NO Is Not the Same as GSNO in the Regulation of Fe Deficiency Responses by Dicot Plants
Abstract
1. Introduction
2. NO and GSNO in Plants
3. Role of NO and GSNO in the Responses of Plants to Mineral Stresses
4. Fe Deficiency Responses in Dicot Plants
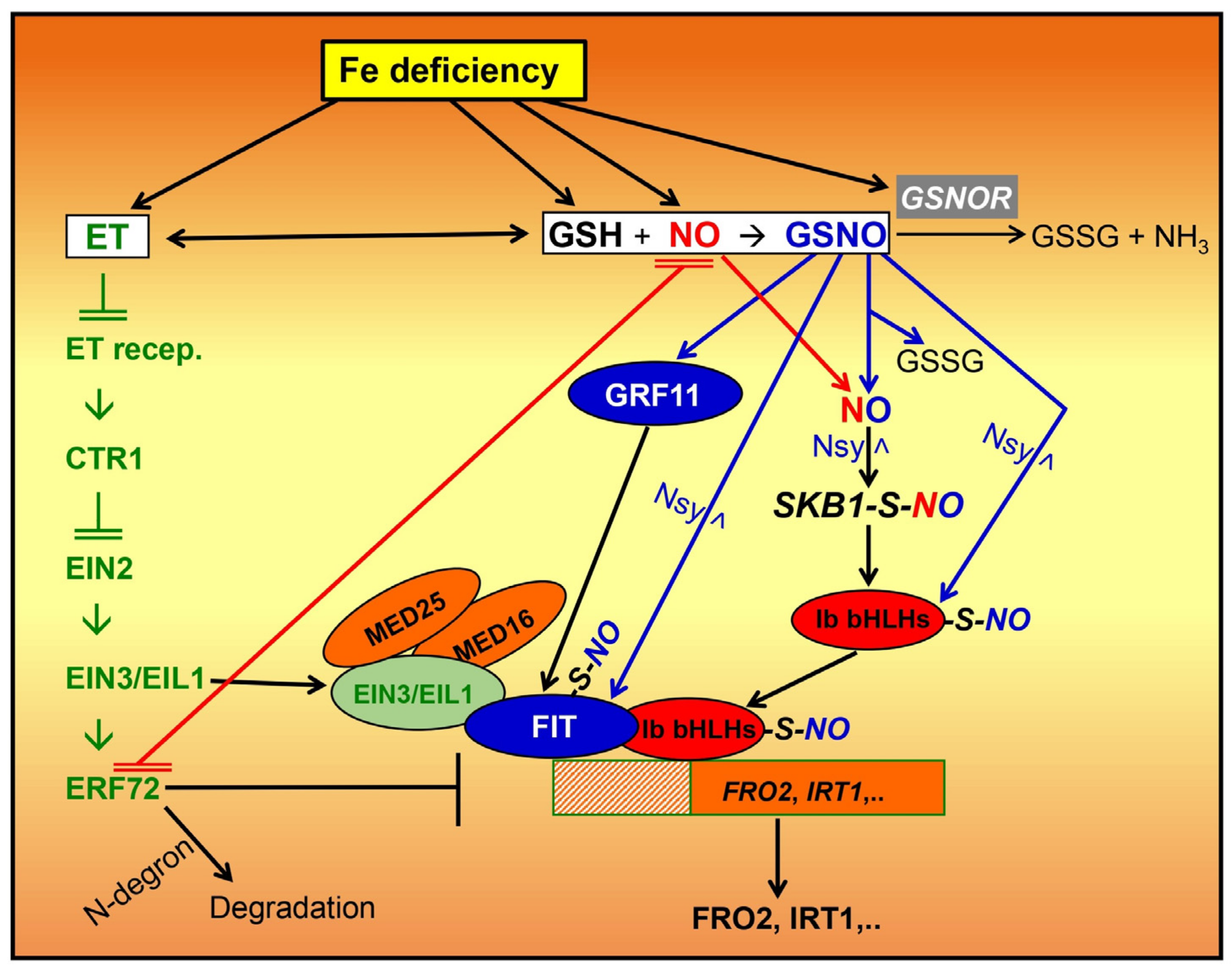
5. Role of NO and GSNO in the Regulation of Fe Deficiency Responses by Dicot Plants
6. Interactions of NO and GSNO with Ethylene and Auxin in the Regulation of Fe Deficiency Responses by Dicot Plants
7. Why Is NO Not the Same as GSNO in the Regulation of Fe Deficiency Responses?
8. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Briat, J.-F.; Dubos, C.; Gaymard, F. Iron Nutrition, Biomass Production, and Plant Product Quality. Trends Plant. Sci. 2015, 20, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Vélez-Bermúdez, I.C.; Schmidt, W. Plant Strategies to Mine Iron from Alkaline Substrates. Plant. Soil. 2023, 483, 1–25. [Google Scholar] [CrossRef]
- Lucena, C.; Romera, F.J.; García, M.J.; Alcántara, E.; Pérez-Vicente, R. Ethylene Participates in the Regulation of Fe Deficiency Responses in Strategy I Plants and in Rice. Front. Plant Sci. 2015, 6, 1056. [Google Scholar] [CrossRef] [PubMed]
- Liang, G. Iron Uptake, Signaling, and Sensing in Plants. Plant Commun. 2022, 3, 100349. [Google Scholar] [CrossRef] [PubMed]
- Brumbarova, T.; Bauer, P.; Ivanov, R. Molecular Mechanisms Governing Arabidopsis Iron Uptake. Trends Plant Sci. 2015, 20, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, B.; Bauer, P. FIT, a Regulatory Hub for Iron Deficiency and Stress Signaling in Roots, and FIT-Dependent and -Independent Gene Signatures. J. Exp. Bot. 2020, 71, 1694–1705. [Google Scholar] [CrossRef]
- García, M.J.; Lucena, C.; Romera, F.J.; Alcántara, E.; Pérez-Vicente, R. Ethylene and Nitric Oxide Involvement in the Up-Regulation of Key Genes Related to Iron Acquisition and Homeostasis in Arabidopsis. J. Exp. Bot. 2010, 61, 3885–3899. [Google Scholar] [CrossRef]
- Romera, F.J.; García, M.J.; Alcántara, E.; Pérez-Vicente, R. Latest Findings about the Interplay of Auxin, Ethylene and Nitric Oxide in the Regulation of Fe Deficiency Responses by Strategy I Plants. Plant Signal. Behav. 2011, 6, 167–170. [Google Scholar] [CrossRef]
- Romera, F.J.; Lucena, C.; García, M.J.; Alcántara, E.; Pérez-Vicente, R. The Role of Ethylene and Other Signals in the Regulation of Fe Deficiency Responses by Dicot Plants. In Stress Signaling in Plants: Genomics and Proteomics Perspective; Springer International Publishing: Cham, Switzerland, 2017; Volume 2, pp. 277–300. [Google Scholar]
- Koen, E.; Szymańska, K.; Klinguer, A.; Dobrowolska, G.; Besson-Bard, A.; Wendehenne, D. Nitric Oxide and Glutathione Impact the Expression of Iron Uptake- and Iron Transport-Related Genes as Well as the Content of Metals in A Thaliana Plants Grown under Iron Deficiency. Plant Signal. Behav. 2012, 7, 1246–1250. [Google Scholar] [CrossRef]
- Shanmugam, V.; Wang, Y.-W.; Tsednee, M.; Karunakaran, K.; Yeh, K.-C. Glutathione Plays an Essential Role in Nitric Oxide-Mediated Iron-Deficiency Signaling and Iron-Deficiency Tolerance in Arabidopsis. Plant J. 2015, 84, 464–477. [Google Scholar] [CrossRef]
- Shee, R.; Ghosh, S.; Khan, P.; Sahid, S.; Roy, C.; Shee, D.; Paul, S.; Datta, R. Glutathione Regulates Transcriptional Activation of Iron Transporters via S-Nitrosylation of BHLH Factors to Modulate Subcellular Iron Homoeostasis. Plant Cell Environ. 2022, 45, 2176–2190. [Google Scholar] [CrossRef] [PubMed]
- García, M.J.; Suárez, V.; Romera, F.J.; Alcántara, E.; Pérez-Vicente, R. A New Model Involving Ethylene, Nitric Oxide and Fe to Explain the Regulation of Fe-Acquisition Genes in Strategy I Plants. Plant Physiol. Biochem. 2011, 49, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Datta, R.; Kumar, D.; Sultana, A.; Hazra, S.; Bhattacharyya, D.; Chattopadhyay, S. Glutathione Regulates ACC Synthase Transcription via WRKY33 and ACC Oxidase by Modulating MRNA Stability to Induce Ethylene Synthesis during Stress. Plant Physiol. 2015, 169, 2963–2981. [Google Scholar] [CrossRef] [PubMed]
- García, M.J.; Corpas, F.J.; Lucena, C.; Alcántara, E.; Pérez-Vicente, R.; Zamarreño, Á.M.; Bacaicoa, E.; García-Mina, J.M.; Bauer, P.; Romera, F.J. A Shoot Fe Signaling Pathway Requiring the OPT3 Transporter Controls GSNO Reductase and Ethylene in Arabidopsis thaliana Roots. Front. Plant Sci. 2018, 9, 1325. [Google Scholar] [CrossRef]
- Kailasam, S.; Wang, Y.; Lo, J.-C.; Chang, H.-F.; Yeh, K.-C. S-Nitrosoglutathione Works Downstream of Nitric Oxide to Mediate Iron-Deficiency Signaling in Arabidopsis. Plant J. 2018, 94, 157–168. [Google Scholar] [CrossRef]
- Corpas, F.J.; Leterrier, M.; Valderrama, R.; Airaki, M.; Chaki, M.; Palma, J.M.; Barroso, J.B. Nitric Oxide Imbalance Provokes a Nitrosative Response in Plants under Abiotic Stress. Plant Sci. 2011, 181, 604–611. [Google Scholar] [CrossRef]
- Lindermayr, C. Crosstalk between Reactive Oxygen Species and Nitric Oxide in Plants: Key Role of S-Nitrosoglutathione Reductase. Free Radic. Biol. Med. 2018, 122, 110–115. [Google Scholar] [CrossRef]
- Feng, J.; Chen, L.; Zuo, J. Protein S-Nitrosylation in Plants: Current Progresses and Challenges. J. Integr. Plant Biol. 2019, 61, 1206–1223. [Google Scholar] [CrossRef]
- Tewari, R.K.; Horemans, N.; Watanabe, M. Evidence for a Role of Nitric Oxide in Iron Homeostasis in Plants. J. Exp. Bot. 2021, 72, 990–1006. [Google Scholar] [CrossRef]
- Kolbert, Z.; Feigl, G.; Freschi, L.; Poór, P. Gasotransmitters in Action: Nitric Oxide-Ethylene Crosstalk during Plant Growth and Abiotic Stress Responses. Antioxidants 2019, 8, 167. [Google Scholar] [CrossRef]
- Domingos, P.; Prado, A.M.; Wong, A.; Gehring, C.; Feijo, J.A. Nitric Oxide: A Multitasked Signaling Gas in Plants. Mol. Plant 2015, 8, 506–520. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, E.E.; Hüther, P.; Forné, I.; Georgii, E.; Han, Y.; Hell, R.; Wirtz, M.; Imhof, A.; Becker, C.; Durner, J.; et al. GSNOR Contributes to Demethylation and Expression of Transposable Elements and Stress-Responsive Genes. Antioxidants 2021, 10, 1128. [Google Scholar] [CrossRef] [PubMed]
- Mahawar, L.; Ramasamy, K.P.; Pandey, A.; Prasad, S.M. Iron Deficiency in Plants: An Update on Homeostasis and Its Regulation by Nitric Oxide and Phytohormones. Plant Growth Regul. 2022, 100, 283–299. [Google Scholar] [CrossRef]
- Pande, A.; Mun, B.G.; Rahim, W.; Khan, M.; Lee, D.S.; Lee, G.M.; Al Azzawi, T.N.I.; Hussain, A.; Kim, C.K.; Yun, B.W. Phytohormonal Regulation Through Protein S-Nitrosylation Under Stress. Front. Plant Sci. 2022, 13, 865542. [Google Scholar] [CrossRef]
- Fancy, N.N.; Bahlmann, A.; Loake, G.J. Nitric Oxide Function in Plant Abiotic Stress. Plant Cell Environ. 2017, 40, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Sahay, S.; Gupta, M. An Update on Nitric Oxide and Its Benign Role in Plant Responses under Metal Stress. Nitric Oxide 2017, 67, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Nabi, R.B.S.; Tayade, R.; Hussain, A.; Kulkarni, K.P.; Imran, Q.M.; Mun, B.G.; Yun, B.W. Nitric Oxide Regulates Plant Responses to Drought, Salinity, and Heavy Metal Stress. Environ. Exp. Bot. 2019, 161, 120–133. [Google Scholar] [CrossRef]
- Zhou, X.; Joshi, S.; Khare, T.; Patil, S.; Shang, J.; Kumar, V. Nitric Oxide, Crosstalk with Stress Regulators and Plant Abiotic Stress Tolerance. Plant Cell Rep. 2021, 40, 1395–1414. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Ali, S.; Al Azzawi, T.N.I.; Yun, B.-W. Nitric Oxide Acts as a Key Signaling Molecule in Plant Development under Stressful Conditions. Int. J. Mol. Sci. 2023, 24, 4782. [Google Scholar] [CrossRef]
- Kumar, D.; Ohri, P. Say “NO” to Plant Stresses: Unravelling the Role of Nitric Oxide under Abiotic and Biotic Stress. Nitric Oxide 2023, 130, 36–57. [Google Scholar] [CrossRef]
- Wang, X.; Du, H.; Ma, M.; Rennenberg, H. The Dual Role of Nitric Oxide (NO) in Plant Responses to Cadmium Exposure. Sci. Total Environ. 2023, 892, 164597. [Google Scholar] [CrossRef]
- Singhal, R.K.; Jatav, H.S.; Aftab, T.; Pandey, S.; Mishra, U.N.; Chauhan, J.; Chand, S.; Indu; Saha, D.; Dadarwal, B.K.; et al. Roles of Nitric Oxide in Conferring Multiple Abiotic Stress Tolerance in Plants and Crosstalk with Other Plant Growth Regulators. J. Plant Growth Regul. 2021, 40, 2303–2328. [Google Scholar] [CrossRef]
- Graziano, M.; Lamattina, L. Nitric Oxide Accumulation Is Required for Molecular and Physiological Responses to Iron Deficiency in Tomato Roots. Plant J. 2007, 52, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Marcos, M.; Sanz, L.; Lewis, D.R.; Muday, G.K.; Lorenzo, O. Nitric Oxide Causes Root Apical Meristem Defects and Growth Inhibition While Reducing PIN-FORMED 1 (PIN1)-Dependent Acropetal Auxin Transport. Proc. Natl. Acad. Sci. USA 2011, 108, 18506–18511. [Google Scholar] [CrossRef]
- Yun, B.; Skelly, M.J.; Yin, M.; Yu, M.; Mun, B.; Lee, S.; Hussain, A.; Spoel, S.H.; Loake, G.J. Nitric Oxide and S-nitrosoglutathione Function Additively during Plant Immunity. New Phytol. 2016, 211, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Jahan, B.; Rasheed, F.; Sehar, Z.; Fatma, M.; Iqbal, N.; Masood, A.; Anjum, N.A.; Khan, N.A. Coordinated Role of Nitric Oxide, Ethylene, Nitrogen, and Sulfur in Plant Salt Stress Tolerance. Stresses 2021, 1, 181–199. [Google Scholar] [CrossRef]
- Kovacs, I.; Ageeva, A.; König, E.E.; Lindermayr, C. S-Nitrosylation of Nuclear Proteins: New Pathways in Regulation of Gene Expression. Adv. Bot. Res. 2016, 77, 15–39. [Google Scholar] [CrossRef]
- Malik, S.I.; Hussain, A.; Yun, B.-W.; Spoel, S.H.; Loake, G.J. GSNOR-Mediated de-Nitrosylation in the Plant Defence Response. Plant Sci. 2011, 181, 540–544. [Google Scholar] [CrossRef]
- Innocenti, G.; Pucciariello, C.; Le Gleuher, M.; Hopkins, J.; De Stefano, M.; Delledonne, M.; Puppo, A.; Baudouin, E.; Frendo, P. Glutathione Synthesis Is Regulated by Nitric Oxide in Medicago truncatula Roots. Planta 2007, 225, 1597–1602. [Google Scholar] [CrossRef]
- Corpas, F.J.; Alché, J.D.; Barroso, J.B. Current Overview of S-Nitrosoglutathione (GSNO) in Higher Plants. Front. Plant Sci. 2013, 4, 126. [Google Scholar] [CrossRef]
- Mioto, P.T.; Rodríguez-Ruiz, M.; Mot, A.C.; Zuccarelli, R.; Corpas, F.J.; Freschi, L.; Mercier, H. Alternative Fluorimetric-Based Method to Detect and Compare Total S-Nitrosothiols in Plants. Nitric Oxide 2017, 68, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, M.; Chaki, M.; Airaki, M.; Valderrama, R.; Palma, J.M.; Barroso, J.B.; Corpas, F.J. Function of S-Nitrosoglutathione Reductase (GSNOR) in Plant Development and under Biotic/Abiotic Stress. Plant Signal. Behav. 2011, 6, 789–793. [Google Scholar] [CrossRef]
- Xu, S.; Guerra, D.; Lee, U.; Vierling, E. S-Nitrosoglutathione Reductases Are Low-Copy Number, Cysteine-Rich Proteins in Plants That Control Multiple Developmental and Defense Responses in Arabidopsis. Front. Plant Sci. 2013, 4, 430. [Google Scholar] [CrossRef] [PubMed]
- Kubienová, L.; Tichá, T.; Jahnová, J.; Luhová, L.; Mieslerová, B.; Petřivalský, M. Effect of Abiotic Stress Stimuli on S-Nitrosoglutathione Reductase in Plants. Planta 2014, 239, 139–146. [Google Scholar] [CrossRef]
- Jahnová, J.; Luhová, L.; Petřivalský, M. S-Nitrosoglutathione Reductase—The Master Regulator of Protein S-Nitrosation in Plant NO Signaling. Plants 2019, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Ventimiglia, L.; Mutus, B. The Physiological Implications of S-Nitrosoglutathione Reductase (GSNOR) Activity Mediating NO Signalling in Plant Root Structures. Antioxidants 2020, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Sun, C.; Lin, X.; Busch, W. The Emerging Role of GSNOR in Oxidative Stress Regulation. Trends Plant Sci. 2021, 26, 156–168. [Google Scholar] [CrossRef]
- Kubienová, L.; Kopečný, D.; Tylichová, M.; Briozzo, P.; Skopalová, J.; Šebela, M.; Navrátil, M.; Tâche, R.; Luhová, L.; Barroso, J.B.; et al. Structural and Functional Characterization of a Plant S-Nitrosoglutathione Reductase from Solanum lycopersicum. Biochimie 2013, 95, 889–902. [Google Scholar] [CrossRef]
- Kwon, E.; Feechan, A.; Yun, B.W.; Hwang, B.H.; Pallas, J.A.; Kang, J.G.; Loake, G.J. AtGSNOR1 Function Is Required for Multiple Developmental Programs in Arabidopsis. Planta 2012, 236, 887–900. [Google Scholar] [CrossRef]
- Guan, M.Y.; Zhu, Y.X.; Liu, X.X.; Jin, C.W. Induction of S-Nitrosoglutathione Reductase Reduces Root Cadmium Uptake by Inhibiting Iron-Regulated Transporter 1. Plant Soil. 2019, 438, 251–262. [Google Scholar] [CrossRef]
- Hussain, A.; Yun, B.W.; Kim, J.H.; Gupta, K.J.; Hyung, N.I.; Loake, G.J. Novel and Conserved Functions of S-Nitrosoglutathione Reductase in Tomato. J. Exp. Bot. 2019, 70, 4877–4886. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Sun, S.; Yang, W.; Zhang, L.; Liu, S.; Gong, B.; Shi, Q. Overexpression of S-Nitrosoglutathione Reductase Alleviated Iron-Deficiency Stress by Regulating Iron Distribution and Redox Homeostasis. J. Plant Physiol. 2019, 237, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Terrile, M.C.; París, R.; Calderón-Villalobos, L.I.A.; Iglesias, M.J.; Lamattina, L.; Estelle, M.; Casalongué, C.A. Nitric Oxide Influences Auxin Signaling through S-Nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 Auxin Receptor. Plant J. 2012, 70, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Freschi, L. Nitric Oxide and Phytohormone Interactions: Current Status and Perspectives. Front. Plant Sci. 2013, 4, 394. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, X.X.; He, X.L.; Liu, L.J.; Wu, H.; Tang, C.X.; Zhang, Y.S.; Jin, C.W. Ethylene and Nitric Oxide Interact to Regulate the Magnesium Deficiency-Induced Root Hair Development in Arabidopsis. New Phytol. 2017, 213, 1242–1256. [Google Scholar] [CrossRef]
- Liu, M.; Wei, J.W.; Liu, W.; Gong, B. S-Nitrosylation of ACO Homolog 4 Improves Ethylene Synthesis and Salt Tolerance in Tomato. New Phytol. 2023, 239, 159–173. [Google Scholar] [CrossRef]
- Lynch, J.P.; St. Clair, S.B. Mineral Stress: The Missing Link in Understanding How Global Climate Change Will Affect Plants in Real World Soils. Field Crops Res. 2004, 90, 101–115. [Google Scholar] [CrossRef]
- Pető, A.; Lehotai, N.; Feigl, G.; Tugyi, N.; Ördög, A.; Gémes, K.; Tari, I.; Erdei, L.; Kolbert, Z. Nitric Oxide Contributes to Copper Tolerance by Influencing ROS Metabolism in Arabidopsis. Plant Cell Rep. 2013, 32, 1913–1923. [Google Scholar] [CrossRef]
- Li, B.; Sun, L.; Huang, J.; Göschl, C.; Shi, W.; Chory, J.; Busch, W. GSNOR Provides Plant Tolerance to Iron Toxicity via Preventing Iron-Dependent Nitrosative and Oxidative Cytotoxicity. Nat. Commun. 2019, 10, 3896. [Google Scholar] [CrossRef]
- Pan, C.; Li, X.; Yao, S.; Luo, S.; Liu, S.; Wang, A.; Xiao, D.; Zhan, J.; He, L. S-Nitrosated Proteomic Analysis Reveals the Regulatory Roles of Protein S-Nitrosation and S-Nitrosoglutathione Reductase during Al-Induced PCD in Peanut Root Tips. Plant Sci. 2021, 308, 110931. [Google Scholar] [CrossRef]
- Kolbert, Z.; Ördög, A. Involvement of Nitric Oxide (NO) in Plant Responses to Metalloids. J. Hazard. Mater. 2021, 420, 126606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Jia, L.; Chu, H.; Wu, D.; Peng, X.; Liu, X.; Zhang, J.; Zhao, J.; Chen, K.; Zhao, L. Arabidopsis CaM1 and CaM4 Promote Nitric Oxide Production and Salt Resistance by Inhibiting S-Nitrosoglutathione Reductase via Direct Binding. PLoS Genet. 2016, 12, e1006255. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Bhatla, S.C. Nitric Oxide Regulates Lateral Root Formation through Modulation of ACC Oxidase Activity in Sunflower Seedlings under Salt Stress. Plant Signal. Behav. 2018, 13, e1473683. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, M.; Airaki, M.; Palma, J.M.; Chaki, M.; Barroso, J.B.; Corpas, F.J. Arsenic Triggers the Nitric Oxide (NO) and S-Nitrosoglutathione (GSNO) Metabolism in Arabidopsis. Environ. Pollut. 2012, 166, 136–143. [Google Scholar] [CrossRef]
- Meng, Z.B.; Chen, L.Q.; Suo, D.; Li, G.X.; Tang, C.X.; Zheng, S.J. Nitric Oxide Is the Shared Signalling Molecule in Phosphorus- and Iron-Deficiency-Induced Formation of Cluster Roots in White Lupin (Lupinus albus). Ann. Bot. 2012, 109, 1055–1064. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, H.; Fang, X.; Zhang, Y.; Jin, C. Auxin Acts Downstream of Ethylene and Nitric Oxide to Regulate Magnesium Deficiency-Induced Root Hair Development in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1452–1465. [Google Scholar] [CrossRef]
- Buet, A.; Galatro, A.; Ramos-Artuso, F.; Simontacchi, M. Nitric Oxide and Plant Mineral Nutrition: Current Knowledge. J. Exp. Bot. 2019, 70, 4461–4476. [Google Scholar] [CrossRef]
- Galatro, A.; Ramos-Artuso, F.; Luquet, M.; Buet, A.; Simontacchi, M. An Update on Nitric Oxide Production and Role Under Phosphorus Scarcity in Plants. Front. Plant Sci. 2020, 11, 413. [Google Scholar] [CrossRef]
- Siddiqui, M.H.; Alamri, S.; Alsubaie, Q.D.; Ali, H.M.; Khan, M.N.; Al-Ghamdi, A.; Ibrahim, A.A.; Alsadon, A. Exogenous Nitric Oxide Alleviates Sulfur Deficiency-Induced Oxidative Damage in Tomato Seedlings. Nitric Oxide 2020, 94, 95–107. [Google Scholar] [CrossRef]
- García, M.J.; Lucena, C.; Romera, F.J. Ethylene and Nitric Oxide Involvement in the Regulation of Fe and P Deficiency Responses in Dicotyledonous Plants. Int. J. Mol. Sci. 2021, 22, 4904. [Google Scholar] [CrossRef]
- Gao, F.; Dubos, C. Transcriptional Integration of Plant Responses to Iron Availability. J. Exp. Bot. 2021, 72, 2056–2070. [Google Scholar] [CrossRef]
- Waters, B.M.; Lucena, C.; Romera, F.J.; Jester, G.G.; Wynn, A.N.; Rojas, C.L.; Alcántara, E.; Pérez-Vicente, R. Ethylene Involvement in the Regulation of the H+-ATPase CsHA1 Gene and of the New Isolated Ferric Reductase CsFRO1 and Iron Transporter CsIRT1 Genes in Cucumber Plants. Plant Physiol. Biochem. 2007, 45, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.H.; Schmidt, W. Mobilization of Iron by Plant-Borne Coumarins. Trends Plant. Sci. 2017, 22, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Lingam, S.; Mohrbacher, J.; Brumbarova, T.; Potuschak, T.; Fink-Straube, C.; Blondet, E.; Genschik, P.; Bauer, P. Interaction between the BHLH Transcription Factor FIT and Ethylene Insensitive3/Ethylene Insensitive3-LIKE1 Reveals Molecular Linkage between the Regulation of Iron Acquisition and Ethylene Signaling in Arabidopsis. Plant Cell 2011, 23, 1815–1829. [Google Scholar] [CrossRef]
- Meiser, J.; Lingam, S.; Bauer, P. Posttranslational Regulation of the Iron Deficiency Basic Helix-Loop-Helix Transcription Factor FIT Is Affected by Iron and Nitric Oxide. Plant Physiol. 2011, 157, 2154–2166. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Chen, W.W.; Chen, L.Q.; Qin, C.; Jin, C.W.; Shi, Y.Z.; Zheng, S.J. The 14-3-3 Protein GENERAL REGULATORY FACTOR11 (GRF11) Acts Downstream of Nitric Oxide to Regulate Iron Acquisition in Arabidopsis thaliana. New Phytol. 2013, 197, 815–824. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Conde, J.V.; Berckhan, S.; Prasad, G.; Mendiondo, G.M.; Holdsworth, M.J. Group VII Ethylene Response Factors Coordinate Oxygen and Nitric Oxide Signal Transduction and Stress Responses in Plants. Plant Physiol. 2015, 169, 23–31. [Google Scholar] [CrossRef]
- Liu, W.; Li, Q.; Wang, Y.; Wu, T.; Yang, Y.; Zhang, X.; Han, Z.; Xu, X. Ethylene Response Factor AtERF72 Negatively Regulates Arabidopsis thaliana Response to Iron Deficiency. Biochem. Biophys. Res. Commun. 2017, 491, 862–868. [Google Scholar] [CrossRef]
- Binder, B.M. Ethylene Signaling in Plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef]
- León, J.; Costa-Broseta, Á.; Castillo, M.C. RAP2.3 Negatively Regulates Nitric Oxide Biosynthesis and Related Responses through a Rheostat-like Mechanism in Arabidopsis. J. Exp. Bot. 2020, 71, 3157–3171. [Google Scholar] [CrossRef]
- Rodríguez-Celma, J.; Connorton, J.M.; Kruse, I.; Green, R.T.; Franceschetti, M.; Chen, Y.T.; Cui, Y.; Ling, H.Q.; Yeh, K.C.; Balk, J. Arabidopsis BRUTUS-LIKE E3 Ligases Negatively Regulate Iron Uptake by Targeting Transcription Factor FIT for Recycling. Proc. Natl. Acad. Sci. USA 2019, 116, 17584–17591. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kai Lu, C.; Yang Li, C.; Hua Lei, R.; Na Pu, M.; Hui Zhao, J.; Peng, F.; Qian Ping, H.; Wang, D.; Liang, G. IRON MAN Interacts with BRUTUS to Maintain Iron Homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2109063118. [Google Scholar] [CrossRef] [PubMed]
- Lichtblau, D.M.; Schwarz, B.; Baby, D.; Endres, C.; Sieberg, C.; Bauer, P. The Iron Deficiency-Regulated Small Protein Effector FEP3/IRON MAN1 Modulates Interaction of BRUTUS-LIKE1 with BHLH Subgroup IVc and POPEYE Transcription Factors. Front. Plant Sci. 2022, 13, 930049. [Google Scholar] [CrossRef]
- Grillet, L.; Lan, P.; Li, W.; Mokkapati, G.; Schmidt, W. IRON MAN Is a Ubiquitous Family of Peptides That Control Iron Transport in Plants. Nat. Plants 2018, 4, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Lei, G.J.; Yamaji, N.; Nakagawa, N.; Ma, J.F. The Putative Peptide Gene FEP1 Regulates Iron Deficiency Response in Arabidopsis. Plant Cell Physiol. 2018, 59, 1739–1752. [Google Scholar] [CrossRef] [PubMed]
- García, M.J.; Angulo, M.; Romera, F.J.; Lucena, C.; Pérez-Vicente, R. A Shoot Derived Long Distance Iron Signal May Act Upstream of the IMA Peptides in the Regulation of Fe Deficiency Responses in Arabidopsis thaliana Roots. Front. Plant Sci. 2022, 13, 971773. [Google Scholar] [CrossRef]
- Romera, F.J.; Alcantara, E. Iron-Deficiency Stress Responses in Cucumber (Cucumis Sativus L.) Roots. A Possible Role for Ethylene? Plant Physiol. 1994, 105, 1133–1138. [Google Scholar] [CrossRef]
- Romera, F.J.; Alcántara, E. Ethylene Involvement in the Regulation of Fe-Deficiency Stress Responses by Strategy I Plants. Funct. Plant Biol. 2004, 31, 315–328. [Google Scholar] [CrossRef]
- Li, W.; Lan, P. The Understanding of the Plant Iron Deficiency Responses in Strategy I Plants and the Role of Ethylene in This Process by Omic Approaches. Front. Plant Sci. 2017, 8, 40. [Google Scholar] [CrossRef]
- Angulo, M.; García, M.J.; Alcántara, E.; Pérez-Vicente, R.; Romera, F.J. Comparative Study of Several Fe Deficiency Responses in the Arabidopsis thaliana Ethylene Insensitive Mutants Ein2-1 and Ein2-5. Plants 2021, 10, 262. [Google Scholar] [CrossRef]
- García, M.J.; Romera, F.J.; Lucena, C.; Alcántara, E.; Pérez-Vicente, R. Ethylene and the Regulation of Physiological and Morphological Responses to Nutrient Deficiencies. Plant Physiol. 2015, 169, 51–60. [Google Scholar] [CrossRef]
- Iqbal, N.; Gautam, H.; Khan, M.I.R.; Per, T.S.; Khan, N.A.; Umar, S. Crosstalk between Ethylene and Mineral Nutrients in Regulation of Morphophysiological Traits and Nutrients Homeostasis in Plants. In The Plant Hormone Ethylene; Elsevier: Amsterdam, The Netherlands, 2023; pp. 191–209. [Google Scholar]
- Ma, B.; Ma, T.; Xian, W.; Hu, B.; Chu, C. Interplay between Ethylene and Nitrogen Nutrition: How Ethylene Orchestrates Nitrogen Responses in Plants. J. Integr. Plant Biol. 2023, 65, 399–407. [Google Scholar] [CrossRef]
- Romera, F.J.; Smith, A.P.; Pérez-Vicente, R. Editorial: Ethylene’s Role in Plant Mineral Nutrition. Front. Plant Sci. 2016, 7, 911. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Yang, J.L.; Qin, C.; Jin, C.W.; Mo, J.H.; Ye, T.; Zheng, S.J. Nitric Oxide Acts Downstream of Auxin to Trigger Root Ferric-Chelate Reductase Activity in Response to Iron Deficiency in Arabidopsis. Plant Physiol. 2010, 154, 810–819. [Google Scholar] [CrossRef]
- Jin, C.W.; Du, S.T.; Shamsi, I.H.; Luo, B.F.; Lin, X.Y. NO Synthase-Generated NO Acts Downstream of Auxin in Regulating Fe-Deficiency-Induced Root Branching That Enhances Fe-Deficiency Tolerance in Tomato Plants. J. Exp. Bot. 2011, 62, 3875–3884. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Dong, Y.; Xu, L.; Liu, S.; Bai, X. Role of Exogenous Nitric Oxide in Alleviating Iron Deficiency-Induced Peanut Chlorosis on Calcareous Soil. J. Plant Interact. 2014, 9, 450–459. [Google Scholar] [CrossRef]
- Buet, A.; Simontacchi, M. Nitric Oxide and Plant Iron Homeostasis. Ann. N. Y. Acad. Sci. 2015, 1340, 39–46. [Google Scholar] [CrossRef]
- Song, Y.L.; Dong, Y.J.; Tian, X.Y.; Wang, W.W.; He, Z.L. Effects of Nitric Oxide and Fe Supply on Recovery of Fe Deficiency Induced Chlorosis in Peanut Plants. Biol. Plant 2017, 61, 155–168. [Google Scholar] [CrossRef]
- Song, Y.; Dong, Y.; Tian, X.; Wang, W.; He, Z. Mechanisms of Exogenous Nitric Oxide and 24-Epibrassinolide Alleviating Chlorosis of Peanut Plants Under Iron Deficiency. Pedosphere 2018, 28, 926–942. [Google Scholar] [CrossRef]
- Kabir, A.H.; Ela, E.J.; Bagchi, R.; Rahman, M.A.; Peiter, E.; Lee, K.-W. Nitric Oxide Acts as an Inducer of Strategy-I Responses to Increase Fe Availability and Mobilization in Fe-Starved Broccoli (Brassica oleracea Var. Oleracea). Plant Physiol. Biochem. 2023, 194, 182–192. [Google Scholar] [CrossRef]
- Liu, X.X.; He, X.L.; Jin, C.W. Roles of Chemical Signals in Regulation of the Adaptive Responses to Iron Deficiency. Plant Signal. Behav. 2016, 11, e1179418. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.Q.; Jin, C.W.; Fan, S.K.; Mao, Q.Q.; Sun, C.L.; Yu, Y.; Lin, X.Y. Elevation of NO Production Increases Fe Immobilization in the Fe-Deficiency Roots Apoplast by Decreasing Pectin Methylation of Cell Wall. Sci. Rep. 2015, 5, 10746. [Google Scholar] [CrossRef]
- Zhu, X.F.; Wang, B.; Song, W.F.; Zheng, S.J.; Shen, R.F. Putrescine Alleviates Iron Deficiency via NO-Dependent Reutilization of Root Cell-Wall Fe in Arabidopsis. Plant Physiol. 2015, 170, 558–567. [Google Scholar] [CrossRef]
- Fan, H.; Zhang, Z.; Wang, N.; Cui, Y.; Sun, H.; Liu, Y.; Wu, H.; Zheng, S.; Bao, S.; Ling, H.-Q. SKB1/PRMT5-Mediated Histone H4R3 Dimethylation of Ib Subgroup BHLH Genes Negatively Regulates Iron Homeostasis in Arabidopsis thaliana. Plant J. 2014, 77, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Huang, X.; Chen, L.; Sun, X.; Lu, C.; Zhang, L.; Wang, Y.; Zuo, J. Site-Specific Nitrosoproteomic Identification of Endogenously S-Nitrosylated Proteins in Arabidopsis. Plant Physiol. 2015, 167, 1731–1746. [Google Scholar] [CrossRef]
- Ye, L.; Li, L.; Wang, L.; Wang, S.; Li, S.; Du, J.; Zhang, S.; Shou, H. MPK3/MPK6 Are Involved in Iron Deficiency-Induced Ethylene Production in Arabidopsis. Front. Plant Sci. 2015, 6, 953. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Song, H.; Li, B.; Kronzucker, H.J.; Shi, W. Auxin Resistant1 and PIN-FORMED2 Protect Lateral Root Formation in Arabidopsis under Iron Stress. Plant Physiol. 2015, 169, 2608–2623. [Google Scholar] [CrossRef]
- Li, G.; Xu, W.; Kronzucker, H.J.; Shi, W. Ethylene Is Critical to the Maintenance of Primary Root Growth and Fe Homeostasis under Fe Stress in Arabidopsis. J. Exp. Bot. 2015, 66, 2041–2054. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.; Singh Saini, H.; Dahiya, A.; Ritu Saini, C. Iron Treatment Enhances the Levels of Reduced Glutathione, Oxidized Glutathione and Glutathione Reductase Activity in Rice (Oryza sativa L.). J. Pharmacogn. Phytochem. 2017, 6, 1321–1328. [Google Scholar]
- Zhang, L.; Li, G.; Wang, M.; Di, D.; Sun, L.; Kronzucker, H.J.; Shi, W. Excess Iron Stress Reduces Root Tip Zone Growth through Nitric Oxide-Mediated Repression of Potassium Homeostasis in Arabidopsis. New Phytol. 2018, 219, 259–274. [Google Scholar] [CrossRef]
- Ramírez, L.; Bartoli, C.G.; Lamattina, L. Glutathione and Ascorbic Acid Protect Arabidopsis Plants against Detrimental Effects of Iron Deficiency. J. Exp. Bot. 2013, 64, 3169–3178. [Google Scholar] [CrossRef]
- Rai, S.; Singh, P.K.; Mankotia, S.; Swain, J.; Satbhai, S.B. Iron Homeostasis in Plants and Its Crosstalk with Copper, Zinc, and Manganese. Plant Stress 2021, 1, 100008. [Google Scholar] [CrossRef]
- Zamioudis, C.; Korteland, J.; Van Pelt, J.A.; Hamersveld, M.; Dombrowski, N.; Bai, Y.; Hanson, J.; Van Verk, M.C.; Ling, H.; Schulze-Lefert, P.; et al. Rhizobacterial Volatiles and Photosynthesis-related Signals Coordinate MYB72 Expression in Arabidopsis Roots during Onset of Induced Systemic Resistance and Iron-deficiency Responses. Plant J. 2015, 84, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Romera, F.J.; García, M.J.; Lucena, C.; Martínez-Medina, A.; Aparicio, M.A.; Ramos, J.; Alcántara, E.; Angulo, M.; Pérez-Vicente, R. Induced Systemic Resistance (ISR) and Fe Deficiency Responses in Dicot Plants. Front. Plant Sci. 2019, 10, 287. [Google Scholar] [CrossRef]
- Pescador, L.; Fernandez, I.; Pozo, M.J.; Romero-Puertas, M.C.; Pieterse, C.M.J.; Martínez-Medina, A. Nitric Oxide Signalling in Roots Is Required for MYB72-Dependent Systemic Resistance Induced by Trichoderma Volatile Compounds in Arabidopsis. J. Exp. Bot. 2021, 73, 584–595. [Google Scholar] [CrossRef]
- Aparicio, M.A.; Lucena, C.; García, M.J.; Ruiz-Castilla, F.J.; Jiménez-Adrián, P.; López-Berges, M.S.; Prieto, P.; Alcántara, E.; Pérez-Vicente, R.; Ramos, J.; et al. The Nonpathogenic Strain of Fusarium oxysporum FO12 Induces Fe Deficiency Responses in Cucumber (Cucumis sativus L.) Plants. Planta 2023, 257, 50. [Google Scholar] [CrossRef]
- Asgher, M.; Per, T.S.; Masood, A.; Fatma, M.; Freschi, L.; Corpas, F.J.; Khan, N.A. Nitric Oxide Signaling and Its Crosstalk with Other Plant Growth Regulators in Plant Responses to Abiotic Stress. Environ. Sci. Pollut. Res. 2017, 24, 2273–2285. [Google Scholar] [CrossRef] [PubMed]
- Oláh, D.; Feigl, G.; Molnár, Á.; Ördög, A.; Kolbert, Z. Strigolactones Interact With Nitric Oxide in Regulating Root System Architecture of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 1019. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Rodríguez-Ruiz, M.; Muñoz-Vargas, M.A.; Palma, J.M. Nitric Oxide and Hydrogen Sulfide Share Regulatory Functions in Higher Plant Events. Biocell 2022, 46, 1–5. [Google Scholar] [CrossRef]
- Kohli, S.K.; Khanna, K.; Bhardwaj, R.; Corpas, F.J.; Ahmad, P. Nitric Oxide, Salicylic Acid and Oxidative Stress: Is It a Perfect Equilateral Triangle? Plant Physiol. Biochem. 2022, 184, 56–64. [Google Scholar] [CrossRef]
- Rasheed, F.; Mir, I.R.; Sehar, Z.; Fatma, M.; Gautam, H.; Khan, S.; Anjum, N.A.; Masood, A.; Sofo, A.; Khan, N.A. Nitric Oxide and Salicylic Acid Regulate Glutathione and Ethylene Production to Enhance Heat Stress Acclimation in Wheat Involving Sulfur Assimilation. Plants 2022, 11, 3131. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.K.; Bhat, J.A.; Ahmad, P.; Allakhverdiev, S.I. Polyamines and Nitric Oxide Crosstalk in Plant Development and Abiotic Stress Tolerance. Funct. Plant Biol. 2023, 50, i. [Google Scholar] [CrossRef] [PubMed]
- García, M.J.; Romera, F.J.; Stacey, M.G.; Stacey, G.; Villar, E.; Alcántara, E.; Pérez-Vicente, R. Shoot to Root Communication Is Necessary to Control the Expression of Iron-Acquisition Genes in Strategy I Plants. Planta 2013, 237, 65–75. [Google Scholar] [CrossRef]
- Yang, Y.; Ou, B.; Zhang, J.; Si, W.; Gu, H.; Qin, G.; Qu, L.-J. The Arabidopsis Mediator Subunit MED16 Regulates Iron Homeostasis by Associating with EIN3/EIL1 through Subunit MED25. Plant J. 2014, 77, 838–851. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine Salvage and S-Adenosylmethionine: Essential Links between Sulfur, Ethylene and Polyamine Biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The Pivotal Role of Ethylene in Plant Growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef]
- Zhu, X.F.; Zhu, C.Q.; Wang, C.; Dong, X.Y.; Shen, R.F. Nitric Oxide Acts Upstream of Ethylene in Cell Wall Phosphorus Reutilization in Phosphorus-Deficient Rice. J. Exp. Bot. 2017, 68, 753–760. [Google Scholar] [CrossRef]
- Melo, N.K.G.; Bianchetti, R.E.; Lira, B.S.; Oliveira, P.M.R.; Zuccarelli, R.; Dias, D.L.O.; Demarco, D.; Peres, L.E.P.; Rossi, M.; Freschi, L. Nitric Oxide, Ethylene, and Auxin Crosstalk Mediates Greening and Plastid Development in Deetiolating Tomato Seedlings. Plant Physiol. 2016, 170, 2278–2294. [Google Scholar] [CrossRef]
- Hartman, S.; Liu, Z.; van Veen, H.; Vicente, J.; Reinen, E.; Martopawiro, S.; Zhang, H.; van Dongen, N.; Bosman, F.; Bassel, G.W.; et al. Ethylene-Mediated Nitric Oxide Depletion Pre-Adapts Plants to Hypoxia Stress. Nat. Commun. 2019, 10, 4020. [Google Scholar] [CrossRef]
- Wünsche, H.; Baldwin, I.T.; Wu, J. S-Nitrosoglutathione Reductase (GSNOR) Mediates the Biosynthesis of Jasmonic Acid and Ethylene Induced by Feeding of the Insect Herbivore Manduca sexta and Is Important for Jasmonate-Elicited Responses in Nicotiana attenuata. J. Exp. Bot. 2011, 62, 4605–4616. [Google Scholar] [CrossRef]
- Rustérucci, C.; Espunya, M.C.; Díaz, M.; Chabannes, M.; Martínez, M.C. S-Nitrosoglutathione Reductase Affords Protection against Pathogens in Arabidopsis, Both Locally and Systemically. Plant Physiol. 2007, 143, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Pommerrenig, B.; Feussner, K.; Zierer, W.; Rabinovych, V.; Klebl, F.; Feussner, I.; Sauera, N. Phloem-Specific Expression of Yang Cycle Genes and Identification of Novel Yang Cycle Enzymes in Plantago and Arabidopsis. Plant Cell. 2011, 23, 1904–1919. [Google Scholar] [CrossRef]
- Yang, L.; Ji, J.; Wang, H.; Harris-Shultz, K.R.; Abd Allah, E.F.; Luo, Y.; Guan, Y.; Hu, X. Carbon Monoxide Interacts with Auxin and Nitric Oxide to Cope with Iron Deficiency in Arabidopsis. Front. Plant Sci. 2016, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Feng, F.; Liu, J.; Zhao, Q. The Interaction between Auxin and Nitric Oxide Regulates Root Growth in Response to Iron Deficiency in Rice. Front. Plant Sci. 2017, 8, 2169. [Google Scholar] [CrossRef]
- Shi, Y.F.; Wang, D.L.; Wang, C.; Culler, A.H.; Kreiser, M.A.; Suresh, J.; Cohen, J.D.; Pan, J.; Baker, B.; Liu, J.Z. Loss of GSNOR1 Function Leads to Compromised Auxin Signaling and Polar Auxin Transport. Mol. Plant 2015, 8, 1350–1365. [Google Scholar] [CrossRef] [PubMed]
- Frungillo, L.; Skelly, M.J.; Loake, G.J.; Spoel, S.H.; Salgado, I. S-Nitrosothiols Regulate Nitric Oxide Production and Storage in Plants through the Nitrogen Assimilation Pathway. Nat. Commun. 2014, 5, 5401. [Google Scholar] [CrossRef] [PubMed]

| Fe Sufficiency | Fe Deficiency | Fe Excess | Plant Species | References | |
|---|---|---|---|---|---|
| ET | + | + + + + | + + + + | Pea Cucumber Squash Arabidopsis (*) | [9,89,108,109,110] |
| GSH | + | + + + + | + + + + | Sugar beet Arabidopsis Rice (**) | [11,15,111] |
| NO | + | + + + + | + + + + | Lupinus Tomato Arabidopsis Peanut | [15,66,101,104,112] |
| GSNO | + + + + | + + | n.d. | Arabidopsis Pea | [11,15,16] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romera, F.J.; García, M.J.; Lucena, C.; Angulo, M.; Pérez-Vicente, R. NO Is Not the Same as GSNO in the Regulation of Fe Deficiency Responses by Dicot Plants. Int. J. Mol. Sci. 2023, 24, 12617. https://doi.org/10.3390/ijms241612617
Romera FJ, García MJ, Lucena C, Angulo M, Pérez-Vicente R. NO Is Not the Same as GSNO in the Regulation of Fe Deficiency Responses by Dicot Plants. International Journal of Molecular Sciences. 2023; 24(16):12617. https://doi.org/10.3390/ijms241612617
Chicago/Turabian StyleRomera, Francisco Javier, María José García, Carlos Lucena, Macarena Angulo, and Rafael Pérez-Vicente. 2023. "NO Is Not the Same as GSNO in the Regulation of Fe Deficiency Responses by Dicot Plants" International Journal of Molecular Sciences 24, no. 16: 12617. https://doi.org/10.3390/ijms241612617
APA StyleRomera, F. J., García, M. J., Lucena, C., Angulo, M., & Pérez-Vicente, R. (2023). NO Is Not the Same as GSNO in the Regulation of Fe Deficiency Responses by Dicot Plants. International Journal of Molecular Sciences, 24(16), 12617. https://doi.org/10.3390/ijms241612617









