Inhibition of Cxcr4 Disrupts Mouse Embryonic Palatal Mesenchymal Cell Migration and Induces Cleft Palate Occurrence
Abstract
1. Introduction
2. Results
2.1. The Migration of the MEPM Cells Was Inhibited by Cxcr4 Knockdown and Was Partially Rescued by the Addition of CXCL12
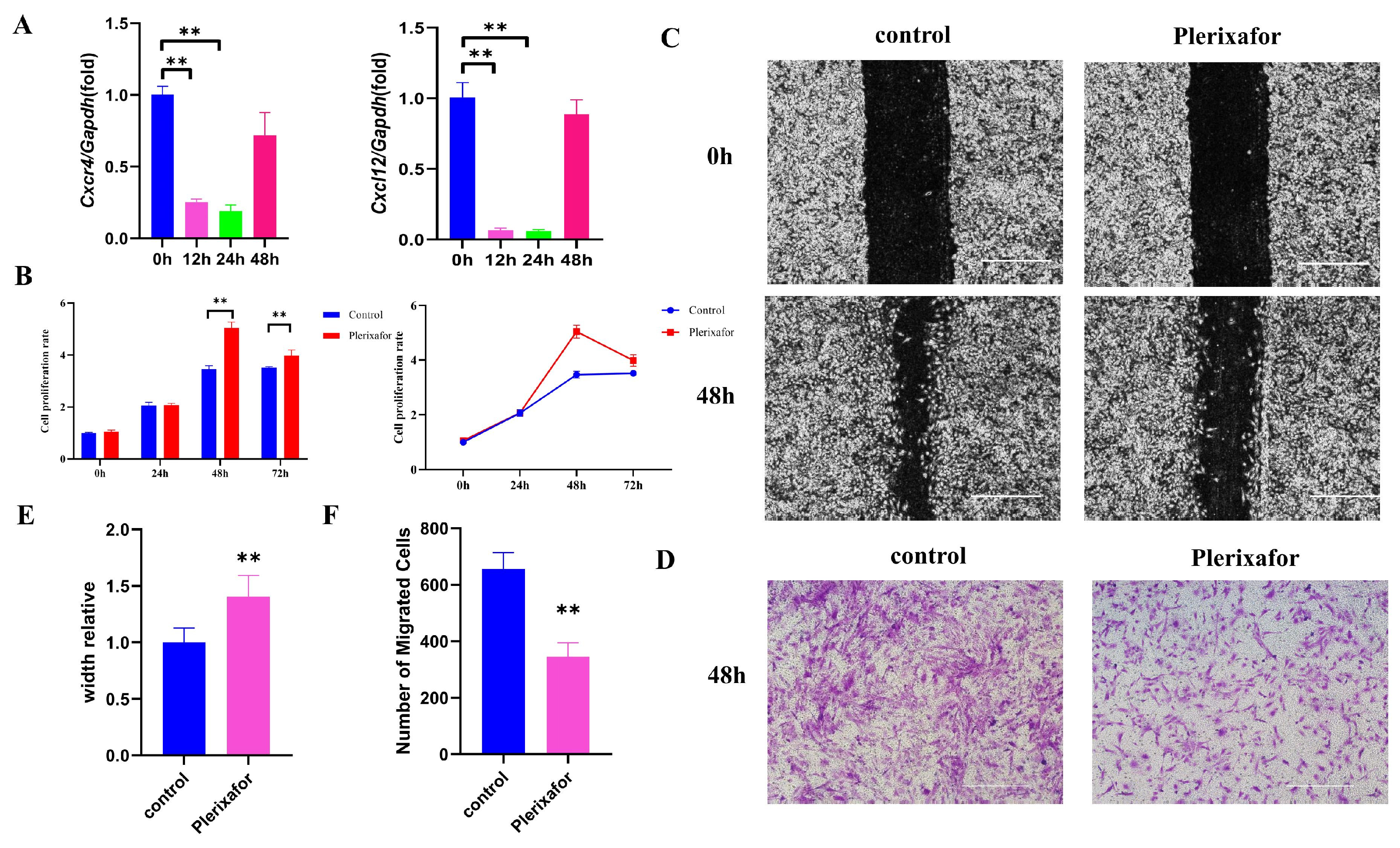
2.2. CXCR4 Inhibitor Plerixafor Blocked the Expression of Cxcr4 and Cxcl12 and Suppressed the Migration of MEPM Cells
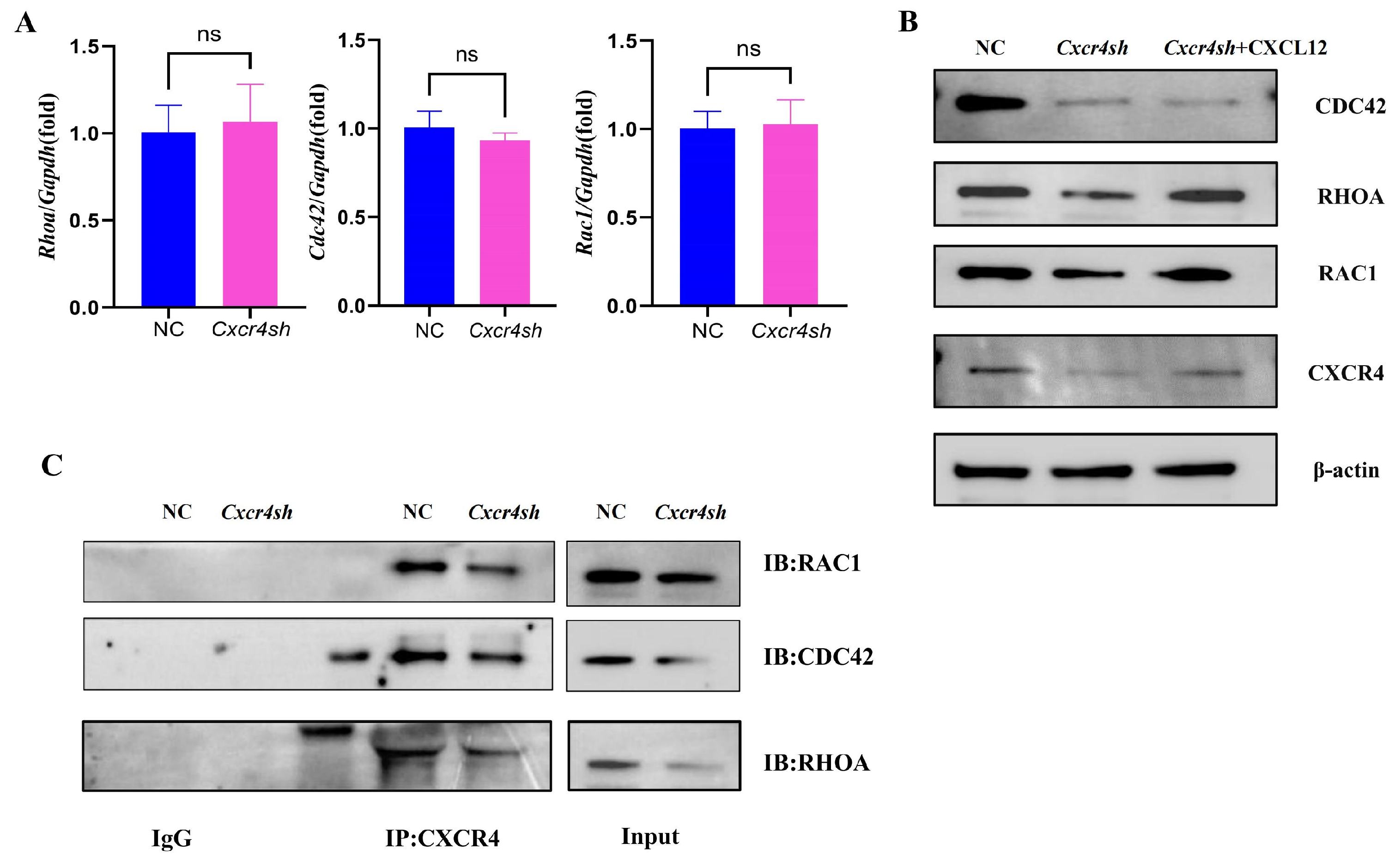
2.3. Cxcr4 Knockdown Reduced RAC1, CDC42, and RHOA Expression by Binding to Them, Respectively
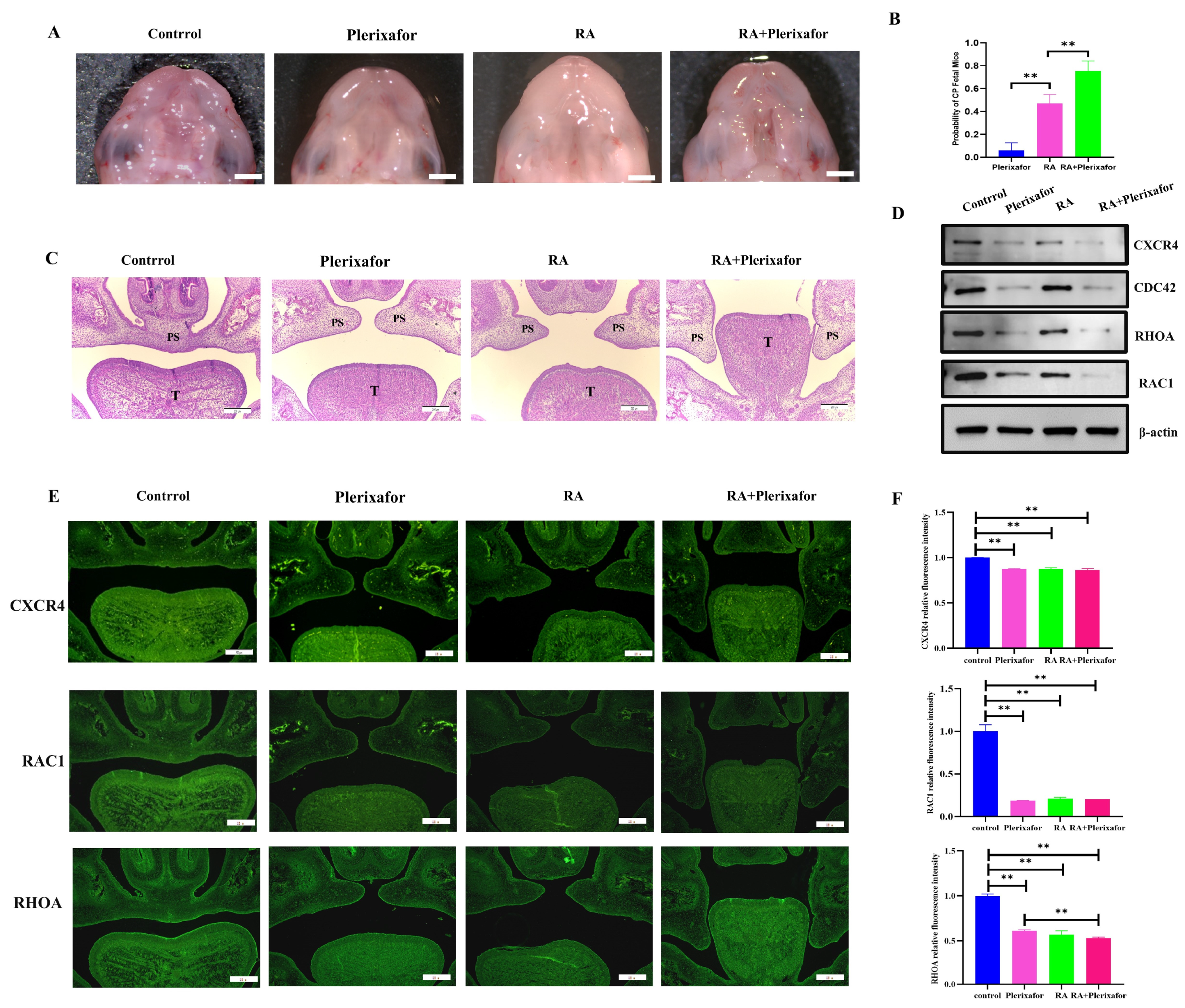
2.4. A Combination of Plerixafor and RA Treatment In Vivo Significantly Increased the Incidence of the Cleft Palate While Reducing the Expression of CXCR4 and Migration-Related Genes
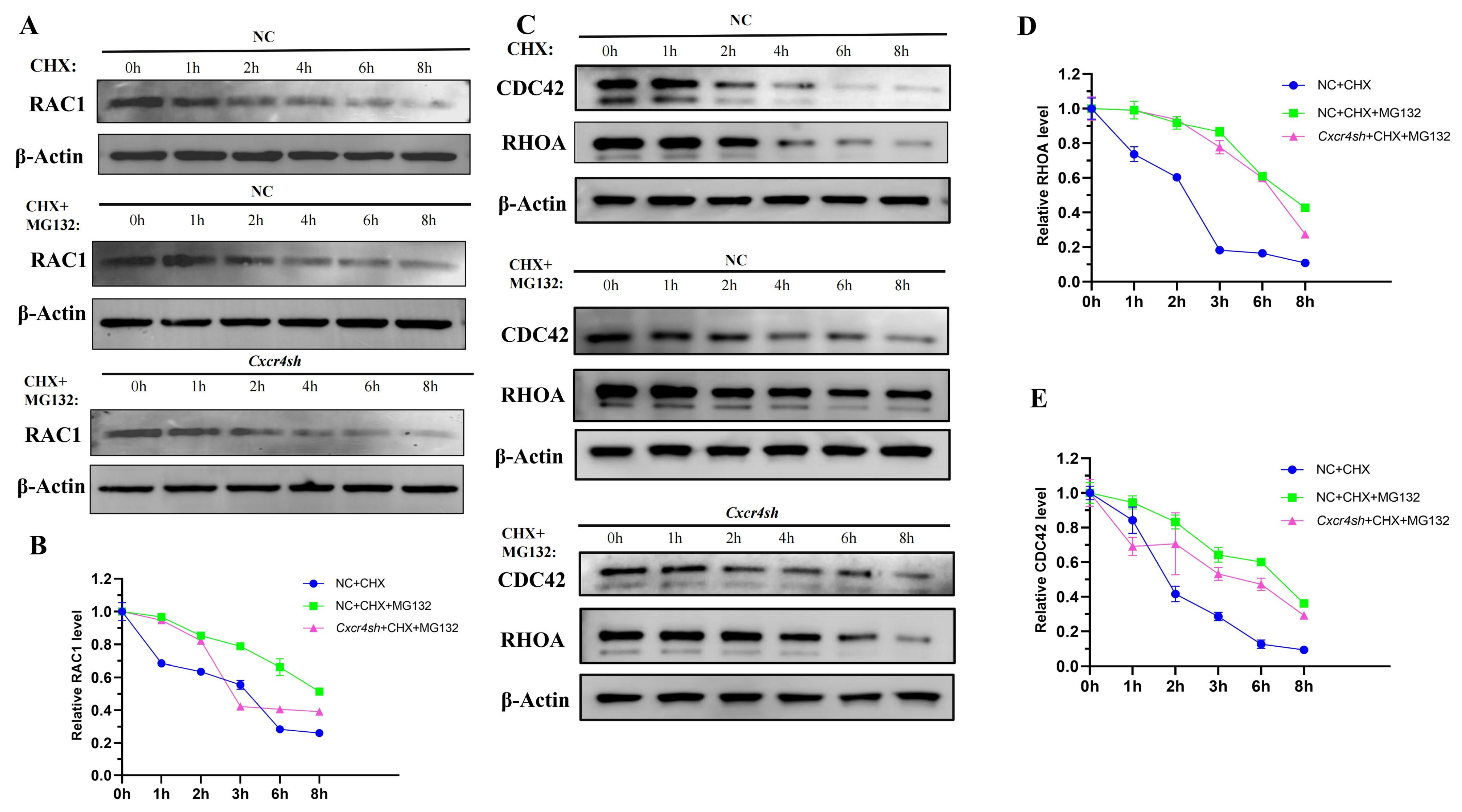
2.5. Knockdown of Cxcr4 Interfered with the Degradation of RAC1, CDC42, and RHOA by Ubiquitin-Proteasome
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Viral Infection
4.3. Real-Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
4.4. Western Blot Analysis
4.5. Scratch Healing Assays
4.6. Transwell Assays
4.7. Alkaline Phosphatase (ALP) Staining
4.8. Alizarin Red Staining (ARS)
4.9. Cell Counting Kit-8 (CCK-8) Assay
4.10. Animals Handling, Dosing, and Embryo Extracting
4.11. Hematoxylin and Eosin (H&E) Staining
4.12. Immunofluorescence (IF) Staining
4.13. Co-Immunoprecipitation, Co-IP
4.14. Cycloheximide and MG132 Treatment
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef]
- Rychlik, D.; Wojcicki, P.; Kozlik, M. Osteoplasty of the alveolar cleft defect. Adv. Clin. Exp. Med. 2012, 21, 255–262. [Google Scholar] [PubMed]
- Bush, J.O.; Jiang, R. Palatogenesis: Morphogenetic and molecular mechanisms of secondary palate development. Development 2012, 139, 231–243. [Google Scholar] [CrossRef]
- Iwaya, C.; Suzuki, A.; Iwata, J. MicroRNAs and Gene Regulatory Networks Related to Cleft Lip and Palate. Int. J. Mol. Sci. 2023, 24, 3552. [Google Scholar] [CrossRef]
- Hammond, N.L.; Dixon, M.J. Revisiting the embryogenesis of lip and palate development. Oral Dis. 2022, 28, 1306–1326. [Google Scholar] [CrossRef] [PubMed]
- Mossey, P.A.; Little, J.; Munger, R.G.; Dixon, M.J.; Shaw, W.C. Cleft lip and palate. Lancet 2009, 374, 1773–1785. [Google Scholar] [CrossRef]
- Jia, S.; Zhou, J.; Fanelli, C.; Wee, Y.; Bonds, J.; Schneider, P.; Mues, G.; D’Souza, R.N. Small-molecule Wnt agonists correct cleft palates in Pax9 mutant mice in utero. Development 2017, 144, 3819–3828. [Google Scholar] [CrossRef] [PubMed]
- Nasroen, S.L.; Maskoen, A.M.; Soedjana, H.; Hilmanto, D.; Gani, B.A. IRF6 rs2235371 as a risk factor for non-syndromic cleft palate only among the Deutero-Malay race in Indonesia and its effect on the IRF6 mRNA expression level. Dent. Med. Probl. 2022, 59, 59–65. [Google Scholar] [CrossRef]
- Paradowska-Stolarz, A.; Mikulewicz, M.; Dus-Ilnicka, I. Current Concepts and Challenges in the Treatment of Cleft Lip and Palate Patients-A Comprehensive Review. J. Pers. Med. 2022, 12, 2089. [Google Scholar] [CrossRef]
- Qiao, W.; Huang, P.; Wang, X.; Meng, L. Susceptibility to DNA damage caused by abrogation of Rad54 homolog B: A putative mechanism for chemically induced cleft palate. Toxicology 2021, 456, 152772. [Google Scholar] [CrossRef]
- Zheng, K.; Ye, Q.N. The involvement of hormone-sensitive lipase in all-trans retinoic acid induced cleft palate. Int. J. Dev. Biol. 2022, 66, 383–389. [Google Scholar] [CrossRef]
- Mandalos, N.P.; Dimou, A.; Gavala, M.A.; Lambraki, E.; Remboutsika, E. Craniofacial Development Is Fine-Tuned by Sox2. Genes 2023, 14, 380. [Google Scholar] [CrossRef] [PubMed]
- Yahya, I.; Morosan-Puopolo, G.; Brand-Saberi, B. The CXCR4/SDF-1 Axis in the Development of Facial Expression and Non-somitic Neck Muscles. Front. Cell Dev. Biol. 2020, 8, 615264. [Google Scholar] [CrossRef] [PubMed]
- Al-Dimassi, S.; Salloum, G.; Saykali, B.; Khoury, O.; Liu, S.; Leppla, S.H.; Abi-Habib, R.; El-Sibai, M. Targeting the MAP kinase pathway in astrocytoma cells using a recombinant anthrax lethal toxin as a way to inhibit cell motility and invasion. Int. J. Oncol. 2016, 48, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Al-Koussa, H.; Atat, O.E.; Jaafar, L.; Tashjian, H.; El-Sibai, M. The Role of Rho GTPases in Motility and Invasion of Glioblastoma Cells. Anal. Cell. Pathol. 2020, 2020, 9274016. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shang, F.F.; Xu, Y.; Belegu, V.; Xia, L.; Zhao, W.; Liu, R.; Wang, W.; Liu, J.; Li, C.Y.; et al. eIF5A1/RhoGDIalpha pathway: A novel therapeutic target for treatment of spinal cord injury identified by a proteomics approach. Sci. Rep. 2015, 5, 16911. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, B.K.; Lou, M.; Zheng, Y.; Lang, R.A. Balanced Rac1 and RhoA activities regulate cell shape and drive invagination morphogenesis in epithelia. Proc. Natl. Acad. Sci. USA 2011, 108, 18289–18294. [Google Scholar] [CrossRef]
- Hanna, S.; El-Sibai, M. Signaling networks of Rho GTPases in cell motility. Cell. Signal. 2013, 25, 1955–1961. [Google Scholar] [CrossRef]
- Rot, A.; von Andrian, U.H. Chemokines in innate and adaptive host defense: Basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 2004, 22, 891–928. [Google Scholar] [CrossRef]
- Wang, X.; Cui, Z.; Chen, X.; Luo, Q.; Jiang, Z.; Liu, X.; Huang, Y.; Jiang, J.; Chen, S.; Qiu, J.; et al. The CXCR4/miR-1910-5p/MMRN2 Axis Is Involved in Corneal Neovascularization by Affecting Vascular Permeability. Investig. Ophthalmol. Vis. Sci. 2023, 64, 10. [Google Scholar] [CrossRef]
- Jaffar, J.; Griffiths, K.; Oveissi, S.; Duan, M.; Foley, M.; Glaspole, I.; Symons, K.; Organ, L.; Westall, G. CXCR4(+) cells are increased in lung tissue of patients with idiopathic pulmonary fibrosis. Respir. Res. 2020, 21, 221. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallstrom, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, A.S.; Daugvilaite, V.; De Filippo, K.; Berg, C.; Mavri, M.; Benned-Jensen, T.; Juzenaite, G.; Hjorto, G.; Rankin, S.; Vabeno, J.; et al. Biased action of the CXCR4-targeting drug plerixafor is essential for its superior hematopoietic stem cell mobilization. Commun. Biol. 2021, 4, 569. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, N.; Zhang, T.T.; Nakanishi, T. Involvement of CXCR4 in Normal and Abnormal Development. Cells 2019, 8, 185. [Google Scholar] [CrossRef]
- Petit, I.; Jin, D.; Rafii, S. The SDF-1-CXCR4 signaling pathway: A molecular hub modulating neo-angiogenesis. Trends Immunol. 2007, 28, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Imadome, K.; Shoji, Y.; Isozaki, T.; Endo, S.; Yamada, S.; Imai, T. Carbon-Ion Irradiation Suppresses Migration and Invasiveness of Human Pancreatic Carcinoma Cells MIAPaCa-2 via Rac1 and RhoA Degradation. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 173–180. [Google Scholar] [CrossRef]
- Shkoukani, M.A.; Lawrence, L.A.; Liebertz, D.J.; Svider, P.F. Cleft palate: A clinical review. Birth Defects Res. C Embryo Today 2014, 102, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Sadri, F.; Rezaei, Z.; Fereidouni, M. The significance of the SDF-1/CXCR4 signaling pathway in the normal development. Mol Biol. Rep. 2022, 49, 3307–3320. [Google Scholar] [CrossRef]
- Masenga, S.K.; Mweene, B.C.; Luwaya, E.; Muchaili, L.; Chona, M.; Kirabo, A. HIV-Host Cell Interactions. Cells 2023, 12, 1351. [Google Scholar] [CrossRef]
- Marayati, R.; Julson, J.; Bownes, L.V.; Quinn, C.H.; Stafman, L.L.; Beierle, A.M.; Markert, H.R.; Hutchins, S.C.; Stewart, J.E.; Crossman, D.K.; et al. PIM3 kinase promotes tumor metastasis in hepatoblastoma by upregulating cell surface expression of chemokine receptor cxcr4. Clin. Exp. Metastasis 2022, 39, 899–912. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Schilling, T.F. Chemokine signaling controls endodermal migration during zebrafish gastrulation. Science 2008, 322, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, L.P.; Borup, R.; Vestergaard, J.; Larsen, L.A.; Lage, K.; Maroun, L.L.; Kjaer, I.; Niemann, C.U.; Andersen, M.; Knudsen, M.A.; et al. Expression analyses of human cleft palate tissue suggest a role for osteopontin and immune related factors in palatal development. Exp. Mol. Med. 2009, 41, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Z.; Chen, Y.; Zhang, Y. Conditional deletion of Bmp2 in cranial neural crest cells recapitulates Pierre Robin sequence in mice. Cell Tissue Res. 2019, 376, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Verheijen, N.; Suttorp, C.M.; van Rheden, R.E.M.; Regan, R.F.; Helmich, M.; Kuijpers-Jagtman, A.M.; Wagener, F. CXCL12-CXCR4 Interplay Facilitates Palatal Osteogenesis in Mice. Front. Cell Dev. Biol. 2020, 8, 771. [Google Scholar] [CrossRef]
- Bryan, B.A.; D’Amore, P.A. What tangled webs they weave: Rho-GTPase control of angiogenesis. Cell. Mol. Life Sci. 2007, 64, 2053–2065. [Google Scholar] [CrossRef]
- Govek, E.E.; Wu, Z.; Acehan, D.; Molina, H.; Rivera, K.; Zhu, X.; Fang, Y.; Tessier-Lavigne, M.; Hatten, M.E. Cdc42 Regulates Neuronal Polarity during Cerebellar Axon Formation and Glial-Guided Migration. iScience 2018, 1, 35–48. [Google Scholar] [CrossRef]
- Al Haddad, M.; El-Rif, R.; Hanna, S.; Jaafar, L.; Dennaoui, R.; Abdellatef, S.; Miskolci, V.; Cox, D.; Hodgson, L.; El-Sibai, M. Differential regulation of rho GTPases during lung adenocarcinoma migration and invasion reveals a novel role of the tumor suppressor StarD13 in invadopodia regulation. Cell Commun. Signal. 2020, 18, 144. [Google Scholar] [CrossRef]
- Zahra, F.T.; Sajib, M.S.; Ichiyama, Y.; Akwii, R.G.; Tullar, P.E.; Cobos, C.; Minchew, S.A.; Doci, C.L.; Zheng, Y.; Kubota, Y.; et al. Endothelial RhoA GTPase is essential for in vitro endothelial functions but dispensable for physiological in vivo angiogenesis. Sci. Rep. 2019, 9, 11666. [Google Scholar] [CrossRef]
- Daniel, S.K.; Seo, Y.D.; Pillarisetty, V.G. The CXCL12-CXCR4/CXCR7 axis as a mechanism of immune resistance in gastrointestinal malignancies. Semin. Cancer Biol. 2020, 65, 176–188. [Google Scholar] [CrossRef]
- Berg, C.; Daugvilaite, V.; Steen, A.; Jorgensen, A.S.; Vabeno, J.; Rosenkilde, M.M. Inhibition of HIV Fusion by Small Molecule Agonists through Efficacy-Engineering of CXCR4. ACS Chem. Biol. 2018, 13, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Kamata, T.; Al Dujaily, E.; Alhamad, S.; So, T.Y.; Margaritaki, O.; Giblett, S.; Pringle, J.H.; Le Quesne, J.; Pritchard, C. Statins mediate anti- and pro-tumourigenic functions by remodelling the tumour microenvironment. Dis. Model. Mech. 2022, 15, dmm049148. [Google Scholar] [CrossRef]
- Ma, X.; Ma, X.; Zhu, L.; Zhao, Y.; Chen, M.; Li, T.; Lin, Y.; Ma, D.; Sun, C.; Han, L. The E3 ubiquitin ligase MG53 inhibits hepatocellular carcinoma by targeting RAC1 signaling. Oncogenesis 2022, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Mialki, R.K.; Dong, S.; Khoo, A.; Mallampalli, R.K.; Zhao, Y.; Zhao, J. A new mechanism of RhoA ubiquitination and degradation: Roles of SCF(FBXL19) E3 ligase and Erk2. Biochim. Biophys. Acta 2013, 1833, 2757–2764. [Google Scholar] [CrossRef] [PubMed]
- Murali, A.; Shin, J.; Yurugi, H.; Krishnan, A.; Akutsu, M.; Carpy, A.; Macek, B.; Rajalingam, K. Ubiquitin-dependent regulation of Cdc42 by XIAP. Cell Death Dis. 2017, 8, e2900. [Google Scholar] [CrossRef] [PubMed]
- Kochhar, D.M. Teratogenicity of retinoic acid. Teratology 2000, 62, 178–180. [Google Scholar] [CrossRef]
- Dong, S.; Zhang, Y.; Huang, H. Involvement of RBP4 in all-trans retinoic acid induced cleft palate. Mol. Med. Rep. 2017, 16, 5915–5923. [Google Scholar] [CrossRef][Green Version]
- Wang, J.; Tannous, B.A.; Poznansky, M.C.; Chen, H. CXCR4 antagonist AMD3100 (plerixafor): From an impurity to a therapeutic agent. Pharmacol. Res. 2020, 159, 105010. [Google Scholar] [CrossRef]
- Brave, M.; Farrell, A.; Ching Lin, S.; Ocheltree, T.; Pope Miksinski, S.; Lee, S.L.; Saber, H.; Fourie, J.; Tornoe, C.; Booth, B.; et al. FDA review summary: Mozobil in combination with granulocyte colony-stimulating factor to mobilize hematopoietic stem cells to the peripheral blood for collection and subsequent autologous transplantation. Oncology 2010, 78, 282–288. [Google Scholar] [CrossRef]
- Wu, Y.; Qiu, J.; Chen, S.; Chen, X.; Zhang, J.; Zhuang, J.; Liu, S.; Yang, M.; Zhou, P.; Chen, H.; et al. Comparison of the Response to the CXCR4 Antagonist AMD3100 during the Development of Retinal Organoids Derived from ES Cells and Zebrafish Retina. Int. J. Mol. Sci. 2022, 23, 7088. [Google Scholar] [CrossRef]
- Jiangbo, Z.; Muquan, Y.; Rongfang, C.; Miaoli, G.; Tianbao, Z. Establishment of mouse model of cleft palate induced by N-methyl-N’-nitro-N-nitrosoguanidine and retinoic acid. Acad. J. Nav. Med. Univ. 2005, 26, 58–60. [Google Scholar] [CrossRef]
- Haiyan, Y.; Kai, L.; Yuya, Z.; Tong, l.; Meiqun, S.; Weiyan, Z. Establishment and significance of an animal model of cleft palate in Kunming mice induced by excessive retinoic acid. J. Bengbu Med. Coll. 2010, 35, 14–16. [Google Scholar] [CrossRef]
- Wang, X.; Peng, X.; Chen, J.; Wang, Y.; Zhao, X.; Li, T.; Du, J. Comparative analysis of mouse embryonic palatal mesenchymal cells isolated by two primary culture methods. Tissue Cell 2022, 76, 101783. [Google Scholar] [CrossRef] [PubMed]
- McCallion, C.; Peters, A.D.; Booth, A.; Rees-Unwin, K.; Adams, J.; Rahi, R.; Pluen, A.; Hutchinson, C.V.; Webb, S.J.; Burthem, J. Dual-action CXCR4-targeting liposomes in leukemia: Function blocking and drug delivery. Blood Adv. 2019, 3, 2069–2081. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; Peng, H.; Liu, R.; Ji, W.; Shi, Z.; Shen, J.; Ma, G.; Zhang, X. Targeted exosome coating gene-chem nanocomplex as “nanoscavenger” for clearing alpha-synuclein and immune activation of Parkinson’s disease. Sci. Adv. 2020, 6, eaba3967. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Chen, J.; Wang, Y.; Wang, X.; Zhao, X.; Zheng, X.; Wang, Z.; Yuan, D.; Du, J. Osteogenic microenvironment affects palatal development through glycolysis. Differentiation 2023, 133, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Han, X.; He, J.; Feng, J.; Jing, J.; Janeckova, E.; Lei, J.; Ho, T.V.; Xu, J.; Chai, Y. KDM6B interacts with TFDP1 to activate P53 signaling in regulating mouse palatogenesis. eLife 2022, 11, e74595. [Google Scholar] [CrossRef]
- Chen, J.; Yao, Y.; Wang, Y.; Wang, X.; Peng, X.; Li, T.; Liu, Y.; Du, J. Autophagy triggered by the ROS/ERK signaling pathway protects mouse embryonic palatal cells from apoptosis induced by nicotine. Environ. Sci. Pollut. Res. Int. 2022, 29, 81909–81922. [Google Scholar] [CrossRef]
- Chen, J.; Yao, Y.; Wang, X.; Wang, Y.; Li, T.; Du, J. Chloroquine regulates the proliferation and apoptosis of palate development on mice embryo by activating P53 through blocking autophagy in vitro. In Vitro Cell. Dev. Biol. Anim. 2022, 58, 558–570. [Google Scholar] [CrossRef]
- Psatha, N.; Sgouramali, E.; Gkountis, A.; Siametis, A.; Baliakas, P.; Constantinou, V.; Athanasiou, E.; Arsenakis, M.; Anagnostopoulos, A.; Papayannopoulou, T.; et al. Superior long-term repopulating capacity of G-CSF+plerixafor-mobilized blood: Implications for stem cell gene therapy by studies in the Hbb(th-3) mouse model. Hum. Gene Ther. Methods 2014, 25, 317–327. [Google Scholar] [CrossRef]
- Feng, J.; Han, X.; Yuan, Y.; Cho, C.K.; Janeckova, E.; Guo, T.; Pareek, S.; Rahman, M.S.; Zheng, B.; Bi, J.; et al. TGF-beta signaling and Creb5 cooperatively regulate Fgf18 to control pharyngeal muscle development. eLife 2022, 11, e80405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ning, Y.; Xiao, Y.; Duan, H.; Qu, G.; Liu, X.; Du, Y.; Jiang, D.; Zhou, J. MAEL contributes to gastric cancer progression by promoting ILKAP degradation. Oncotarget 2017, 8, 113331–113344. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.-X.; Zheng, X.-Y.; Yu, J.-J.; Ma, H.-R.; Hua, C.; Gao, R.-T. EGFR enhances the stemness and progression of oral cancer through inhibiting autophagic degradation of SOX2. Cancer Med. 2020, 9, 1131–1140. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer Sequence (5′-3′) |
|---|---|
| Cxcr4-F | GAAGTGGTCTGGAGACTAT |
| Cxcr4-R | TTGCCGACTATGCCAGTCAAG |
| Cxcl12-F | TGCATCAGTGACGGTAAACCA |
| Cxcl12-R | TTCTTCAGCCGTGCAACAATC |
| Rac1-F | GAGACGGAGCTGTTGGTAAAA |
| Rac1-R | ATAGGCCCAGATTCACTGGTT |
| Cdc42-F | ATTATGACAGACTACGACCGCT |
| Cdc42-R | AGTGGTGAGTTATCTCAGGCA |
| Rhoa-F | CTCTCTTATCCAGACACCGATGT |
| Rhoa-R | TGTGCTCGTCATTCCGAAGG |
| Gapdh-F | AGGTCGGTGTGAACGGATTTG |
| Gapdh-R | TGTAGACCATGTAGTTGAGGTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Zhao, X.; Wang, Y.; Chen, J.; Wang, X.; Peng, X.; Ma, L.; Du, J. Inhibition of Cxcr4 Disrupts Mouse Embryonic Palatal Mesenchymal Cell Migration and Induces Cleft Palate Occurrence. Int. J. Mol. Sci. 2023, 24, 12740. https://doi.org/10.3390/ijms241612740
Zheng X, Zhao X, Wang Y, Chen J, Wang X, Peng X, Ma L, Du J. Inhibition of Cxcr4 Disrupts Mouse Embryonic Palatal Mesenchymal Cell Migration and Induces Cleft Palate Occurrence. International Journal of Molecular Sciences. 2023; 24(16):12740. https://doi.org/10.3390/ijms241612740
Chicago/Turabian StyleZheng, Xiaoyu, Xige Zhao, Yijia Wang, Jing Chen, Xiaotong Wang, Xia Peng, Li Ma, and Juan Du. 2023. "Inhibition of Cxcr4 Disrupts Mouse Embryonic Palatal Mesenchymal Cell Migration and Induces Cleft Palate Occurrence" International Journal of Molecular Sciences 24, no. 16: 12740. https://doi.org/10.3390/ijms241612740
APA StyleZheng, X., Zhao, X., Wang, Y., Chen, J., Wang, X., Peng, X., Ma, L., & Du, J. (2023). Inhibition of Cxcr4 Disrupts Mouse Embryonic Palatal Mesenchymal Cell Migration and Induces Cleft Palate Occurrence. International Journal of Molecular Sciences, 24(16), 12740. https://doi.org/10.3390/ijms241612740





