Augmentation of Cathepsin Isoforms in Diabetic db/db Mouse Kidneys Is Associated with an Increase in Renal MARCKS Expression and Proteolysis
Abstract
:1. Introduction
2. Results
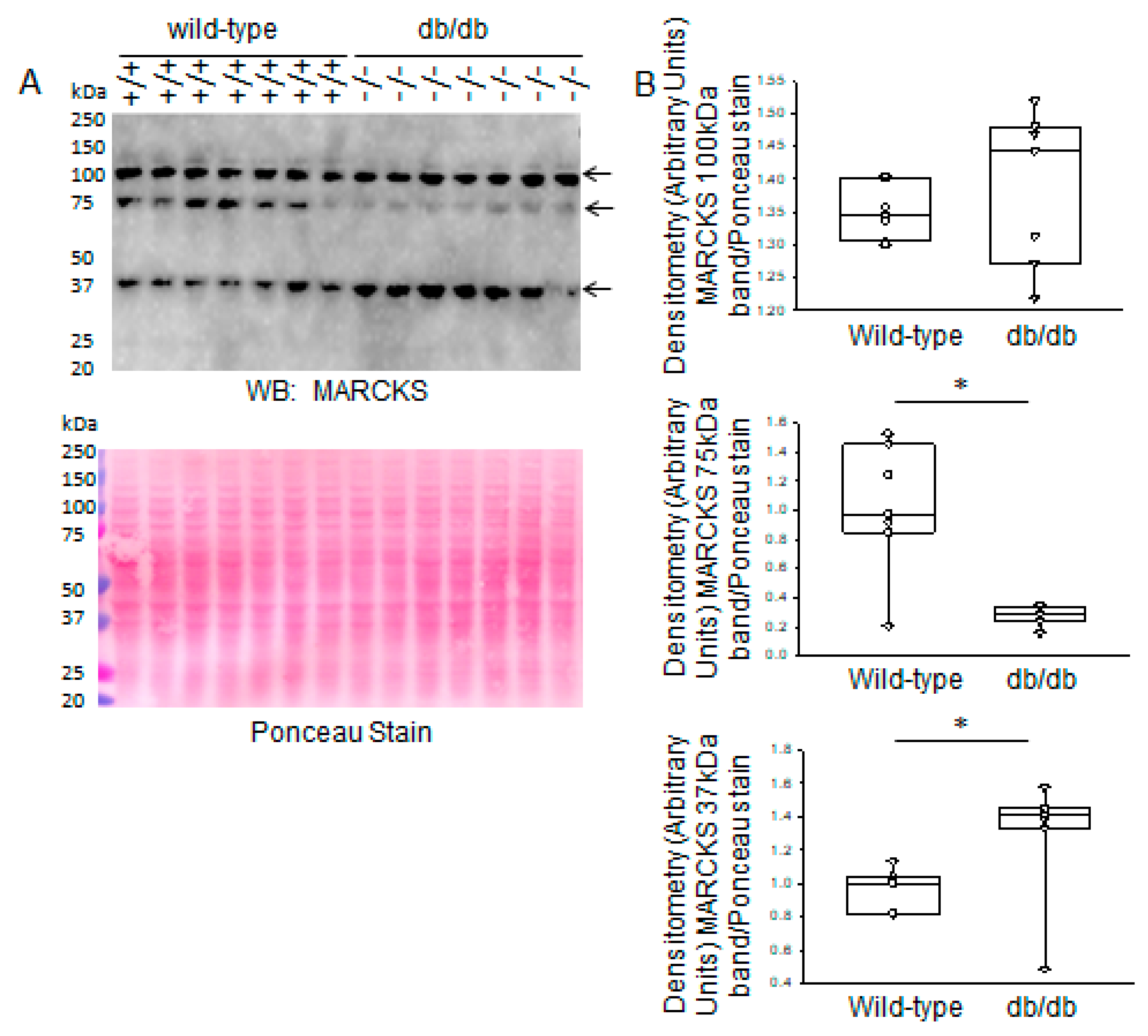
2.1. Increased Proteolysis of MARCKS Proteins in Diabetic db/db Kidneys
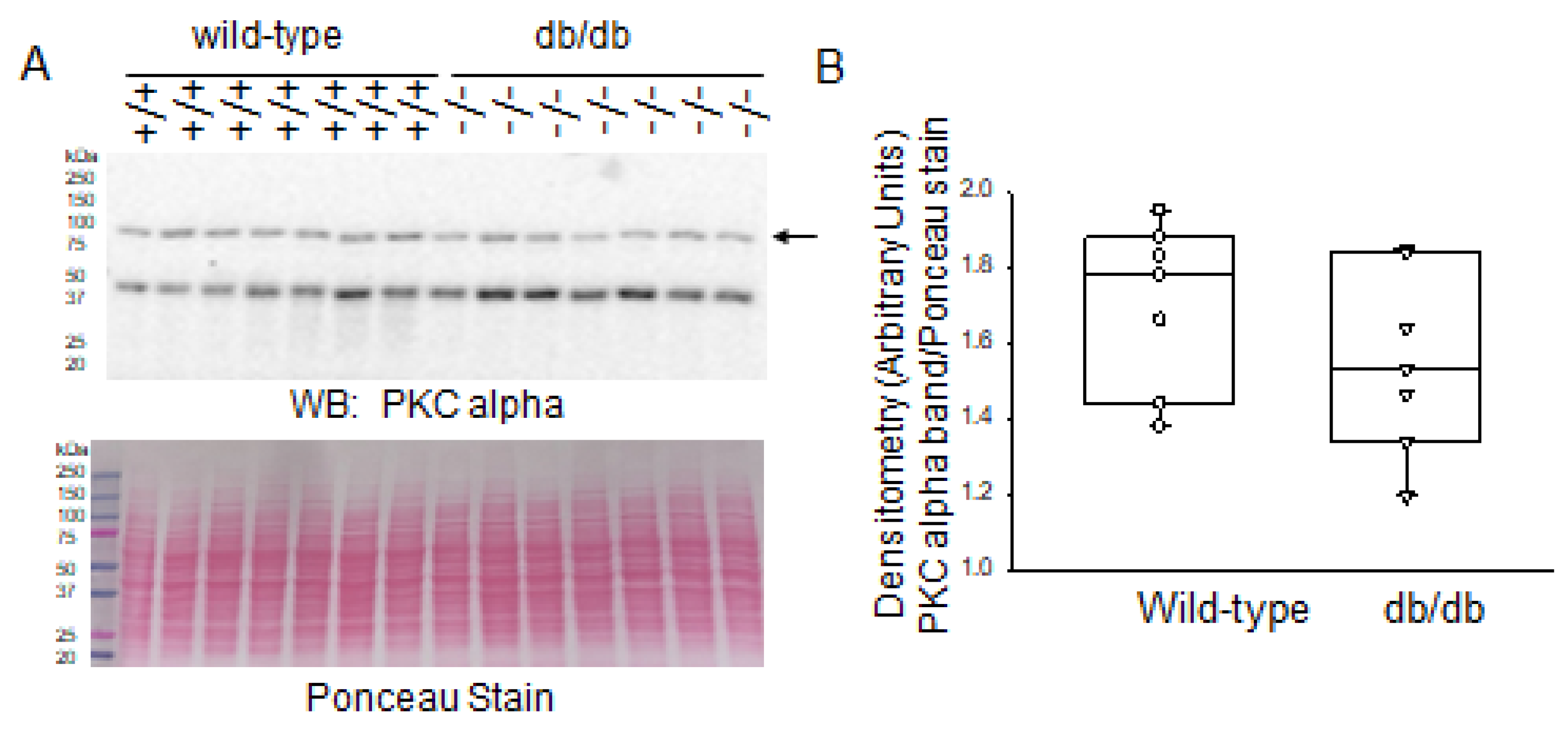
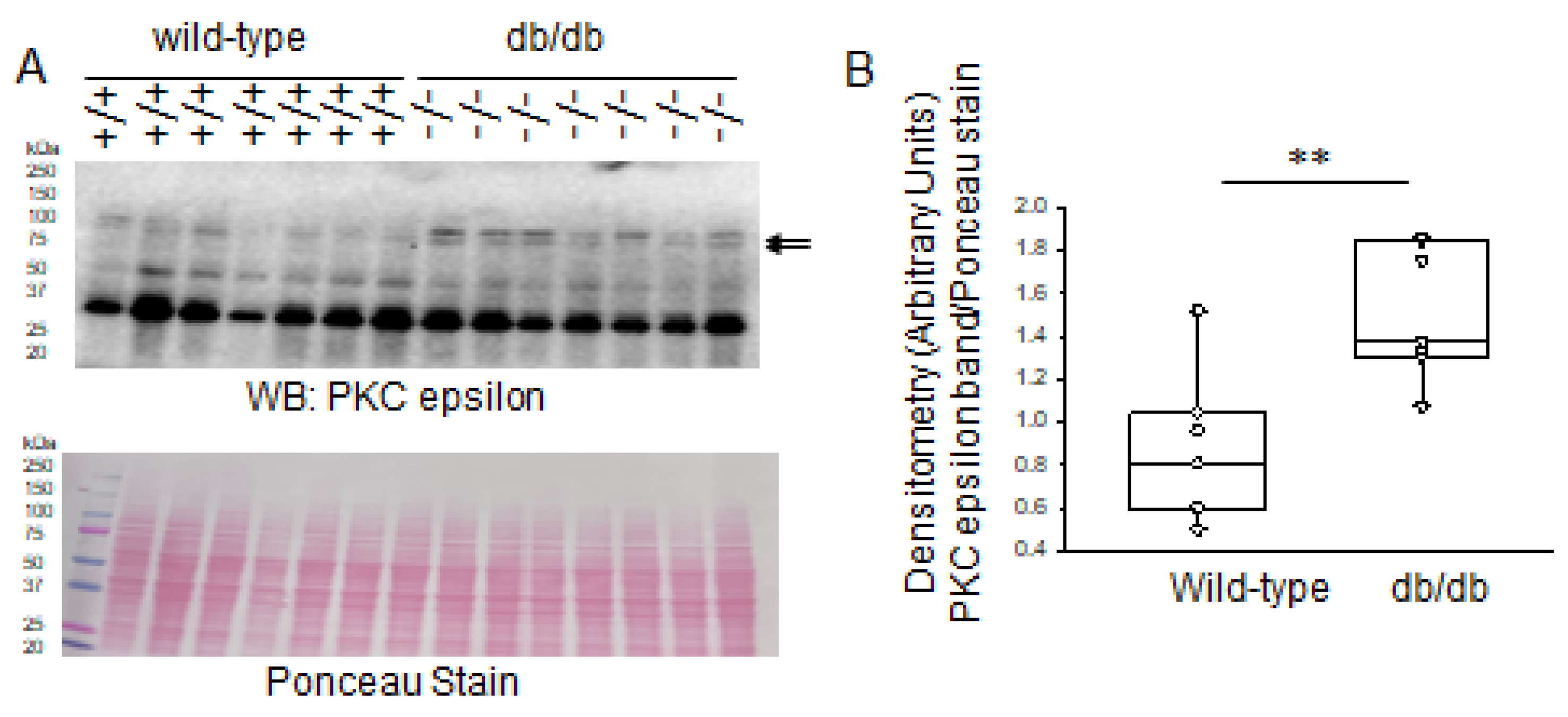
2.2. Increased PKC Isoform Expression in Diabetic db/db Kidneys
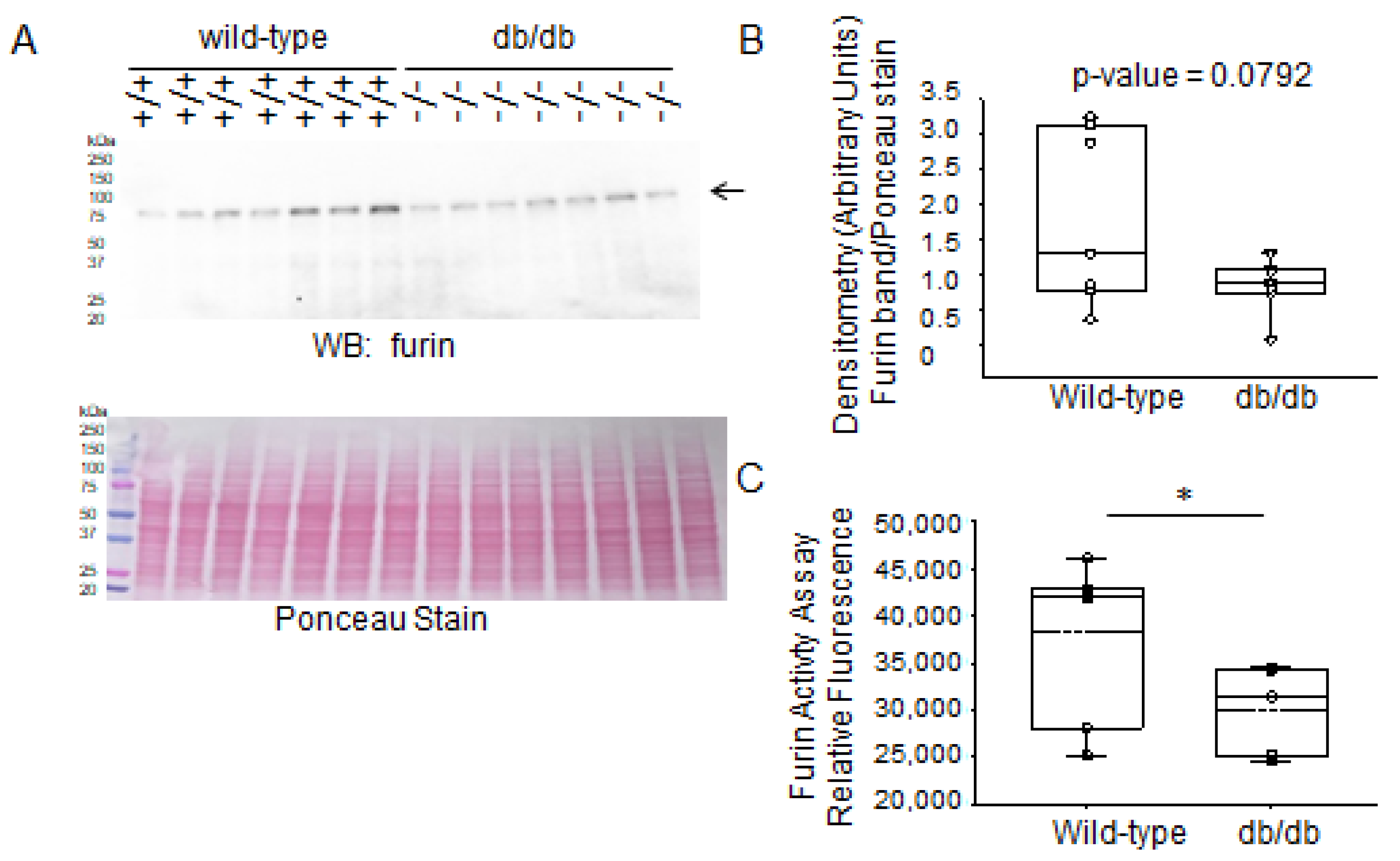
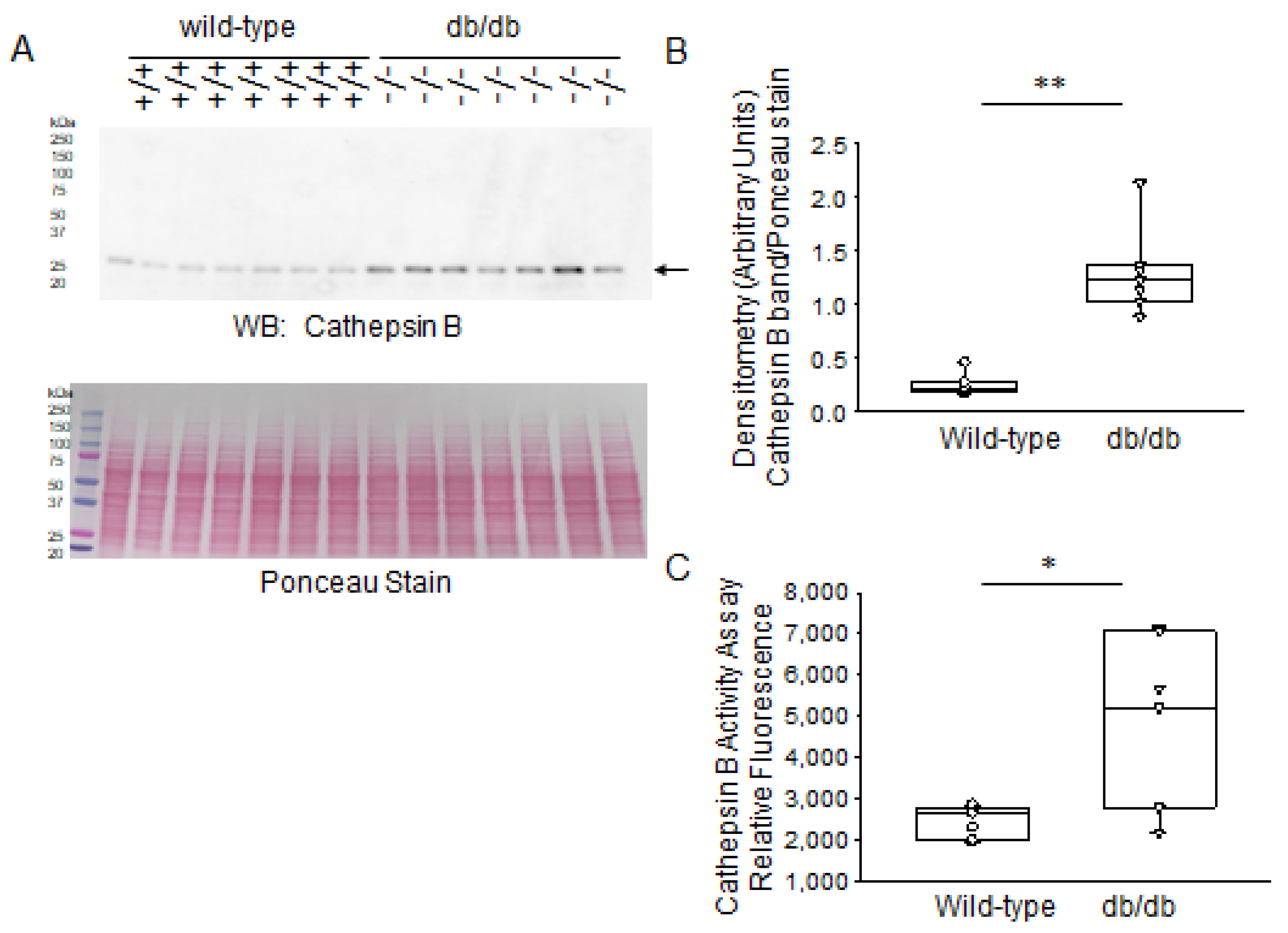
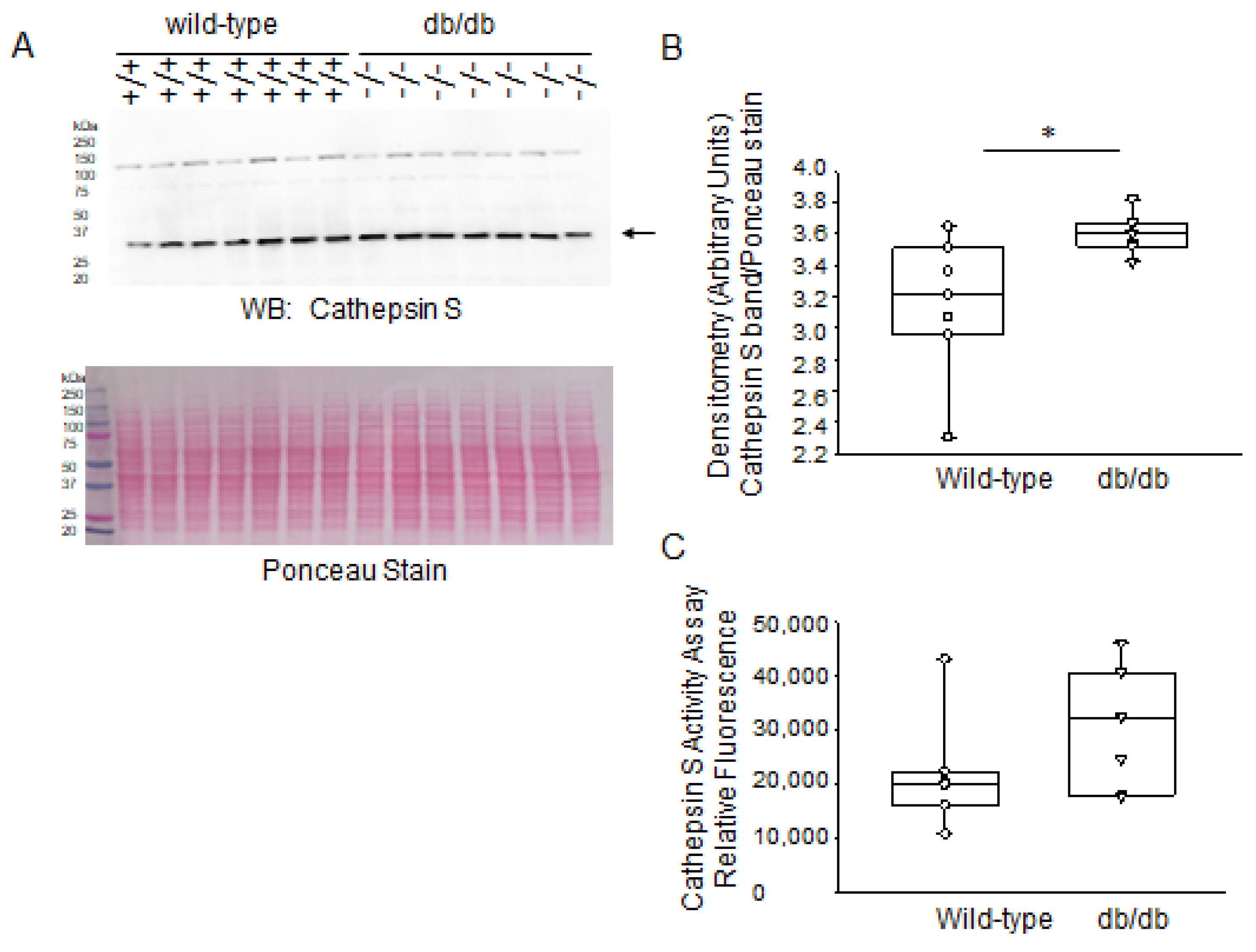
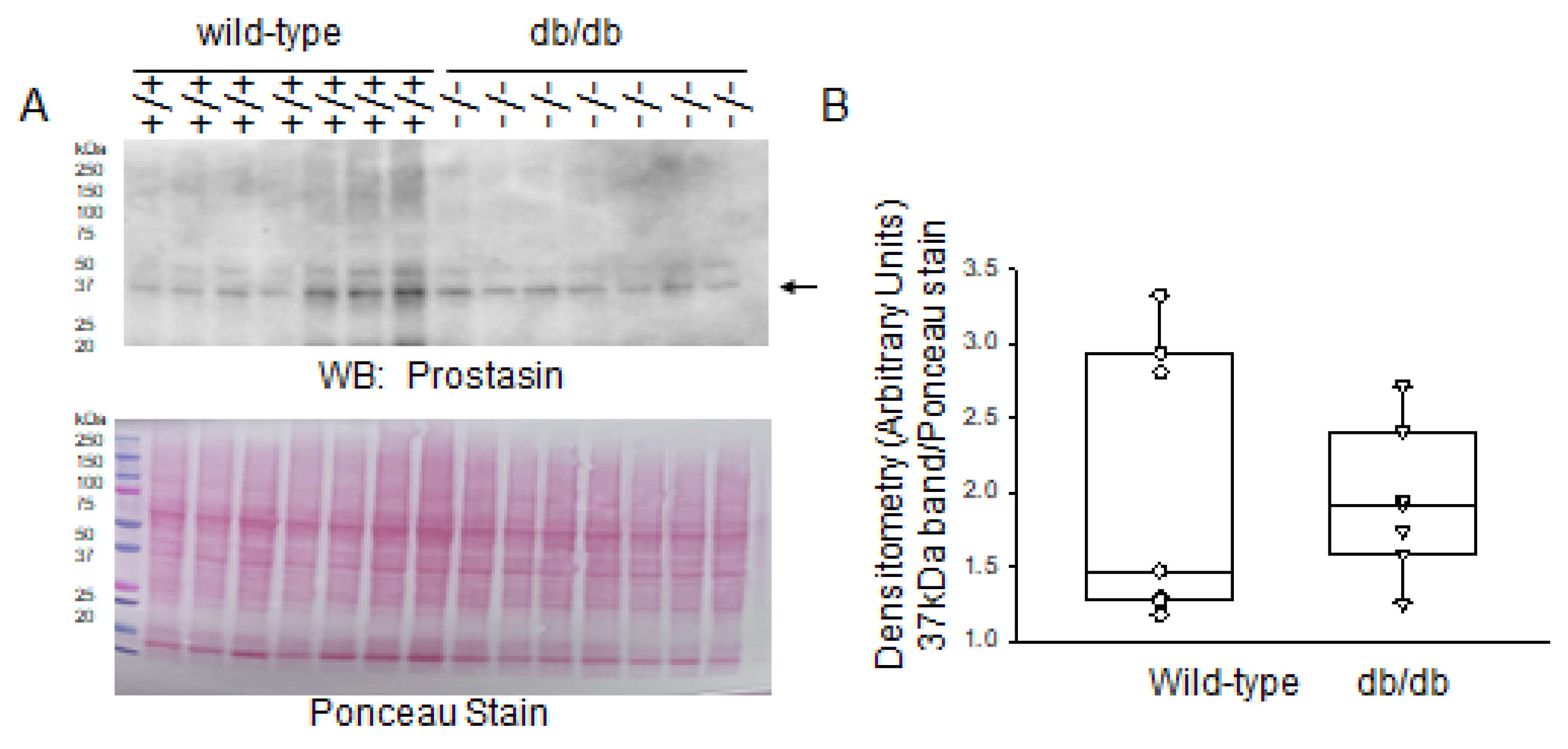
2.3. Expression Levels of Proteases in Diabetic db/db Kidneys Compared to Kidneys of Wild-Type Control Mice
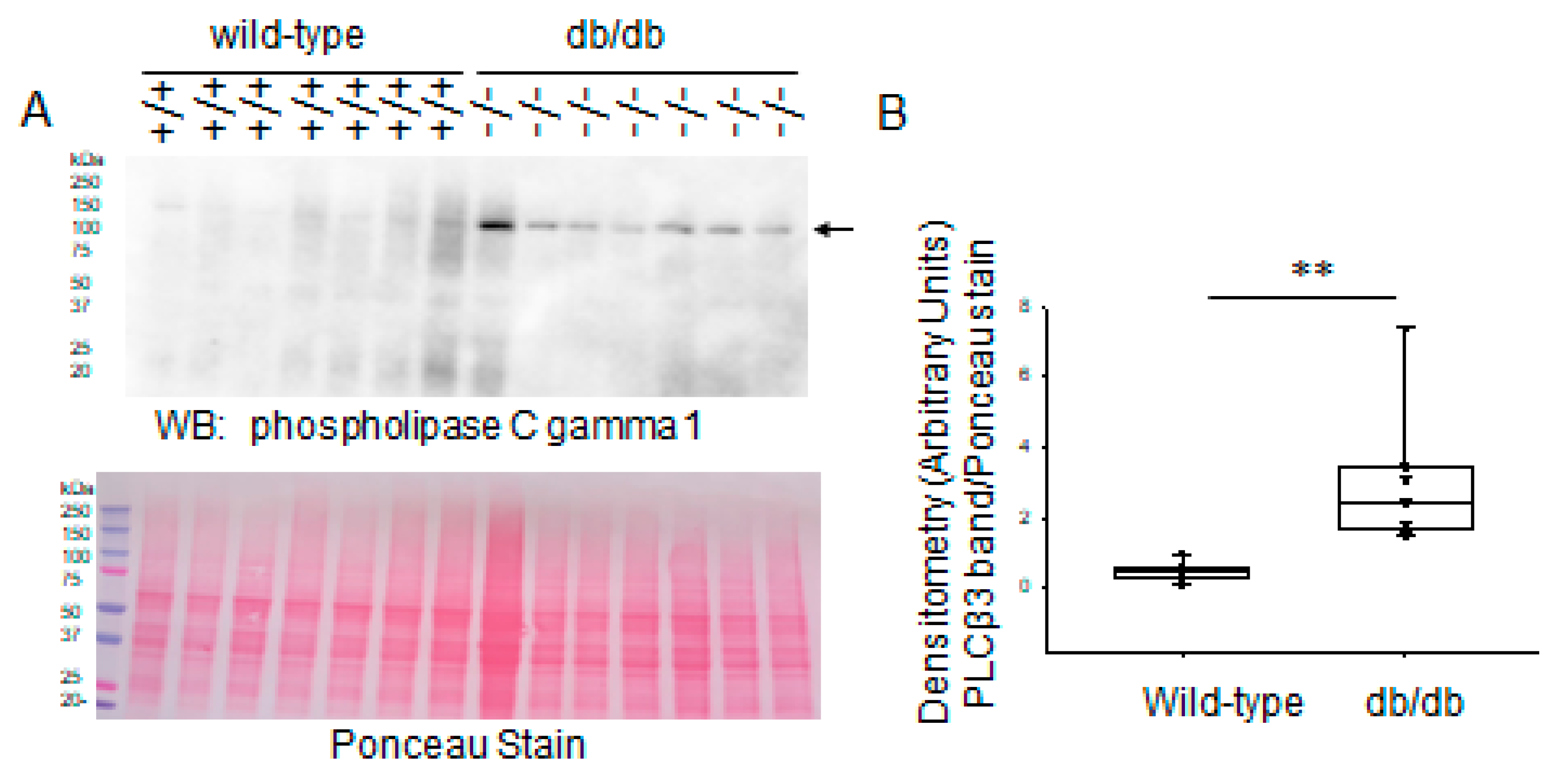
2.4. Expression Levels of Phospholipases in Diabetic db/db Kidneys Compared to Kidneys of Wild-Type Control Mice
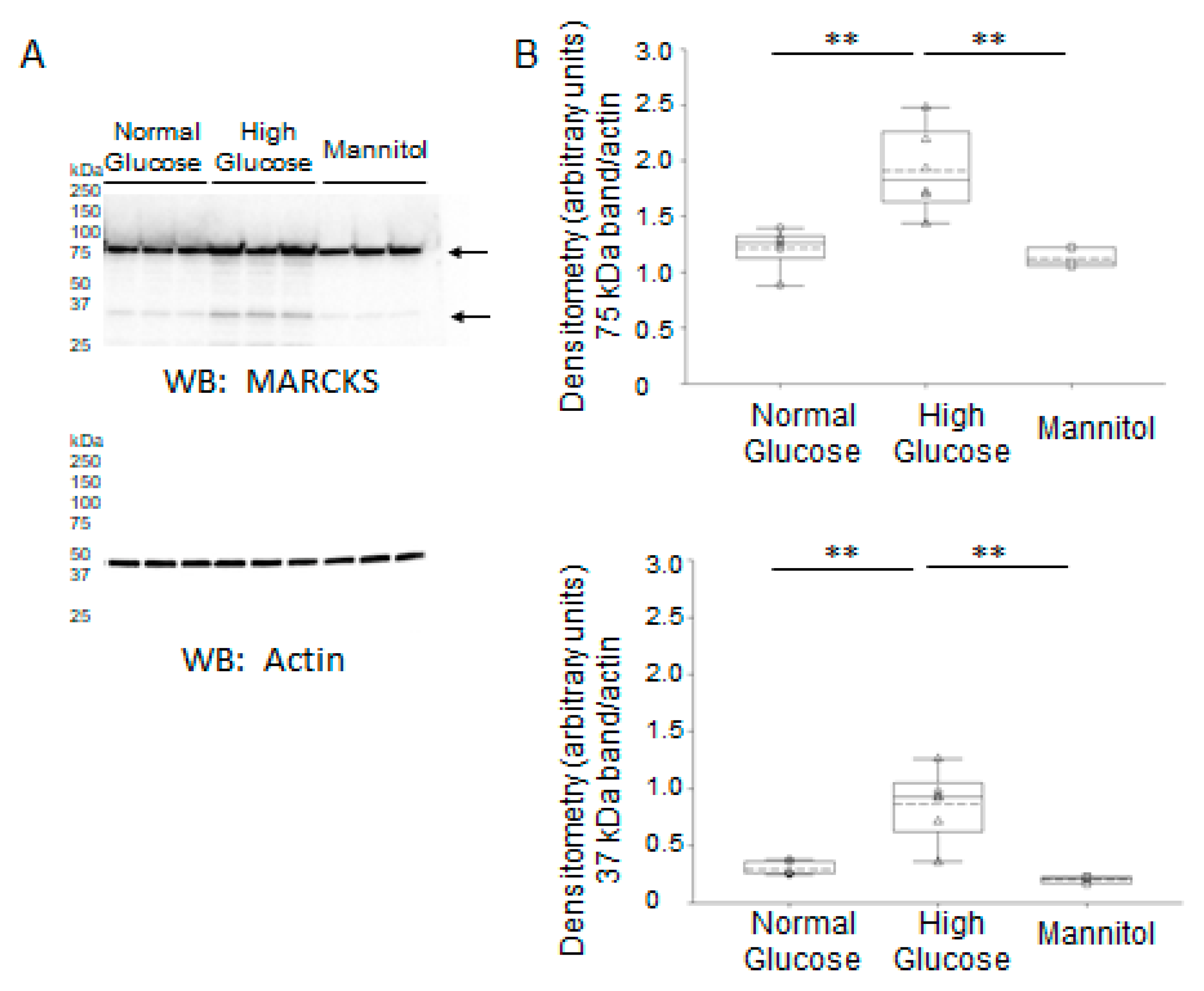
2.5. Expression and Proteolysis of MARCKS Protein in Mouse Cortical Collecting Duct Cells Cultured in Normal Glucose Compared to High Glucose Conditions
3. Discussion
4. Materials and Methods
4.1. Animal Studies
4.2. Tail Bleeds for Measuring Blood Glucose
4.3. Measurement of Urinary Albumin
4.4. Measurement of Urinary Creatinine
4.5. Cathepsin Activity Assays
4.6. Furin Activity Assay
4.7. Tissue Homogenization and BCA Assay
4.8. Western Blot and Densitometric Analysis
4.9. Immunohistochemistry
4.10. Culture and Treatment of mpkCCD Cells
4.11. siRNA Transfection
4.12. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Craven, P.A.; Studer, R.K.; Negrete, H.; DeRubertis, F.R. Protein kinase C in diabetic nephropathy. J. Diabetes Complicat. 1995, 9, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Derubertis, F.R.; Craven, P.A. Activation of protein kinase C in glomerular cells in diabetes. Mechanisms and potential links to the pathogenesis of diabetic glomerulopathy. Diabetes 1994, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.; Yu, M.R.; Choi, Y.J.; Lee, H.B. Activation of protein kinase c-δ and c-ε by oxidative stress in early diabetic rat kidney. Am. J. Kidney Dis. 2001, 38 (Suppl. 1), S204–S207. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.; McGinty, A.; Godson, C. Protein kinases C: Potential targets for intervention in diabetic nephropathy. Curr. Opin. Nephrol. Hypertens 1998, 7, 563–570. [Google Scholar] [CrossRef]
- Meier, M.; Menne, J.; Haller, H. Targeting the protein kinase C family in the diabetic kidney: Lessons from analysis of mutant mice. Diabetologia 2009, 52, 765–775. [Google Scholar] [CrossRef] [Green Version]
- Alli, A.A.; Bao, H.-F.; Alli, A.A.; Aldrugh, Y.; Song, J.Z.; Ma, H.-P.; Yu, L.; Al-Khalili, O.; Eaton, D.C. Phosphatidylinositol phosphate-dependent regulation of Xenopus ENaC by MARCKS protein. Am. J. Physiol.-Ren. Physiol. 2012, 303, F800–F811. [Google Scholar] [CrossRef] [Green Version]
- Alli, A.A.; Bao, H.-F.; Liu, B.-C.; Yu, L.; Aldrugh, S.; Montgomery, D.S.; Ma, H.-P.; Eaton, D.C. Calmodulin and CaMKII modulate ENaC activity by regulating the association of MARCKS and the cytoskeleton with the apical membrane. Am. J. Physiol. Physiol. 2015, 309, F456–F463. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, D.S.; Yu, L.; Ghazi, Z.M.; Thai, T.L.; Al-Khalili, O.; Ma, H.-P.; Eaton, D.C.; Alli, A.A. ENaC activity is regulated by calpain-2 proteolysis of MARCKS proteins. Am. J. Physiol. Physiol. 2017, 313, C42–C53. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Yue, Q.; Moseley, A.; Al-Khalili, O.; Wynne, B.M.; Ma, H.; Wang, L.; Eaton, D.C. Myristoylated alanine-rich C kinase substrate-like protein-1 regulates epithelial sodium channel activity in renal distal convoluted tubule cells. Am. J. Physiol. Physiol. 2020, 319, C589–C604. [Google Scholar] [CrossRef]
- Yue, Q.; Al-Khalili, O.; Moseley, A.; Yoshigi, M.; Wynne, B.M.; Ma, H.; Eaton, D.C. PIP2 Interacts Electrostatically with MARCKS-like Protein-1 and ENaC in Renal Epithelial Cells. Biology 2022, 11, 1694. [Google Scholar] [CrossRef]
- Zhai, Y.-J.; Wu, M.-M.; Linck, V.A.; Zou, L.; Yue, Q.; Wei, S.-P.; Song, C.; Zhang, S.; Williams, C.R.; Song, B.-L.; et al. Intracellular cholesterol stimulates ENaC by interacting with phosphatidylinositol-4,5-bisphosphate and mediates cyclosporine A-induced hypertension. Biochim. et Biophys. Acta BBA -Mol. Basis Dis. 2019, 1865, 1915–1924. [Google Scholar] [CrossRef] [PubMed]
- Tuna, K.M.; Liu, B.-C.; Yue, Q.; Ghazi, Z.M.; Ma, H.-P.; Eaton, D.C.; Alli, A.A. Mal protein stabilizes luminal membrane PLC-β3 and negatively regulates ENaC in mouse cortical collecting duct cells. Am. J. Physiol. Physiol. 2019, 317, F986–F995. [Google Scholar] [CrossRef] [PubMed]
- Lugo, C.I.; Liu, L.P.; Bala, N.; Morales, A.G.; Gholam, M.F.; Abchee, J.C.; Elmoujahid, N.; Elshikha, A.S.; Avdiaj, R.; Searcy, L.A.; et al. Human Alpha-1 Antitrypsin Attenuates ENaC and MARCKS and Lowers Blood Pressure in Hypertensive Diabetic db/db Mice. Biomolecules 2022, 13, 66. [Google Scholar] [CrossRef]
- Arnold, T.P.; Standaert, M.L.; Hernandez, H.; Watson, J.; Mischak, H.; Kazanietz, M.G.; Zhao, L.; Cooper, D.R.; Farese, R.V. Effects of insulin and phorbol esters on MARCKS (myristoylated alanine-rich C-kinase substrate) phosphorylation (and other parameters of protein kinase C activation) in rat adipocytes, rat soleus muscle and BC3H-1 myocytes. Biochem. J. 1993, 295, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Chappell, D.S.; Patel, N.A.; Jiang, K.; Li, P.; Watson, J.E.; Byers, D.M.; Cooper, D.R. Functional involvement of protein kinase C-βII and its substrate, myristoylated alanine-rich C-kinase substrate (MARCKS), in insulin-stimulated glucose transport in L6 rat skeletal muscle cells. Diabetologia 2009, 52, 901–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, N.; Alexander, G.; Park, J.K.; Maasch, C.; Buchwalow, I.; Luft, F.C.; Haller, H. Differential expression of protein kinase C isoforms in streptozotocin-induced diabetic rats. Kidney Int. 1999, 56, 1737–1750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, D.; Cao, T.; Zhang, L.-L.; Fan, G.-C.; Qiu, J.; Peng, T.-Q. Targeted inhibition of calpain in mitochondria alleviates oxidative stress-induced myocardial injury. Acta Pharmacol. Sin. 2021, 42, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Huwiler, A.; Schulze-Lohoff, E.; Fabbro, D.; Pfeilschifter, J. Immunocharacterization of protein kinase C isoenzymes in rat kidney glomeruli, and cultured glomerular epithelial and mesangial cells. Nephron Exp. Nephrol. 1993, 1, 19–25. [Google Scholar]
- Babazono, T.; Kapor-Drezgic, J.; Dlugosz, J.A.; Whiteside, C. Altered expression and subcellular localization of diacylglycerol-sensitive protein kinase C isoforms in diabetic rat glomerular cells. Diabetes 1998, 47, 668–676. [Google Scholar] [CrossRef]
- Kolczynska, K.; Loza-Valdes, A.; Hawro, I.; Sumara, G. Diacylglycerol-evoked activation of PKC and PKD isoforms in regulation of glucose and lipid metabolism: A review. Lipids Health Dis. 2020, 19, 113. [Google Scholar] [CrossRef]
- Park, C.; Zhang, Y.; Zhang, X.; Wu, J.; Chen, L.; Cha, D.; Su, D.; Hwang, M.-T.; Fan, X.; Davis, L.; et al. PPARα agonist fenofibrate improves diabetic nephropathy in db/db mice. Kidney Int. 2006, 69, 1511–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.-W.; Kim, Y.G.; Lee, S.H.; Lee, A.; Kim, D.-J.; Jeong, K.-H.; Lee, K.H.; Hwang, S.J.; Woo, J.S.; Lim, S.J.; et al. Mycophenolate Mofetil Ameliorates Diabetic Nephropathy in db/db Mice. BioMed Res. Int. 2015, 2015, 301627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, H.; Tamura, Y.; Murayama, T.; Minami, M.; Yokode, M. Differential effect of statins on diabetic nephropathy in db/db mice. Int. J. Mol. Med. 2011, 28, 683–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Group | Wild-Type | db/db | p-Value |
|---|---|---|---|
| Urinary albumin (mg/dL) | 0.0588 ± 0.0114 | 0.0669 ± 0.0253 | 0.77 |
| Urinary creatinine (mg/dL) | 9.187 ± 2.206 | 8.327 ± 1.771 | 0.76 |
| Body weight (gm) | 23.484 ± 0.513 | 34.624 ± 2.653 | 0.0053 |
| Blood glucose (mg/dL) | 115.143 ± 14.838 | 536.143 ± 12.695 | 8.581 × 10−11 |
| Assay | Manufacturer | Catalog Number |
|---|---|---|
| Cathepsin B | Abcam | ab65300 |
| Cathepsin S | Abcam | ab65307 |
| Furin | BPS Bioscience | 78040 |
| Albumin | Proteintech | KE00076 |
| Creatinine | Abcam | ab204537 |
| Antibody | Application | Manufacturer | Catalog Number |
|---|---|---|---|
| MARCKS | WB | Abcam | ab72459 |
| Cathepsin B | WB, IHC | Cell Signaling Technologies | 31718 |
| Cathepsin D | WB | Cell Signaling Technologies | 69854 |
| Cathepsin S | WB | Abcam | ab232740 |
| Prostasin | WB, IHC | ThermoFisher Scientific Invitrogen | PA5-27977 |
| Furin | WB | Cell Signaling Technologies | 64709 |
| PKC alpha | WB | Cell Signaling Technologies | 2056 |
| PKC delta | WB | Cell Signaling Technologies | 9616 |
| PKC epsilon | WB | Cell Signaling Technologies | 2683 |
| KLK1 | WB | Boster Biological Technology | PA1709 |
| PLCβ3 | WB | Cell Signaling Technologies | 14247 |
| PLCγ1 | WB, IHC | Cell Signaling Technologies | 5690 |
| MARCKS | IHC | Santa Cruz Biotechnologies | sc-100777 |
| Furin | IHC | Santa Cruz Biotechnologies | sc-133142 |
| Cathepsin S | IHC | Santa Cruz Biotechnologies | sc-271619 |
| Cathepsin D | IHC | Santa Cruz Biotechnologies | sc-377299 |
| PKC delta | IHC | Santa Cruz Biotechnologies | sc-8402 |
| PKC epsilon | IHC | Santa Cruz Biotechnologies | sc-1681 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gholam, M.F.; Bala, N.; Dogan, Y.E.; Alli, A.A. Augmentation of Cathepsin Isoforms in Diabetic db/db Mouse Kidneys Is Associated with an Increase in Renal MARCKS Expression and Proteolysis. Int. J. Mol. Sci. 2023, 24, 12484. https://doi.org/10.3390/ijms241512484
Gholam MF, Bala N, Dogan YE, Alli AA. Augmentation of Cathepsin Isoforms in Diabetic db/db Mouse Kidneys Is Associated with an Increase in Renal MARCKS Expression and Proteolysis. International Journal of Molecular Sciences. 2023; 24(15):12484. https://doi.org/10.3390/ijms241512484
Chicago/Turabian StyleGholam, Mohammed F., Niharika Bala, Yunus E. Dogan, and Abdel A. Alli. 2023. "Augmentation of Cathepsin Isoforms in Diabetic db/db Mouse Kidneys Is Associated with an Increase in Renal MARCKS Expression and Proteolysis" International Journal of Molecular Sciences 24, no. 15: 12484. https://doi.org/10.3390/ijms241512484






