A High-Quality Genome Sequence of the Penicillium oxalicum 5-18 Strain Isolated from a Poplar Plantation Provides Insights into Its Lignocellulose Degradation
Abstract
:1. Introduction
2. Results
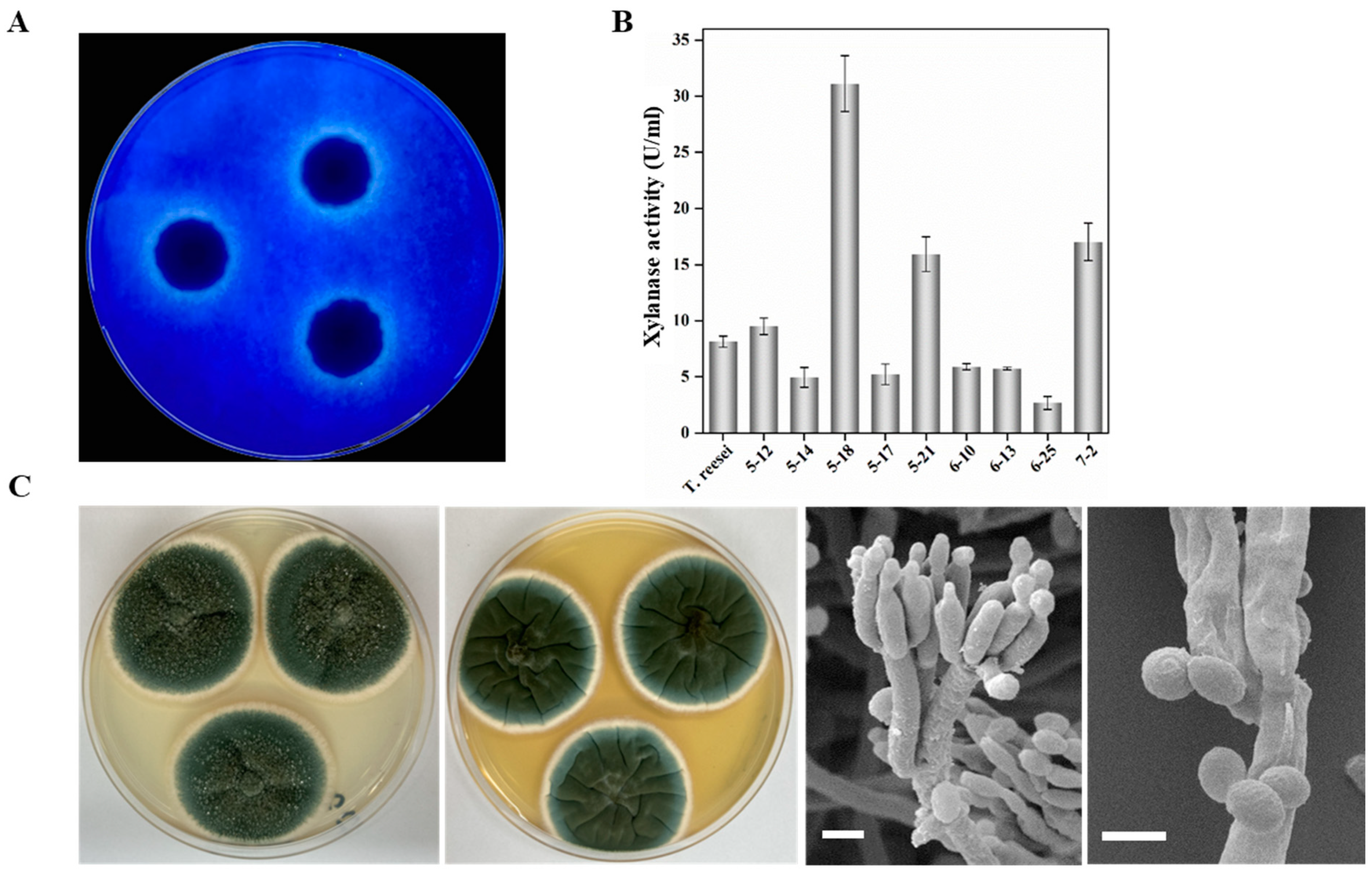
2.1. Isolation, Morphology and Phylogenetic Analysis of High-Efficient Xylan-Degrading Fungus
2.2. General Genome Features and Functional Annotation
2.3. Gene Function Classification
2.4. Carbohydrate-Active Enzymes of P. oxalicum 5-18
2.4.1. Degradation of Cellulose
2.4.2. Degradation of Hemicellulose
2.4.3. Degradation of Other Plant Cell Wall Polymers
3. Discussion
4. Materials and Methods
4.1. Isolation and Screening of Xylan-Degrading Fungi
4.2. Morphological Characteristics of the Isolates
4.3. Molecular Identification and Phylogenetic Analyses
4.4. Genome Sequencing, Assembly, Gene Prediction, and Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paripok, P.; Kazuo, S.; Khanok, R. Improvement of lignocellulosic biomass in planta: A review of feedstocks, biomass recalcitrance, and strategic manipulation of ideal plants designed for ethanol production and processability. Biomass Bioenergy 2013, 58, 390–405. [Google Scholar] [CrossRef]
- Guerriero, G.; Hausman, J.F.; Strauss, J.; Ertan, H.; Siddiqui, K.S. Lignocellulosic biomass: Biosynthesis, degradation, and industrial utilization. Eng. Life Sci. 2016, 16, 1–16. [Google Scholar] [CrossRef]
- Shashi Kant, B.; Sujit Sadashiv, J.; Ashwini Ashok, B.; Ravi Kant, B.; Anil Kumar, P.; Deepak, P.J.; Christopher, V.R.; Yun-Gon, K.; Yung-Hun, Y. Recent developments in pretreatment technologies on lignocellulosic biomass: Effect of key parameters, technological improvements, and challenges. Bioresour. Technol. 2020, 300, 122724. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2021, 50, D571–D577. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Singh, S.; Singh, O.V. Bioconversion of lignocellulosic biomass: Biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 2008, 35, 377. [Google Scholar] [CrossRef]
- Spiridon, I.; Popa, V.I. Hemicelluloses: Major Sources, Properties and Applications. In Monomers, Polymers and Composites from Renewable Resources; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Houfani, A.A.; Anders, N.; Spiess, A.C.; Baldrian, P.; Benallaoua, S. Insights from enzymatic degradation of cellulose and hemicellulose to fermentable sugars—A review. Biomass Bioenergy 2020, 134, 105481. [Google Scholar] [CrossRef]
- Huang, D.; Liu, J.; Qi, Y.; Yang, K.; Xu, Y.; Feng, L. Synergistic hydrolysis of xylan using novel xylanases, β-xylosidases, and an α-L-arabinofuranosidase from Geobacillus thermodenitrificans NG80-2. Appl. Biochem. Biotechnol. 2017, 101, 6023–6037. [Google Scholar] [CrossRef]
- Mltm, P.; Acs, R.; Monti, R.; Terenzi, H.F.; Jorge, J.A.; Amorim, D.S. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005, 67, 577–591. [Google Scholar]
- Del-Cid, A.; Ubilla, P.; Ravanal, M.-C.; Medina, E.; Vaca, I.; Levicán, G.; Eyzaguirre, J.; Chávez, R. Cold-Active Xylanase Produced by Fungi Associated with Antarctic Marine Sponges. Appl. Biochem. Biotechnol. 2013, 172, 524–532. [Google Scholar] [CrossRef]
- Ho, H.L. Batch Submerged Fermentation in Shake Flask Culture and Bioreactor: Influence of Different Agricultural Residuals as the Substrate on the Optimization of Xylanase Production by Bacillus subtilis and Aspergillus brasiliensis. J. Appl. Biotechnol. Bioeng. 2016, 1, 96–104. [Google Scholar]
- Heeger, F.; Bourne, E.C.; Wurzbacher, C.; Funke, E.; Lipzen, A.; He, G.; Ng, V.; Grigoriev, I.V.; Schlosser, D.; Monaghan, M.T. Evidence for Lignocellulose-Decomposing Enzymes in the Genome and Transcriptome of the Aquatic Hyphomycete Clavariopsis aquatica. J. Fungi 2021, 7, 854. [Google Scholar] [CrossRef]
- Grossart, H.P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, Z.; Cui, F.; Ping, L.; Qiu, C.; Li, G.; Yan, L. Isolation and identification of a newly isolated Alternaria sp. ND-16 and characterization of xylanase. Appl. Biochem. Biotechnol. 2009, 157, 36–49. [Google Scholar] [CrossRef] [PubMed]
- da Cruz Kerber, C.M.; Rasbold, L.M.; Heinen, P.R.; Henn, C.; Maller, A.; da Conceicao Silva, J.L.; Garcia Simao, R.d.C.; Simoes, M.R.; Kadowaki, M.K. Production of Hemicellulolytic Enzymes by a Novel Trichoderma koningiopsis 2OI2A1M and Its Application in the Saccharification of Barley Bagasse. Waste Biomass Valorization 2021, 12, 5949–5958. [Google Scholar] [CrossRef]
- Ravindran, R.; Williams, G.A.; Jaiswal, A.K. Spent Coffee Waste as a Potential Media Component for Xylanase Production and Potential Application in Juice Enrichment. Foods 2019, 8, 585. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.D.B.; Gomes, T.C.; Ullah, S.F.; Ticona, A.R.P.; Hamann, P.R.V.; Noronha, E.F. Biochemical Properties of Carbohydrate-Active Enzymes Synthesized by Penicillium chrysogenum Using Corn Straw as Carbon Source. Waste Biomass Valorization 2020, 11, 2455–2466. [Google Scholar] [CrossRef]
- Ogunyewo, O.A.; Randhawa, A.; Joshi, M.; Jain, K.K.; Wadekar, P.; Odaneth, A.A.; Lali, A.M.; Yazdani, S.S. Engineered Penicillium funiculosum produces potent lignocellulolytic enzymes for saccharification of various pretreated biomasses. Process Biochem. 2020, 92, 49–60. [Google Scholar] [CrossRef]
- Sunkar, B.; Kannoju, B.; Bhukya, B. Optimized Production of Xylanase by Penicillium purpurogenum and Ultrasound Impact on Enzyme Kinetics for the Production of Monomeric Sugars from Pretreated Corn Cobs. Front. Microbiol. 2020, 11, 772. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, L.; Wei, X.; Zou, G.; Qin, Y.; Ma, L.; Li, J.; Zheng, H.; Wang, S.; Wang, C.; et al. Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS ONE 2013, 8, e55185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, C.; Li, Z.; Xu, Y.; Yang, P.; Gao, L.; Yan, Q.; Li, S.; Wang, Y.; Qu, Y.; Song, X. Introduction of heterologous transcription factors and their target genes into Penicillium oxalicum leads to increased lignocellulolytic enzyme production. Appl. Microbiol. Biotechnol. 2019, 103, 2675–2687. [Google Scholar] [CrossRef]
- Qu, Y.B.; Gao, P.J.; Wang, Z.N. Screening of catabolite repression-resistant mutants of cellulase producing Penicillium spp. Mycosystema 1984, 3, 238–243. [Google Scholar]
- Gong, W.; Zhang, H.; Liu, S.; Zhang, L.; Gao, P.; Chen, G.; Wang, L. Comparative Secretome Analysis of Aspergillus niger, Trichoderma reesei, and Penicillium oxalicum during Solid-State Fermentation. Appl. Biochem. Biotechnol. 2015, 177, 1252–1271. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Bae, H. The diversity of fungal genome. Biol. Proced. Online 2015, 17, 8. [Google Scholar] [CrossRef] [Green Version]
- Chi, B.B.; Lu, Y.N.; Yin, P.C.; Liu, H.Y.; Chen, H.Y.; Shan, Y. Sequencing and Comparative Genomic Analysis of a Highly Metal-Tolerant Penicillium janthinellum P1 Provide Insights into Its Metal Tolerance. Front. Microbiol. 2021, 12, 663217. [Google Scholar] [CrossRef]
- Huttner, S.; Nguyen, T.T.; Granchi, Z.; Chin, A.W.T.; Ahren, D.; Larsbrink, J.; Thanh, V.N.; Olsson, L. Combined genome and transcriptome sequencing to investigate the plant cell wall degrading enzyme system in the thermophilic fungus Malbranchea cinnamomea. Biotechnol. Biofuels 2017, 10, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [Green Version]
- Steindorff, A.S.; Serra, L.A.; Formighieri, E.F.; de Faria, F.P.; Pocas-Fonseca, M.J.; de Almeida, J.R.M. Insights into the Lignocellulose-Degrading Enzyme System of Humicola grisea var. thermoidea Based on Genome and Transcriptome Analysis. Microbiol. Spectr. 2021, 9, e0108821. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Challacombe, J.; Morgenstern, I.; Hibbett, D.; Schmoll, M.; Kubicek, C.P.; Ferreira, P.; Ruiz-Duenas, F.J.; Martinez, A.T.; Kersten, P.; et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. USA 2009, 106, 1954–1959. [Google Scholar] [CrossRef] [PubMed]
- Chavez, R.; Bull, P.; Eyzaguirre, J. The xylanolytic enzyme system from the genus Penicillium. J. Biotechnol. 2006, 123, 413–433. [Google Scholar] [CrossRef]
- Muhammad, M.; Beaucamp, A.; Mario, C.; Maurice, N.C. Cellulose: Characteristics and applications for rechargeable batteries. Int. J. Biol. Macromol. 2022, 219, 788–803. [Google Scholar] [CrossRef]
- Jiang, S.; Zheng, X.; Li, L. De novo assembly of Auricularia polytricha transcriptome and discovery of genes involved in the degradation of lignocellulose. Biotechnol. Appl. Biochem. 2021, 68, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Béguin, P.; Aubert, J.P. The biological degradation of cellulose. FEMS Microbiol. Rev. 1994, 13, 25–58. [Google Scholar] [CrossRef] [PubMed]
- Percival Zhang, Y.H.; Himmel, M.E.; Mielenz, J.R. Outlook for cellulase improvement: Screening and selection strategies. Biotechnol. Adv. 2006, 24, 452–481. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, S.; Sun, H.; Jiang, Z.; Zhou, Z.; Han, X.; Zhou, Y.; Sun, H.; Zhou, W.; Mao, J. Enzyme Production Potential of Penicillium oxalicum M1816 and Its Application in Ferulic Acid Production. Foods 2021, 10, 2577. [Google Scholar] [CrossRef]
- Anna, E.; Thomas, H. Xylan and xylan derivatives—Biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedures and properties. Macromol. Rapid Commun. 2000, 21, 542–556. [Google Scholar]
- Nguyen, S.T.C.; Freund, H.L.; Kasanjian, J.; Berlemont, R. Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy. Appl. Microbiol. Biotechnol. 2018, 102, 1629–1637. [Google Scholar] [CrossRef]
- Vuong, T.V.; Master, E.R. Enzymatic production of 4-O-methyl d-glucaric acid from hardwood xylan. Biotechnol. Biofuels 2020, 13, 51. [Google Scholar] [CrossRef] [Green Version]
- Chimphango, A.F.; Rose, S.H.; van Zyl, W.H.; Görgens, J.F. Production and characterisation of recombinant α-L-arabinofuranosidase for production of xylan hydrogels. Appl. Microbiol. Biotechnol. 2012, 95, 101–112. [Google Scholar] [CrossRef]
- Matsuzawa, T.; Watanabe, M.; Kameda, T.; Kameyama, A.; Yaoi, K. Cooperation between β-galactosidase and an isoprimeverose-producing oligoxyloglucan hydrolase is key for xyloglucan degradation in Aspergillus oryzae. FEBS J. 2019, 286, 3182–3193. [Google Scholar] [CrossRef]
- Vardakou, M.; Dumon, C.; Murray, J.W.; Christakopoulos, P.; Weiner, D.P.; Juge, N.; Lewis, R.J.; Gilbert, H.J.; Flint, J.E. Understanding the structural basis for substrate and inhibitor recognition in eukaryotic GH11 xylanases. J. Mol. Biol. 2008, 375, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhang, Y.; Zhang, F.; Wang, Z.; Zheng, J. α-L-rhamnosidase: Production, properties, and applications. World J. Microbiol. Biotechnol. 2023, 39, 191. [Google Scholar] [CrossRef]
- Kant, S.; Vohra, A.; Gupta, R. Purification and physicochemical properties of polygalacturonase from Aspergillus niger MTCC 3323. Protein Expr. Purif. 2013, 87, 11–16. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, X.Y.; Zhao, Y.; Zhang, H.; Zhou, Y.F.; Gao, J. A Novel PL9 Pectate Lyase from Paenibacillus polymyxa KF-1: Cloning, Expression, and Its Application in Pectin Degradation. Int. J. Mol. Sci. 2019, 20, 3060. [Google Scholar] [CrossRef] [Green Version]
- Marín-Rodríguez, M.C.; Orchard, J.; Seymour, G.B. Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 2002, 53, 2115–2119. [Google Scholar] [CrossRef] [PubMed]
- Shahin, L.; Zhang, L.; Mohnen, D.; Urbanowicz, B.R. Insights into pectin O-acetylation in the plant cell wall: Structure, synthesis, and modification. Cell Surf. 2023, 9, 100099. [Google Scholar] [CrossRef]
- Christiaens, S.; Van Buggenhout, S.; Houben, K.; Jamsazzadeh Kermani, Z.; Moelants, K.R.; Ngouémazong, E.D.; Van Loey, A.; Hendrickx, M.E. Process-Structure-Function Relations of Pectin in Food. Crit. Rev. Food Sci. Nutr. 2016, 56, 1021–1042. [Google Scholar] [CrossRef] [PubMed]
- Christopher, M.; Sreeja-Raju, A.; Sankar, M.; Gokhale, D.V.; Pandey, A.; Sukumaran, R.K. Lignocellulose degradation by Penicillium janthinellum enzymes is influenced by its variable secretome and a unique set of feedstock characteristics. Bioresour. Technol. 2022, 365, 128129. [Google Scholar] [CrossRef]
- Strakowska, J.; Błaszczyk, L.; Chełkowski, J. The significance of cellulolytic enzymes produced by Trichoderma in opportunistic lifestyle of this fungus. J. Basic Microbiol. 2014, 54 (Suppl. 1), S2–S13. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-generation sequencing technologies: An overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Berka, R.M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S.E.; Chapman, J.; Chertkov, O.; Coutinho, P.M.; Cullen, D.; et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 2008, 26, 553–560. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, C. Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnol. Adv. 2009, 27, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Várnai, A.; Mäkelä, M.R.; Djajadi, D.T.; Rahikainen, J.; Hatakka, A.; Viikari, L. Carbohydrate-binding modules of fungal cellulases: Occurrence in nature, function, and relevance in industrial biomass conversion. Adv. Appl. Microbiol. 2014, 88, 103–165. [Google Scholar] [CrossRef]
- Várnai, A.; Siika-Aho, M.; Viikari, L. Carbohydrate-binding modules (CBMs) revisited: Reduced amount of water counterbalances the need for CBMs. Biotechnol. Biofuels 2013, 6, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Y.; Liu, D.; Li, G.; Li, P.; Xu, Y.; Shen, Q.; Zhang, R. Genome-wide transcriptomic analysis of a superior biomass-degrading strain of A. fumigatus revealed active lignocellulose-degrading genes. BMC Genom. 2015, 16, 459. [Google Scholar] [CrossRef] [Green Version]
- de Vries, R.P.; Mäkelä, M.R. Genomic and Postgenomic Diversity of Fungal Plant Biomass Degradation Approaches. Trends Microbiol. 2020, 28, 487–499. [Google Scholar] [CrossRef]
- Chen, H.; Li, S.; Cui, Z.; Feng, T.; Wang, H.; Ni, Z.; Gao, E.; Fang, Z. Synergistic Degradation of Maize Straw Lignin by Manganese Peroxidase from Irpex lacteus. Appl. Biochem. Biotechnol. 2023, 195, 3855–3871. [Google Scholar] [CrossRef]
- Chen, X.Q.; Deng, X.Y.; Shen, W.H.; Jia, M.Y. Preparation and characterization of the spherical nanosized cellulose by the enzymatic hydrolysis of pulp fibers. Carbohydr. Polym. 2018, 181, 879–884. [Google Scholar] [CrossRef]
- Jiang, C.; Song, J.; Cong, H.; Zhang, J.; Yang, Q. Expression and Characterization of a Novel Antifungal Exo-β-1,3-glucanase from Chaetomium cupreum. Appl. Biochem. Biotechnol. 2017, 182, 261–275. [Google Scholar] [CrossRef]
- Ooi, T.; Sato, H.; Matsumoto, K.; Taguchi, S. A unique post-translational processing of an exo-beta-1,3-glucanase of Penicillium sp. KH10 expressed in Aspergillus oryzae. Protein Expr. Purif. 2009, 67, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Juhász, T.; Szengyel, Z.; Réczey, K.; Siika-Aho, M.; Viikari, L. Characterization of cellulases and hemicellulases produced by Trichoderma reesei on various carbon sources. Process Biochem. 2005, 40, 3519–3525. [Google Scholar] [CrossRef]
- Vasina, D.V.; Pavlov, A.R.; Koroleva, O.V. Extracellular proteins of Trametes hirsuta st. 072 induced by copper ions and a lignocellulose substrate. BMC Microbiol. 2016, 16, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G. Use of dinitrosalicyclic acid reagent for determination of reducing sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Houbraken, J.; Samson, R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 2011, 70, 1–51. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, S.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Kwiatkowski, N.P.; Babiker, W.M.; Merz, W.G.; Carroll, K.C.; Zhang, S.X. Evaluation of nucleic acid sequencing of the D1/D2 region of the large subunit of the 28S rDNA and the internal transcribed spacer region using SmartGene IDNS [corrected] software for identification of filamentous fungi in a clinical laboratory. J. Mol. Diagn 2012, 14, 393–401. [Google Scholar] [CrossRef]
- DG, H. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nuclc. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.; Waterhouse, R.M.; Panagiotis, I.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 19, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]




| Genome Assembly | Value |
|---|---|
| Total length (bp) | 30,780,966 |
| Number of contigs | 9 |
| Max (bp) | 5,859,503 |
| Min (bp) | 51,307 |
| Median (bp) | 3,697,257 |
| N50 (bp) | 3,787,046 |
| G+C content (%) | 50.55 |
| BUSCO (%) | 98.8 |
| Protein annotation | |
| Total number of predicted protein-encoding genes | 10,074 |
| Proteins/genes with functional annotation | 10,026 |
| Non-coding RNAs | |
| tRNAs | 180 |
| rRNAs | 53 |
| snRNAs | 33 |
| sRNA | 3 |
| Exon | |
| Number of exon | 31,080 |
| Total exon length (Mb) | 15 |
| Average of exon length (bp) | 471.48 |
| Average of exon number | 3.09 |
| Intron | |
| Number of intron | 21,006 |
| Total intron length (bp) | 2,151,025 |
| Average of intron length (bp) | 102.4 |
| Organisms | Phylum | Scaffold | N50 (bp) | Genome Size (Mb) | G + C (%) | Number of Protein Coding Genes | References |
|---|---|---|---|---|---|---|---|
| Penicillium oxalicum 5-18 | Ascomycota | - | 3,787,046 | 30.78 | 50.55 | 10,074 | In this study |
| Penicillium oxalicum 114-2 | 9 | 157,054 | 30.17 | 50.60 | 10,013 | [26] | |
| Penicillium chrysogenum | 49 | - | 32.22 | 49.00 | 13,911 | [27] | |
| Neurospora crassa | 21 | 656,009 | 41.1 | 48.23 | 10,812 | [27] | |
| Aspergillus nidulans | 8 | 679,860 | 29.83 | 50.37 | 10,455 | [27] | |
| Aspergillus niger | - | - | 33.98 | 50.40 | 10,785 | [27] | |
| Aspergillus oryzae | 12 | 2,324,132 | 37.91 | 48.26 | 12,074 | [28] | |
| Trichoderma reesei | 89 | - | 34.1 | 51.00 | 9143 | [29] | |
| Phanerochaete chrysosporium | Basidiomycota | 1323 | 78,962 | 29.9 | 57.00 | 11,777 | [27] |
| Postia placenta | 1842 | 73,576 | 36.37 | 53.60 | 11,246 | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Zhu, R.; Yu, X.-Y.; Wang, B.-T.; Ruan, H.-H.; Jin, F.-J. A High-Quality Genome Sequence of the Penicillium oxalicum 5-18 Strain Isolated from a Poplar Plantation Provides Insights into Its Lignocellulose Degradation. Int. J. Mol. Sci. 2023, 24, 12745. https://doi.org/10.3390/ijms241612745
Hu S, Zhu R, Yu X-Y, Wang B-T, Ruan H-H, Jin F-J. A High-Quality Genome Sequence of the Penicillium oxalicum 5-18 Strain Isolated from a Poplar Plantation Provides Insights into Its Lignocellulose Degradation. International Journal of Molecular Sciences. 2023; 24(16):12745. https://doi.org/10.3390/ijms241612745
Chicago/Turabian StyleHu, Shuang, Rui Zhu, Xing-Ye Yu, Bao-Teng Wang, Hong-Hua Ruan, and Feng-Jie Jin. 2023. "A High-Quality Genome Sequence of the Penicillium oxalicum 5-18 Strain Isolated from a Poplar Plantation Provides Insights into Its Lignocellulose Degradation" International Journal of Molecular Sciences 24, no. 16: 12745. https://doi.org/10.3390/ijms241612745
APA StyleHu, S., Zhu, R., Yu, X.-Y., Wang, B.-T., Ruan, H.-H., & Jin, F.-J. (2023). A High-Quality Genome Sequence of the Penicillium oxalicum 5-18 Strain Isolated from a Poplar Plantation Provides Insights into Its Lignocellulose Degradation. International Journal of Molecular Sciences, 24(16), 12745. https://doi.org/10.3390/ijms241612745






