Resistance to ABT-737 in Chronic Lymphocytic Leukemia. Blood 2009, 113, 4403–4413. [Google Scholar] [CrossRef]
- Bresin, A.; Callegari, E.; D’Abundo, L.; Cattani, C.; Bassi, C.; Zagatti, B.; Narducci, M.G.; Caprini, E.; Pekarsky, Y.; Croce, C.M.; et al. MiR-181b as a Therapeutic Agent for Chronic Lymphocytic Leukemia in the Eμ-TCL1 Mouse Model. Oncotarget 2015, 6, 19807–19818. [Google Scholar] [CrossRef]
- Di Marco, M.; Veschi, S.; Lanuti, P.; Ramassone, A.; Pacillo, S.; Pagotto, S.; Pepe, F.; George-William, J.N.; Curcio, C.; Marchisio, M.; et al. Enhanced Expression of miR-181b in b Cells of Cll Improves the Anti-Tumor Cytotoxic t Cell Response. Cancers 2021, 13, 257. [Google Scholar] [CrossRef] [PubMed]
| Family Name | Members | Targets | Therapeutic Strategies | |
|---|---|---|---|---|
| miR-15a/16-1 | miR-15a miR-16-1 | All members | BCL-2 [35] MCL-1, JUN [36] ROR-1 [38] TP53 [39] | In vivo delivery of miR-15a and miR-16-1 (preclinical) [41,42] |
| miR-29 | miR-29a miR-29b-1 miR-29b-2 miR-29c | miR-29b | TCL-1 [46] MCL-1 [50] TRAF4 [52] | Immuno-nanoparticle-based miR-29b (preclinical) [54] |
| miR-34 | miR-34a miR-34b miR-34c | All members | TP53 [57] | miR-34a liposomal mimic, MRX34. (phase 1 study, prematurely terminated after the emergency of severe side-effects) [76] |
| miR-34a | CDK4 CDK6 CCND1 CCNE2 MET FOXP1 [65] | |||
| miR-34b/c | ZAP70 [40] | |||
| miR-17-92 cluster | miR-17 miR-18a miR-19a miR-19b-1 miR-20a miR-20b miR-92a-1 | miR-17 miR-19 | PTEN [82] | AntagomiR17 (preclinical) [88] |
| miR-92a-1 miR-19 | Bim [82] | |||
| miR-17 miR-20a miR-20b miR-17 miR-18a miR-19b-1 miR-92a-1 | ZBTB4 TP53INP1 MYC [84] | |||
| miR-155 | miR-155 | SHIP1 [121] BUB1 CENP-F ZW10 [122] | Cobomarsen, MRG-106 (phase 1 clinical trial) [125] | |
| miR-181 | miR-181a-1 miR-181a-2 miR-181b-1 miR-181b-2 miR-181c miR-181d | miR-181b | TCL-1 MCL-1 BCL-2 [127] c-FOS [141] | In vivo administration of miR-181b (preclinical) [140] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Autore, F.; Ramassone, A.; Stirparo, L.; Pagotto, S.; Fresa, A.; Innocenti, I.; Visone, R.; Laurenti, L. Role of microRNAs in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2023, 24, 12471. https://doi.org/10.3390/ijms241512471
Autore F, Ramassone A, Stirparo L, Pagotto S, Fresa A, Innocenti I, Visone R, Laurenti L. Role of microRNAs in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences. 2023; 24(15):12471. https://doi.org/10.3390/ijms241512471
Chicago/Turabian StyleAutore, Francesco, Alice Ramassone, Luca Stirparo, Sara Pagotto, Alberto Fresa, Idanna Innocenti, Rosa Visone, and Luca Laurenti. 2023. "Role of microRNAs in Chronic Lymphocytic Leukemia" International Journal of Molecular Sciences 24, no. 15: 12471. https://doi.org/10.3390/ijms241512471
APA StyleAutore, F., Ramassone, A., Stirparo, L., Pagotto, S., Fresa, A., Innocenti, I., Visone, R., & Laurenti, L. (2023). Role of microRNAs in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences, 24(15), 12471. https://doi.org/10.3390/ijms241512471








