Neuroprotective Potential of L-Glutamate Transporters in Human Induced Pluripotent Stem Cell-Derived Neural Cells against Excitotoxicity
Abstract
1. Introduction
2. Results
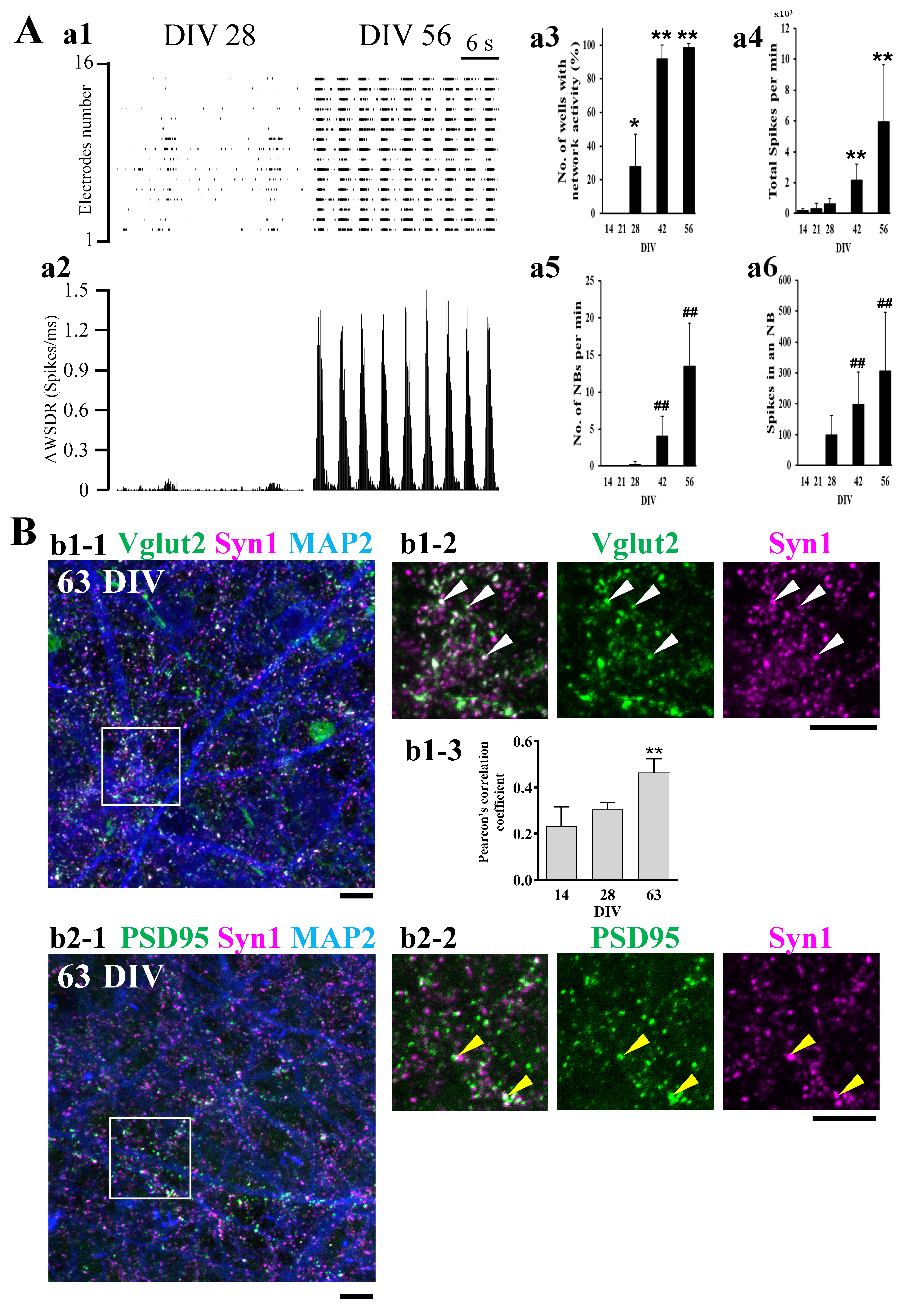
2.1. Functional Maturation of hiPSC-Derived Neural Networks
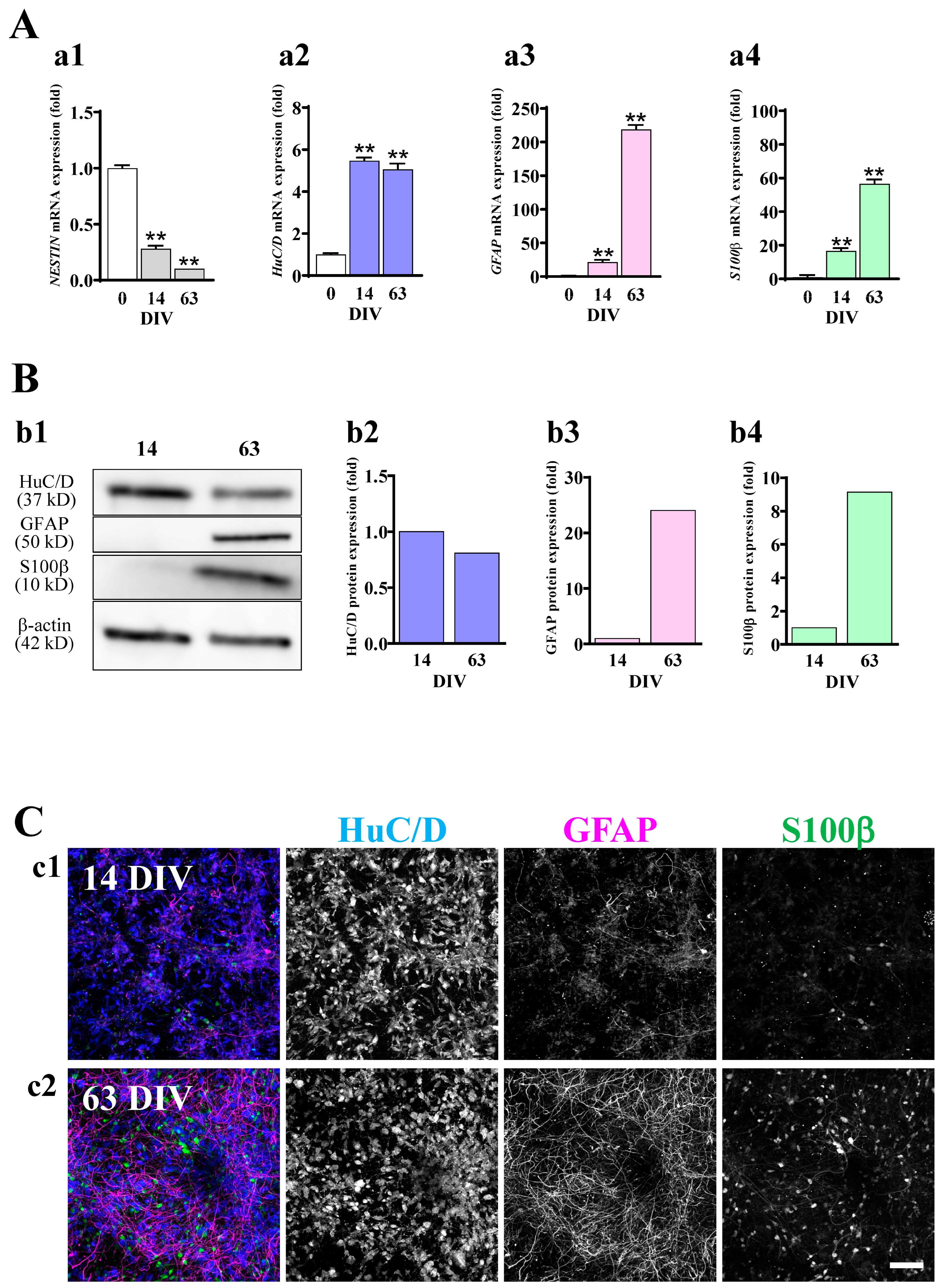
2.2. Astrocytes Are Differentiated in hiPSC-Derived Neural Cell Cultures
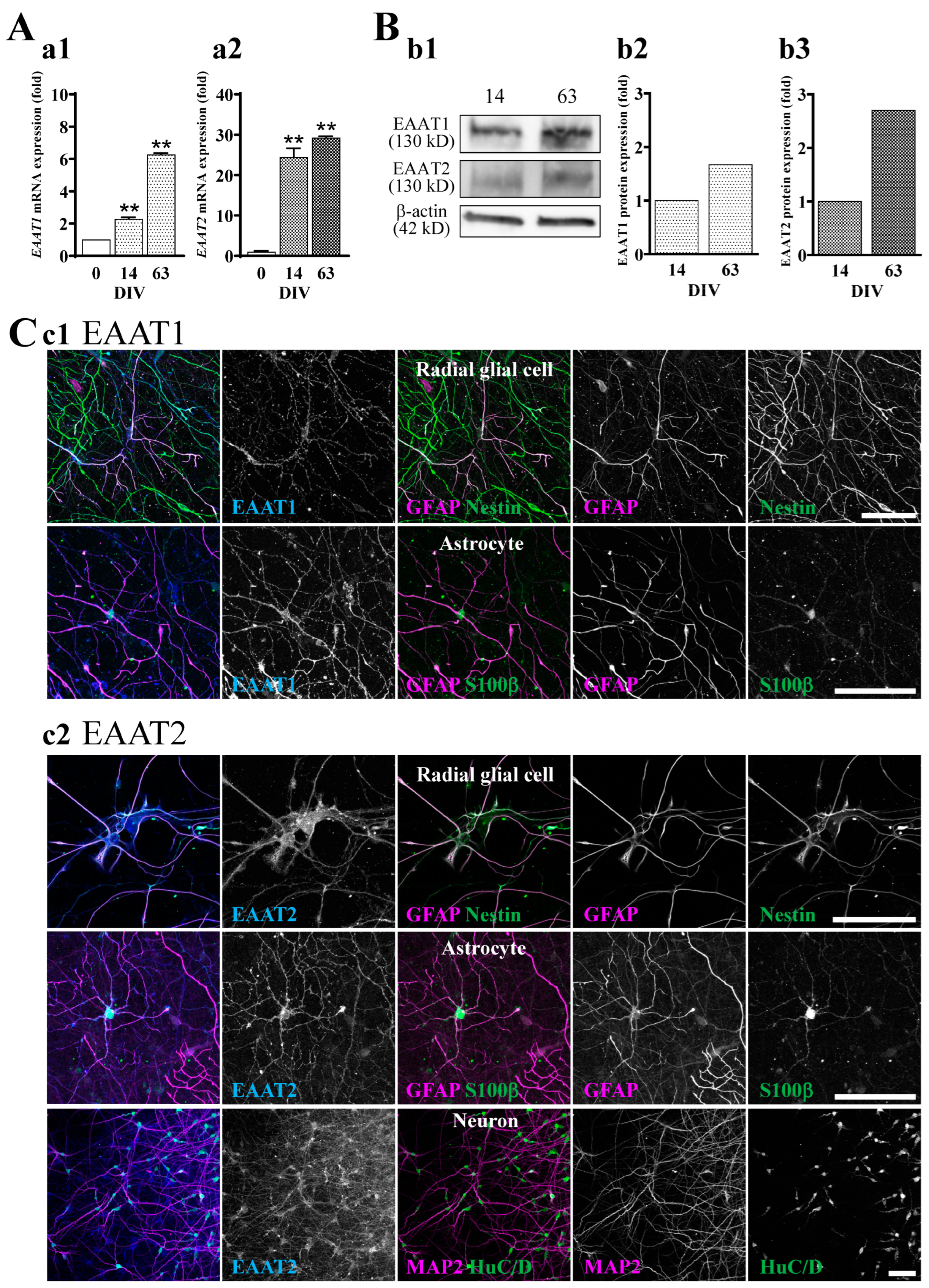
2.3. The Expression of EAAT1 and EAAT2 in hiPSC-Derived Neural Cells
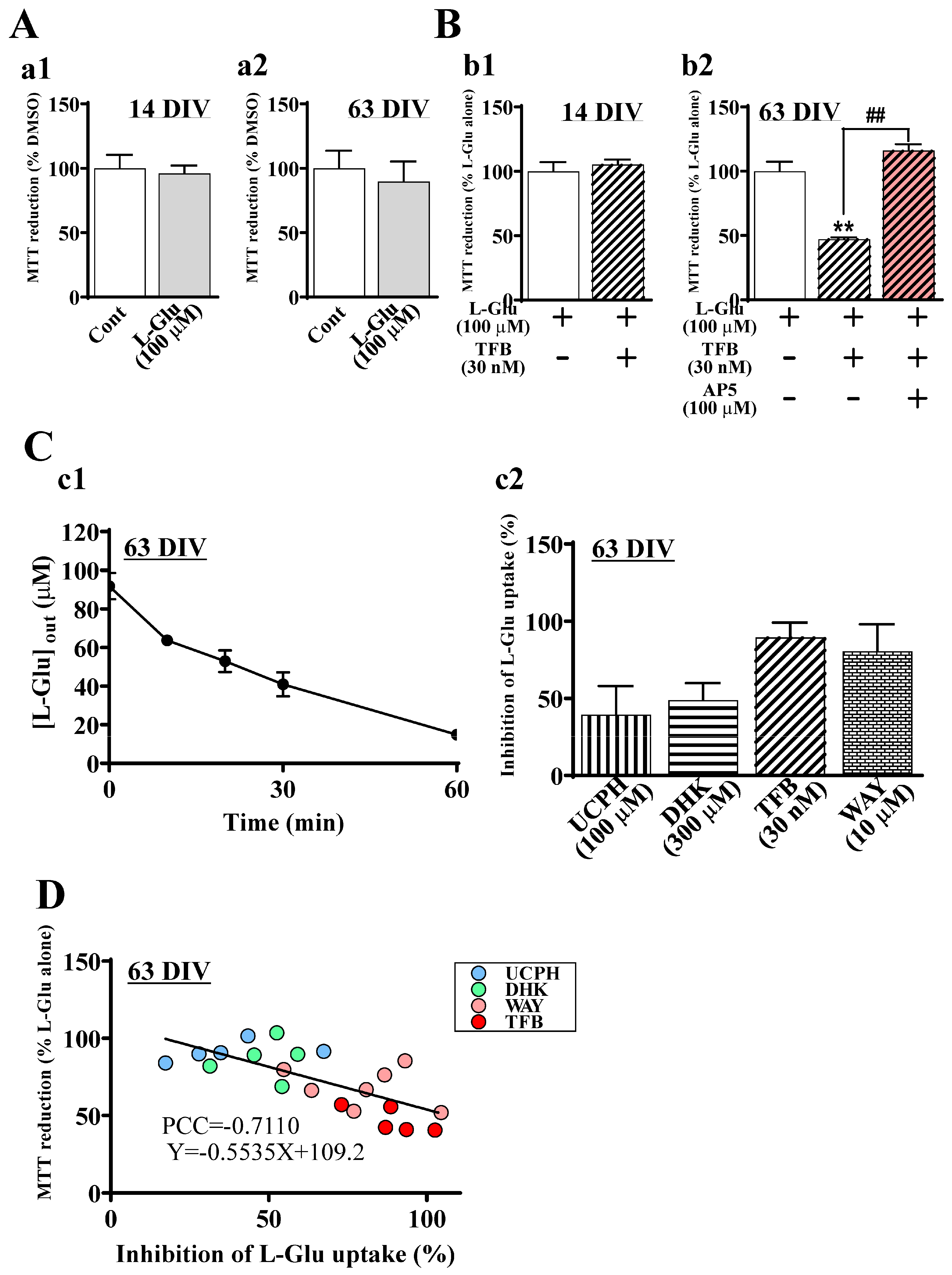
2.4. The Roles of EAATs in the Sensitivity of hiPSC-Derived Neurons to Excitotoxicity
3. Discussion
3.1. Patterns of the Expression of Developmental Markers and L-Glu Transporters
3.2. EAAT Subtypes Contribute to Tolerance to Excitotoxicity in hiPSC-Derived Neurons
3.3. The Significance of Our Data for the CNS Safety/Toxicity Assessment System at Preclinical Stage
4. Materials and Methods
4.1. Materials
4.2. Culture of hiPSC-Derived Neurons
4.3. Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.4. Western Blotting
4.5. Immunocytochemistry Scale
4.6. MEA Recording and Data Analysis (Extracellular Recording, Burst Analysis)
4.7. MTT Reduction Assays
4.8. Measurement of the Extracellular L-Glu Concentration (L-Glu Uptake Assay)
4.9. Drug Treatment
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bonaventura, G.; Iemmolo, R.; Attaguile, G.A.; La Cognata, V.; Pistone, B.S.; Raudino, G.; D’Agata, V.; Cantarella, G.; Barcellona, M.L.; Cavallaro, S. iPSCs: A Preclinical Drug Research Tool for Neurological Disorders. Int. J. Mol. Sci. 2021, 22, 4596. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Brown, D.; Alexander, R.; March, R.; Morgan, P.; Satterthwaite, G.; Pangalos, M.N. Lessons learned from the fate of AstraZeneca’s drug pipeline: A five-dimensional framework. Nat. Rev. Drug Discov. 2014, 13, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Onakpoya, I.J.; Heneghan, C.J.; Aronson, J.K. Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: A systematic review of the world literature. BMC Med. 2016, 14, 10. [Google Scholar] [CrossRef]
- Arrowsmith, J.; Miller, P. Trial watch: Phase II and phase III attrition rates 2011–2012. Nat. Rev. Drug Discov. 2013, 12, 569. [Google Scholar] [CrossRef]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Michaelis, E.K. Molecular biology of glutamate receptors in the central nervous system and their role in excitotoxicity, oxidative stress and aging. Prog. Neurobiol. 1998, 54, 369–415. [Google Scholar] [CrossRef]
- Kritis, A.A.; Stamoula, E.G.; Paniskaki, K.A.; Vavilis, T.D. Researching glutamate—Induced cytotoxicity in different cell lines: A comparative/collective analysis/study. Front. Cell. Neurosci. 2015, 9, 91. [Google Scholar] [CrossRef]
- Sato, K.; Takahashi, K.; Shigemoto-Mogami, Y.; Chujo, K.; Sekino, Y. Glypican 6 Enhances N-Methyl-D-Aspartate Receptor Function in Human-Induced Pluripotent Stem Cell-Derived Neurons. Front. Cell. Neurosci. 2016, 10, 259. [Google Scholar] [CrossRef]
- Bauersachs, H.G.; Bengtson, C.P.; Weiss, U.; Hellwig, A.; García-Vilela, C.; Zaremba, B.; Kaessmann, H.; Pruunsild, P.; Bading, H. N-methyl-d-aspartate Receptor-mediated Preconditioning Mitigates Excitotoxicity in Human Induced Pluripotent Stem Cell-derived Brain Organoids. Neuroscience 2022, 484, 83–97. [Google Scholar] [CrossRef]
- Daneshgar, N.; Leidinger, M.R.; Le, S.; Hefti, M.; Prigione, A.; Dai, D.F. Activated microglia and neuroinflammation as a pathogenic mechanism in Leigh syndrome. Front. Neurosci. 2022, 16, 1068498. [Google Scholar] [CrossRef] [PubMed]
- Danbolt, N.C. Glutamate uptake. Prog. Neurobiol. 2001, 65, 1–105. [Google Scholar] [CrossRef]
- Zerangue, N.; Kavanaugh, M.P. Flux coupling in a neuronal glutamate transporter. Nature 1996, 383, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Oberheim, N.A.; Takano, T.; Han, X.; He, W.; Lin, J.H.; Wang, F.; Xu, Q.; Wyatt, J.D.; Pilcher, W.; Ojemann, J.G.; et al. Uniquely hominid features of adult human astrocytes. J. Neurosci. 2009, 29, 3276–3287. [Google Scholar] [CrossRef] [PubMed]
- Bigarreau, J.; Rouach, N.; Perrier, A.L.; Mouthon, F.; Charvériat, M. Modeling and Targeting Neuroglial Interactions with Human Pluripotent Stem Cell Models. Int. J. Mol. Sci. 2022, 23, 1684. [Google Scholar] [CrossRef]
- Gilyarov, A.V. Nestin in central nervous system cells. Neurosci. Behav. Physiol. 2008, 38, 165–169. [Google Scholar] [CrossRef]
- Akamatsu, W.; Okano, H.J.; Osumi, N.; Inoue, T.; Nakamura, S.; Sakakibara, S.; Miura, M.; Matsuo, N.; Darnell, R.B.; Okano, H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc. Natl. Acad. Sci. USA 1999, 96, 9885–9890. [Google Scholar] [CrossRef]
- Zimmer, D.B.; Cornwall, E.H.; Landar, A.; Song, W. The S100 protein family: History, function, and expression. Brain Res. Bull. 1995, 37, 417–429. [Google Scholar] [CrossRef]
- Eng, L.F.; Ghirnikar, R.S.; Lee, Y.L. Glial fibrillary acidic protein: GFAP-thirty-one years (1969–2000). Neurochem. Res. 2000, 25, 1439–1451. [Google Scholar] [CrossRef]
- Lucas, D.R.; Newhouse, J.P. The toxic effect of sodium L-glutamate on the inner layers of the retina. AMA Arch. Ophthalmol. 1957, 58, 193–201. [Google Scholar] [CrossRef]
- Olney, J.W.; Ho, O.L.; Rhee, V. Cytotoxic effects of acidic and sulphur containing amino acids on the infant mouse central nervous system. Exp. Brain Res. 1971, 14, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Olney, J.W.; Sharpe, L.G. Brain lesions in an infant rhesus monkey treated with monsodium glutamate. Science 1969, 166, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.W. Glutamate neurotoxicity and diseases of the nervous system. Neuron 1988, 1, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Tymianski, M. Glutamate receptors, neurotoxicity and neurodegeneration. Pflug. Arch. Eur. J. Physiol. 2010, 460, 525–542. [Google Scholar] [CrossRef] [PubMed]
- Maragakis, N.J.; Rothstein, J.D. Glutamate transporters in neurologic disease. Arch. Neurol. 2001, 58, 365–370. [Google Scholar] [CrossRef]
- Rosenberg, P.A.; Amin, S.; Leitner, M. Glutamate uptake disguises neurotoxic potency of glutamate agonists in cerebral cortex in dissociated cell culture. J. Neurosci. 1992, 12, 56–61. [Google Scholar] [CrossRef]
- Choi, D.W.; Maulucci-Gedde, M.; Kriegstein, A.R. Glutamate neurotoxicity in cortical cell culture. J. Neurosci. 1987, 7, 357–368. [Google Scholar] [CrossRef]
- Abe, K.; Matsuki, N. Measurement of cellular 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction activity and lactate dehydrogenase release using MTT. Neurosci. Res. 2000, 38, 325–329. [Google Scholar] [CrossRef]
- Shimamoto, K.; Sakai, R.; Takaoka, K.; Yumoto, N.; Nakajima, T.; Amara, S.G.; Shigeri, Y. Characterization of novel L-threo-beta-benzyloxyaspartate derivatives, potent blockers of the glutamate transporters. Mol. Pharmacol. 2004, 65, 1008–1015. [Google Scholar] [CrossRef]
- Abrahamsen, B.; Schneider, N.; Erichsen, M.N.; Huynh, T.H.; Fahlke, C.; Bunch, L.; Jensen, A.A. Allosteric modulation of an excitatory amino acid transporter: The subtype-selective inhibitor UCPH-101 exerts sustained inhibition of EAAT1 through an intramonomeric site in the trimerization domain. J. Neurosci. 2013, 33, 1068–1087. [Google Scholar] [CrossRef]
- Arriza, J.L.; Fairman, W.A.; Wadiche, J.I.; Murdoch, G.H.; Kavanaugh, M.P.; Amara, S.G. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J. Neurosci. 1994, 14, 5559–5569. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, J.; McIlvain, H.B.; Carrick, T.A.; Jow, B.; Lu, Q.; Kowal, D.; Lin, S.; Greenfield, A.; Grosanu, C.; Fan, K.; et al. Characterization of novel aryl-ether, biaryl, and fluorene aspartic acid and diaminopropionic acid analogs as potent inhibitors of the high-affinity glutamate transporter EAAT2. Mol. Pharmacol. 2005, 68, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Marín-Padilla, M. Prenatal development of fibrous (white matter), protoplasmic (gray matter), and layer I astrocytes in the human cerebral cortex: A Golgi study. J. Comp. Neurol. 1995, 357, 554–572. [Google Scholar] [CrossRef] [PubMed]
- Noctor, S.C.; Flint, A.C.; Weissman, T.A.; Dammerman, R.S.; Kriegstein, A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 2001, 409, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Malatesta, P.; Hartfuss, E.; Götz, M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 2000, 127, 5253–5263. [Google Scholar] [CrossRef]
- Bar-Peled, O.; Ben-Hur, H.; Biegon, A.; Groner, Y.; Dewhurst, S.; Furuta, A.; Rothstein, J.D. Distribution of glutamate transporter subtypes during human brain development. J. Neurochem. 1997, 69, 2571–2580. [Google Scholar] [CrossRef]
- DeSilva, T.M.; Borenstein, N.S.; Volpe, J.J.; Kinney, H.C.; Rosenberg, P.A. Expression of EAAT2 in neurons and protoplasmic astrocytes during human cortical development. J. Comp. Neurol. 2012, 520, 3912–3932. [Google Scholar] [CrossRef]
- Lundin, A.; Delsing, L.; Clausen, M.; Ricchiuto, P.; Sanchez, J.; Sabirsh, A.; Ding, M.; Synnergren, J.; Zetterberg, H.; Brolen, G.; et al. Human iPS-Derived Astroglia from a Stable Neural Precursor State Show Improved Functionality Compared with Conventional Astrocytic Models. Stem Cell Rep. 2018, 10, 1030–1045. [Google Scholar] [CrossRef]
- Santos, R.; Vadodaria, K.C.; Jaeger, B.N.; Mei, A.; Lefcochilos-Fogelquist, S.; Mendes, A.P.D.; Erikson, G.; Shokhirev, M.; Randolph-Moore, L.; Fredlender, C.; et al. Differentiation of Inflammation-Responsive Astrocytes from Glial Progenitors Generated from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2017, 8, 1757–1769. [Google Scholar] [CrossRef]
- Krencik, R.; Weick, J.P.; Liu, Y.; Zhang, Z.J.; Zhang, S.C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011, 29, 528–534. [Google Scholar] [CrossRef]
- Holmseth, S.; Dehnes, Y.; Huang, Y.H.; Follin-Arbelet, V.V.; Grutle, N.J.; Mylonakou, M.N.; Plachez, C.; Zhou, Y.; Furness, D.N.; Bergles, D.E.; et al. The density of EAAC1 (EAAT3) glutamate transporters expressed by neurons in the mammalian CNS. J. Neurosci. 2012, 32, 6000–6013. [Google Scholar] [CrossRef] [PubMed]
- Furness, D.N.; Dehnes, Y.; Akhtar, A.Q.; Rossi, D.J.; Hamann, M.; Grutle, N.J.; Gundersen, V.; Holmseth, S.; Lehre, K.P.; Ullensvang, K.; et al. A quantitative assessment of glutamate uptake into hippocampal synaptic terminals and astrocytes: New insights into a neuronal role for excitatory amino acid transporter 2 (EAAT2). Neuroscience 2008, 157, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Danbolt, N.C.; Furness, D.N.; Zhou, Y. Neuronal vs glial glutamate uptake: Resolving the conundrum. Neurochem. Int. 2016, 98, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Watase, K.; Manabe, T.; Yamada, K.; Watanabe, M.; Takahashi, K.; Iwama, H.; Nishikawa, T.; Ichihara, N.; Kikuchi, T.; et al. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science 1997, 276, 1699–1702. [Google Scholar] [CrossRef]
- Gegelashvili, G.; Danbolt, N.C.; Schousboe, A. Neuronal soluble factors differentially regulate the expression of the GLT1 and GLAST glutamate transporters in cultured astroglia. J. Neurochem. 1997, 69, 2612–2615. [Google Scholar] [CrossRef]
- Gegelashvili, G.; Dehnes, Y.; Danbolt, N.C.; Schousboe, A. The high-affinity glutamate transporters GLT1, GLAST, and EAAT4 are regulated via different signalling mechanisms. Neurochem. Int. 2000, 37, 163–170. [Google Scholar] [CrossRef]
- Plachez, C.; Danbolt, N.C.; Récasens, M. Transient expression of the glial glutamate transporters GLAST and GLT in hippocampal neurons in primary culture. J. Neurosci. Res. 2000, 59, 587–593. [Google Scholar] [CrossRef]
- Martinez-Lozada, Z.; Guillem, A.M.; Robinson, M.B. Transcriptional Regulation of Glutamate Transporters: From Extracellular Signals to Transcription Factors. Adv. Pharmacol. 2016, 76, 103–145. [Google Scholar] [CrossRef]
- Lau, A.C.; Cui, H.; Tymianski, M. The use of propidium iodide to assess excitotoxic neuronal death in primary mixed cortical cultures. Methods Mol. Biol. 2007, 399, 15–29. [Google Scholar] [CrossRef]
- Semkova, I.; Krieglstein, J. Neuroprotection mediated via neurotrophic factors and induction of neurotrophic factors. Brain Res. Brain Res. Rev. 1999, 30, 176–188. [Google Scholar] [CrossRef]
- Bullock, R.; Zauner, A.; Woodward, J.J.; Myseros, J.; Choi, S.C.; Ward, J.D.; Marmarou, A.; Young, H.F. Factors affecting excitatory amino acid release following severe human head injury. J. Neurosurg. 1998, 89, 507–518. [Google Scholar] [CrossRef] [PubMed]
- O’Byrne, M.; Tipton, K.; McBean, G.; Kollegger, H. Assessment of neurotoxicity and “neuroprotection”. J. Neural. Transm. Suppl. 1997, 50, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Takaki, J.; Fujimori, K.; Miura, M.; Suzuki, T.; Sekino, Y.; Sato, K. L-glutamate released from activated microglia downregulates astrocytic L-glutamate transporter expression in neuroinflammation: The ‘collusion’ hypothesis for increased extracellular L-glutamate concentration in neuroinflammation. J. Neuroinflamm. 2012, 9, 275. [Google Scholar] [CrossRef] [PubMed]
- Hama, H.; Hioki, H.; Namiki, K.; Hoshida, T.; Kurokawa, H.; Ishidate, F.; Kaneko, T.; Akagi, T.; Saito, T.; Saido, T.; et al. ScaleS: An optical clearing palette for biological imaging. Nat. Neurosci. 2015, 18, 1518–1529. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, N.; Odawara, A.; Katoh, H.; Okuyama, N.; Yokoi, R.; Suzuki, I. Detection of synchronized burst firing in cultured human induced pluripotent stem cell-derived neurons using a 4-step method. Biochem. Biophys. Res. Commun. 2018, 497, 612–618. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, K.; Ishibashi, Y.; Chujo, K.; Suzuki, I.; Sato, K. Neuroprotective Potential of L-Glutamate Transporters in Human Induced Pluripotent Stem Cell-Derived Neural Cells against Excitotoxicity. Int. J. Mol. Sci. 2023, 24, 12605. https://doi.org/10.3390/ijms241612605
Takahashi K, Ishibashi Y, Chujo K, Suzuki I, Sato K. Neuroprotective Potential of L-Glutamate Transporters in Human Induced Pluripotent Stem Cell-Derived Neural Cells against Excitotoxicity. International Journal of Molecular Sciences. 2023; 24(16):12605. https://doi.org/10.3390/ijms241612605
Chicago/Turabian StyleTakahashi, Kanako, Yuto Ishibashi, Kaori Chujo, Ikuro Suzuki, and Kaoru Sato. 2023. "Neuroprotective Potential of L-Glutamate Transporters in Human Induced Pluripotent Stem Cell-Derived Neural Cells against Excitotoxicity" International Journal of Molecular Sciences 24, no. 16: 12605. https://doi.org/10.3390/ijms241612605
APA StyleTakahashi, K., Ishibashi, Y., Chujo, K., Suzuki, I., & Sato, K. (2023). Neuroprotective Potential of L-Glutamate Transporters in Human Induced Pluripotent Stem Cell-Derived Neural Cells against Excitotoxicity. International Journal of Molecular Sciences, 24(16), 12605. https://doi.org/10.3390/ijms241612605



