The Role of Cytokine-Inducible SH2 Domain-Containing Protein (CISH) in the Regulation of Basal and Cytokine-Mediated Myelopoiesis
Abstract
:1. Introduction
2. Results
2.1. Role of CISH in Basal Myelopoiesis
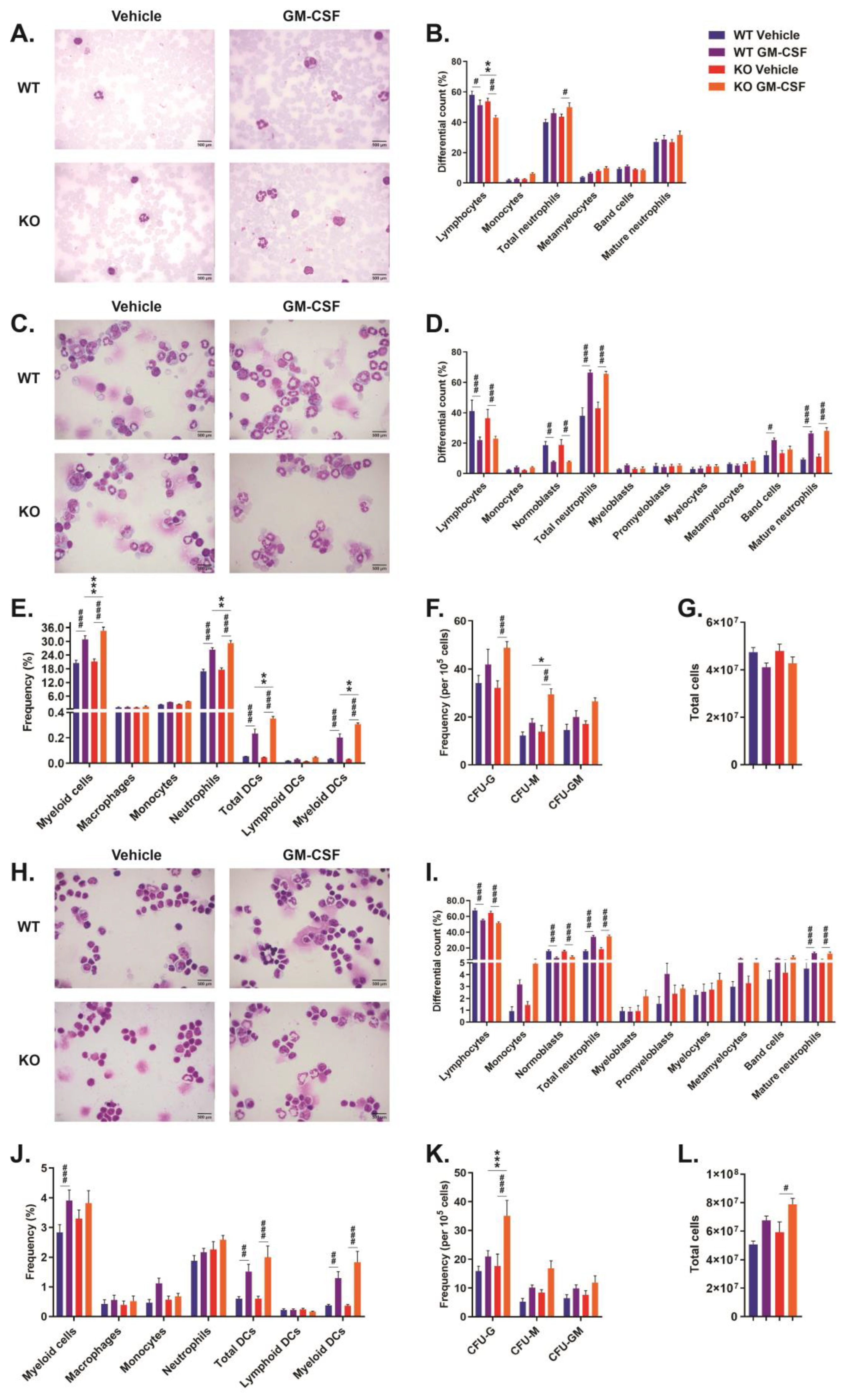
2.2. Role of CISH in GM-CSF-Induced Myelopoiesis
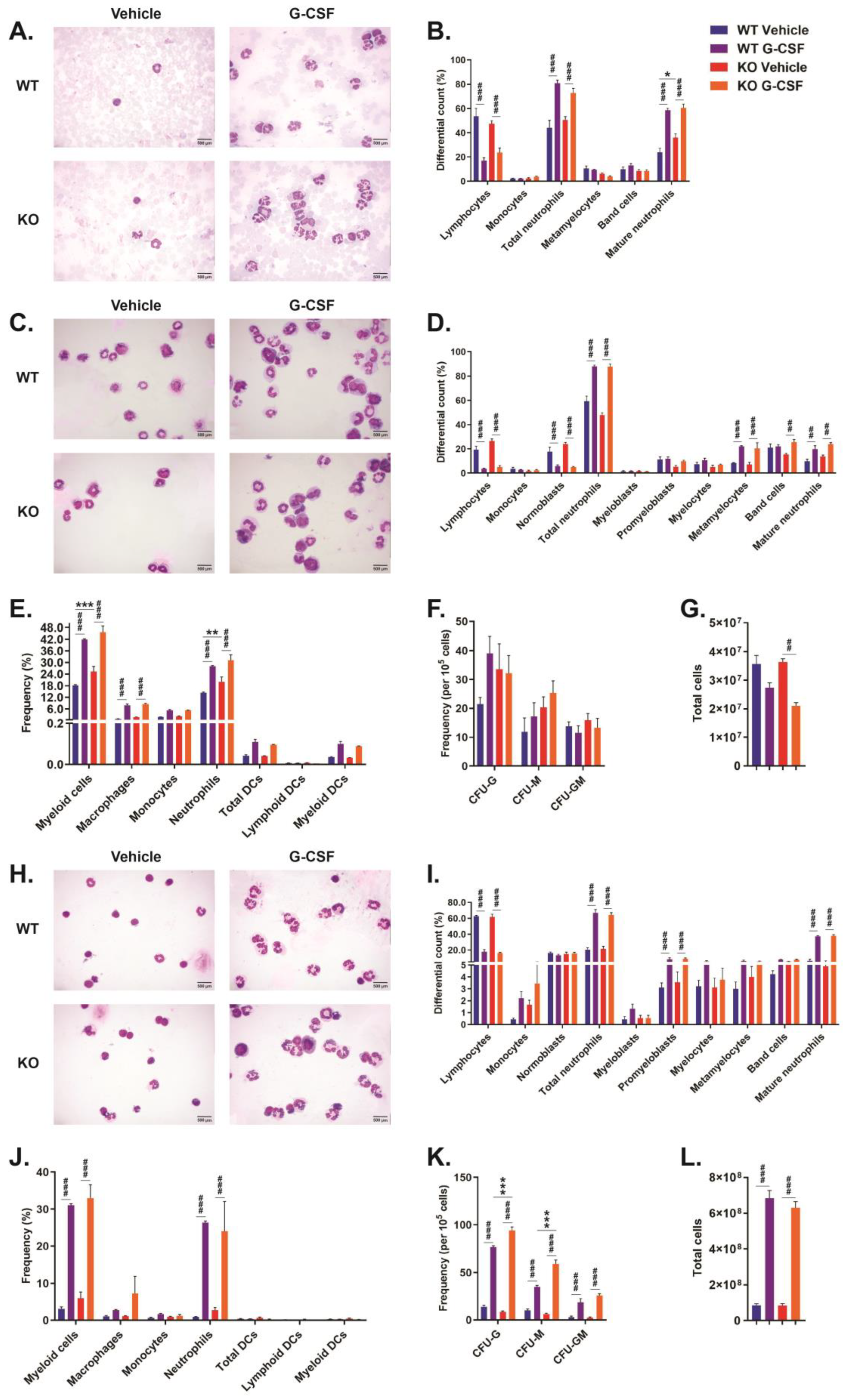
2.3. Role of CISH in G-CSF-Induced Myelopoiesis
3. Discussion
4. Materials and Methods
4.1. Animal Husbandry
4.2. Tissue Collection and Histochemistry
4.3. FACS Analysis
4.4. Colony-Forming Assays
4.5. Electropheoretic Mobility Shift Assays
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trengove, M.C.; Ward, A.C. SOCS proteins in development and disease. Am. J. Exp. Clin. Immunol. 2013, 2, 1–29. [Google Scholar]
- Sobah, M.L.; Liongue, C.; Ward, A.C. SOCS proteins in immunity, inflammatory diseases and immune-related cancer. Front. Med. 2021, 8, 727987. [Google Scholar] [CrossRef]
- Yoshimura, A.; Ohkubo, T.; Kiguchi, T.; Jenkins, N.; Gilbert, D.; Copeland, N.; Hara, T.; Miyajima, A. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 1995, 14, 2816–2826. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.O.; Zhang, H.; Kim, B.S.; Niu, X.; Peng, J.; Chen, Y.; Kerketta, R.; Lee, Y.-H.; Chang, S.H.; Corry, D.B.; et al. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat. Immunol. 2013, 14, 732–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delconte, R.B.; Kolesnik, T.B.; Dagley, L.F.; Rautela, J.; Shi, W.; Putz, E.M.; Stannard, K.; Zhang, J.-G.; Teh, C.; Firth, M.; et al. CIS is a potent checkpoint in NK cell-mediated tumor immunity. Nat. Immunol. 2016, 17, 816–824. [Google Scholar] [CrossRef]
- Li, S.; Chen, S.; Xu, X.; Sundstedt, A.; Paulsson, K.M.; Anderson, P.; Karlsson, S.; Sjögren, H.-O.; Wang, P. Cytokine-induced Src homology 2 protein (CIS) promotes T cell receptor-mediated proliferation and prolongs survival of activated T cells. J. Exp. Med. 2000, 191, 985–994. [Google Scholar] [CrossRef]
- Matsumoto, A.; Seki, Y.; Kubo, M.; Ohtsuka, S.; Suzuki, A.; Hayashi, I.; Tsuji, K.; Nakahata, T.; Okabe, M.; Yamada, S.; et al. Suppression of STAT5 functions in liver, mammary glands, and T cells in cytokine-inducible SH2 protein-1 (CIS1) transgenic mice. Mol. Cell. Biol. 1999, 19, 6396–6407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Wu, X.; Wu, D.; Jiang, R.-L.; Castillo, E.F.; Chock, C.J.; Zhou, Q.; Liu, M.; Dong, C.; Yang, X.O. Treg expression of CIS suppresses allergic airway inflammation through antagonizing an autonomous TH2 program. Mucosal. Immunol. 2020, 13, 293–302. [Google Scholar] [CrossRef]
- Palmer, D.C.; Guittard, G.C.; Franco, Z.; Crompton, J.G.; Eil, R.L.; Patel, S.J.; Ji, Y.; Van Panhuys, N.; Klebanoff, C.A.; Sukumar, M.; et al. Cish actively silences TCR signaling in CD8+ T cells to maintain tumor tolerance. J. Exp. Med. 2015, 212, 2095–2113. [Google Scholar] [CrossRef]
- Louis, C.; Souza-Fonseca-Guimaraes, F.; Yang, Y.; D’Silva, D.; Kratina, T.; Dagley, L.; Hediyeh-Zadeh, S.; Rautela, J.; Masters, S.L.; Davis, M.J.; et al. NK cell-derived GM-CSF potentiates inflammatory arthritis and is negatively regulated by CIS. J. Exp. Med. 2020, 217, e20191421. [Google Scholar] [CrossRef]
- Miah, M.A.; Yoon, C.H.; Kim, J.; Jang, J.; Seong, Y.R.; Bae, Y.S. CISH is induced during DC development and regulates DC-mediated CTL activation. Eur. J. Immunol. 2012, 42, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.G.; Jacob, A.; O’Donnell, L.C.; Agler, A.; Druhan, L.J.; Coggeshall, K.M.; Avalos, B.R. Loss of SHIP and CIS recruitment to the granulocyte colony-stimulating factor receptor contribute to hyperproliferative responses in severe congenital neutropenia/acute myelogenous leukemia. J. Immunol. 2004, 173, 5036–5045. [Google Scholar] [CrossRef] [PubMed]
- Naser, W.; Maymand, S.; Rivera, L.R.; Connor, T.; Liongue, C.; Smith, C.M.; Aston-Mourney, K.; McCulloch, D.R.; McGee, S.L.; Ward, A.C. Cytokine-inducible SH2 domain containing protein contributes to regulation of adiposity, food intake, and glucose metabolism. FASEB J. 2022, 36, e22320. [Google Scholar] [CrossRef] [PubMed]
- Unsworth, A.; Anderson, R.; Haynes, N.; Britt, K. OMIP-032: Two multi-color immunophenotyping panels for assessing the innate and adaptive immune cells in the mouse mammary gland. Cytom. A. 2016, 89, 527–530. [Google Scholar] [CrossRef]
- Croker, B.A.; Metcalf, D.; Robb, L.; Wei, W.; Mifsud, S.; DiRago, L.; A Cluse, L.; Sutherland, K.D.; Hartley, L.; Williams, E.; et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity 2004, 20, 153–165. [Google Scholar] [CrossRef] [Green Version]
- Borriello, F.; Galdiero, M.R.; Varricchi, G.; Loffredo, S.; Spadaro, G.; Marone, G. Innate immune modulation by GM-CSF and IL-3 in health and disease. Int. J. Mol. Sci. 2019, 20, 834. [Google Scholar] [CrossRef] [Green Version]
- Metcalf, D.; Begley, C.G.; Williamson, D.J.; Nice, E.C.; De Lamarter, J.; Mermod, J.J.; Thatcher, D.; Schmidt, A. Hemopoietic responses in mice injected with purified recombinant murine GM-CSF. Exp. Hematol. 1987, 15, 1–9. [Google Scholar]
- Khatami, S.; Brummer, E.; Stevens, D.A. Effects of granulocyte-macrophage colony stimulating factor (GM-CSF) in vivo on cytokine production and proliferation by spleen cells. Clin. Exp. Immunol. 2001, 125, 198–201. [Google Scholar] [CrossRef]
- Bugl, S.; Wirths, S.; Muller, M.R.; Radsak, M.P.; Kopp, H.G. Current insights into neutrophil homeostasis. Ann N. Y. Acad. Sci. 2012, 1266, 171–178. [Google Scholar] [CrossRef]
- Paudel, S.; Ghimire, L.; Jin, L.; Jeansonne, D.; Jeyaseelan, S. Regulation of emergency granulopoiesis during infection. Front. Immunol. 2022, 13, 961601. [Google Scholar] [CrossRef]
- Fujisawa, M.; Kobayashi, Y.; Okabe, T.; Takaku, F.; Komatsu, Y.; Itoh, S. Recombinant human granulocyte colony-stimulating factor induces granulocytosis in vivo. Jpn. J. Cancer Res. 1986, 77, 866–869. [Google Scholar]
- Khor, C.C.; Vannberg, F.O.; Chapman, S.J.; Guo, H.; Wong, S.H.; Walley, A.J.; Vukcevic, D.; Rautanen, A.; Mills, T.C.; Chang, K.C.; et al. CISH and susceptibility to infectious diseases. N. Engl. J. Med. 2010, 362, 2092–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, H.V.; Toan, N.L.; Song, L.H.; Kremsner, P.G.; Kun, J.F.; Tp, V. Association of CISH-292A/T genetic variant with hepatitis B virus infection. Immunogenetics 2012, 64, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Nicola, N.A. Granulocyte colony-stimulating factor and differentiation-induction in myeloid leukemic cells. Int. J. Cell Cloning 1987, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.C.; Hermans, M.H.A.; Smith, L.; van Aesch, Y.M.; Schelen, A.M.; Antonissen, C.; Touw, I.P. Tyrosine-dependent and independent mechanisms of STAT3 activation by the human granulocyte colony-stimulating factor (G-CSF) receptor are differentially utilized depending on G-CSF concentration. Blood 1999, 93, 113–124. [Google Scholar] [CrossRef]

| KO vs. WT | KO+GM-CSF vs. WT+GM-CSF | KO+G-CSF vs. WT+G-CSF | ||
|---|---|---|---|---|
| Blood | Neutrophils | ↑ | − | − |
| Bone marrow | Neutrophils | ↑ | ↑ | − |
| Myeloid DCs | − | ↑↑ | − | |
| CFU-M | − | ↑↑ | − | |
| Spleen | Neutrophils | ↑↑ | ↑ | − |
| CFU-G | − | ↑↑ | ↑ | |
| CFU-M | − | − | ↑↑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naser, W.; Maymand, S.; Dlugolenski, D.; Basheer, F.; Ward, A.C. The Role of Cytokine-Inducible SH2 Domain-Containing Protein (CISH) in the Regulation of Basal and Cytokine-Mediated Myelopoiesis. Int. J. Mol. Sci. 2023, 24, 12757. https://doi.org/10.3390/ijms241612757
Naser W, Maymand S, Dlugolenski D, Basheer F, Ward AC. The Role of Cytokine-Inducible SH2 Domain-Containing Protein (CISH) in the Regulation of Basal and Cytokine-Mediated Myelopoiesis. International Journal of Molecular Sciences. 2023; 24(16):12757. https://doi.org/10.3390/ijms241612757
Chicago/Turabian StyleNaser, Wasan, Saeed Maymand, Daniel Dlugolenski, Faiza Basheer, and Alister C. Ward. 2023. "The Role of Cytokine-Inducible SH2 Domain-Containing Protein (CISH) in the Regulation of Basal and Cytokine-Mediated Myelopoiesis" International Journal of Molecular Sciences 24, no. 16: 12757. https://doi.org/10.3390/ijms241612757
APA StyleNaser, W., Maymand, S., Dlugolenski, D., Basheer, F., & Ward, A. C. (2023). The Role of Cytokine-Inducible SH2 Domain-Containing Protein (CISH) in the Regulation of Basal and Cytokine-Mediated Myelopoiesis. International Journal of Molecular Sciences, 24(16), 12757. https://doi.org/10.3390/ijms241612757







