Inhibition of USP2 Enhances TRAIL-Mediated Cancer Cell Death through Downregulation of Survivin
Abstract
1. Introduction
2. Results
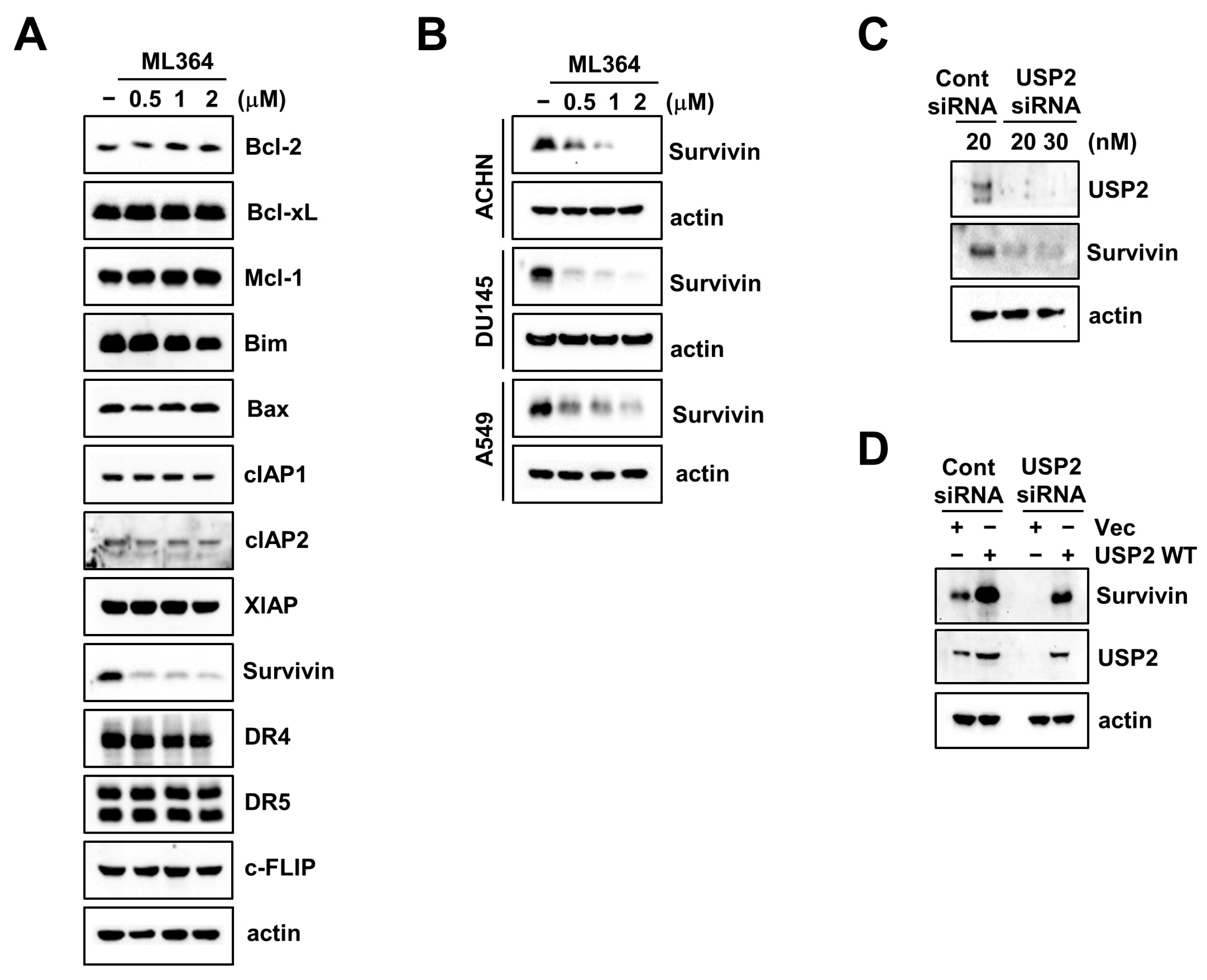
2.1. The USP2 Inhibitor ML364 Induces Downregulation of Survivin Expression
2.2. USP2 Regulates Survivin Stability
2.3. USP2 Interacts with and Deubiquitinates Survivin
2.4. ML364 Sensitizes TRAIL-Mediated Apoptosis
2.5. Downregulation of Survivin Contributes to ML364-Mediated TRAIL Sensitization
2.6. USP2 Plays an Important Role in TRAIL-Mediated Apoptosis
3. Discussion
4. Materials and Methods
4.1. Cells and Chemicals
4.2. Transfection
4.3. Examination of Protein and mRNA Level
4.4. Immunoprecipitation and Ubiquitination Assays
4.5. Certification of Apoptosis
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, F.; Xiao, H.; Wang, M.; Xie, X.; Hu, F. The role of the ubiquitin-proteasome pathway in cancer development and treatment. Front. Biosci. (Landmark Ed) 2014, 19, 886–895. [Google Scholar] [CrossRef]
- Ravid, T.; Hochstrasser, M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 2008, 9, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef]
- Baek, S.H.; Choi, K.S.; Yoo, Y.J.; Cho, J.M.; Baker, R.T.; Tanaka, K.; Chung, C.H. Molecular cloning of a novel ubiquitin-specific protease, UBP41, with isopeptidase activity in chick skeletal muscle. J. Biol. Chem. 1997, 272, 25560–25565. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Hashimoto, M. USP2-Related Cellular Signaling and Consequent Pathophysiological Outcomes. Int. J. Mol. Sci. 2021, 22, 1209. [Google Scholar] [CrossRef] [PubMed]
- Graner, E.; Tang, D.; Rossi, S.; Baron, A.; Migita, T.; Weinstein, L.J.; Lechpammer, M.; Huesken, D.; Zimmermann, J.; Signoretti, S.; et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell 2004, 5, 253–261. [Google Scholar] [CrossRef]
- Calvisi, D.F.; Wang, C.; Ho, C.; Ladu, S.; Lee, S.A.; Mattu, S.; Destefanis, G.; Delogu, S.; Zimmermann, A.; Ericsson, J.; et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 2011, 140, 1071–1083. [Google Scholar] [CrossRef]
- Tao, B.B.; He, H.; Shi, X.H.; Wang, C.L.; Li, W.Q.; Li, B.; Dong, Y.; Hu, G.H.; Hou, L.J.; Luo, C.; et al. Up-regulation of USP2a and FASN in gliomas correlates strongly with glioma grade. J. Clin. Neurosci. 2013, 20, 717–720. [Google Scholar] [CrossRef]
- Boustani, M.R.; Khoshnood, R.J.; Nikpasand, F.; Taleshi, Z.; Ahmadi, K.; Yahaghi, E.; Goudarzi, P.K. Overexpression of ubiquitin-specific protease 2a (USP2a) and nuclear factor erythroid 2-related factor 2 (Nrf2) in human gliomas. J. Neurol. Sci. 2016, 363, 249–252. [Google Scholar] [CrossRef]
- Jeong, P.; Ha, Y.S.; Yun, S.J.; Yoon, H.Y.; Freeman, M.R.; Kim, J.; Kim, W.J. Assess the expression of ubiquitin specific protease USP2a for bladder cancer diagnosis. BMC Urol. 2015, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.I.; Pragani, R.; Fox, J.T.; Shen, M.; Parmar, K.; Gaudiano, E.F.; Liu, L.; Tanega, C.; McGee, L.; Hall, M.D.; et al. Small Molecule Inhibition of the Ubiquitin-specific Protease USP2 Accelerates cyclin D1 Degradation and Leads to Cell Cycle Arrest in Colorectal Cancer and Mantle Cell Lymphoma Models. J. Biol. Chem. 2016, 291, 24628–24640. [Google Scholar] [CrossRef] [PubMed]
- Benassi, B.; Marani, M.; Loda, M.; Blandino, G. USP2a alters chemotherapeutic response by modulating redox. Cell Death Dis. 2013, 4, e812. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, L.; Sun, L.; Hu, X.; Wang, X.; Nie, T.; Li, X.; Wang, P.; Pang, P.; Pang, J.; et al. ML364 exerts the broad-spectrum antivirulence effect by interfering with the bacterial quorum sensing system. Front. Microbiol. 2022, 13, 980217. [Google Scholar] [CrossRef] [PubMed]
- Kretz, A.L.; Trauzold, A.; Hillenbrand, A.; Knippschild, U.; Henne-Bruns, D.; von Karstedt, S.; Lemke, J. TRAILblazing Strategies for Cancer Treatment. Cancers 2019, 11, 456. [Google Scholar] [CrossRef]
- de Miguel, D.; Lemke, J.; Anel, A.; Walczak, H.; Martinez-Lostao, L. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016, 23, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Brodaczewska, K.K.; Szczylik, C.; Fiedorowicz, M.; Porta, C.; Czarnecka, A.M. Choosing the right cell line for renal cell cancer research. Mol. Cancer 2016, 15, 83. [Google Scholar] [CrossRef]
- Motzer, R.J.; Hutson, T.E.; McCann, L.; Deen, K.; Choueiri, T.K. Overall survival in renal-cell carcinoma with pazopanib versus sunitinib. N. Engl. J. Med. 2014, 370, 1769–1770. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef]
- Stevenson, L.F.; Sparks, A.; Allende-Vega, N.; Xirodimas, D.P.; Lane, D.P.; Saville, M.K. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007, 26, 976–986. [Google Scholar] [CrossRef]
- Allende-Vega, N.; Sparks, A.; Lane, D.P.; Saville, M.K. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene 2010, 29, 432–441. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nepal, S.; Shrestha, A.; Park, P.H. Ubiquitin specific protease 2 acts as a key modulator for the regulation of cell cycle by adiponectin and leptin in cancer cells. Mol. Cell. Endocrinol. 2015, 412, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Solomon, L.R.; Pereda-Lopez, A.; Giranda, V.L.; Luo, Y.; Johnson, E.F.; Shoemaker, A.R.; Leverson, J.; Liu, X. Ubiquitin-specific cysteine protease 2a (USP2a) regulates the stability of Aurora-A. J. Biol. Chem. 2011, 286, 38960–38968. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, X.; Wang, Q.; Deng, Y.; Li, K.; Zhang, M.; Zhang, Q.; Zhou, J.; Wang, H.Y.; Bai, P.; et al. USP2a Supports Metastasis by Tuning TGF-beta Signaling. Cell Rep. 2018, 22, 2442–2454. [Google Scholar] [CrossRef]
- Kim, J.; Alavi Naini, F.; Sun, Y.; Ma, L. Ubiquitin-specific peptidase 2a (USP2a) deubiquitinates and stabilizes beta-catenin. Am. J. Cancer Res. 2018, 8, 1823–1836. [Google Scholar]
- Haimerl, F.; Erhardt, A.; Sass, G.; Tiegs, G. Down-regulation of the de-ubiquitinating enzyme ubiquitin-specific protease 2 contributes to tumor necrosis factor-alpha-induced hepatocyte survival. J. Biol. Chem. 2009, 284, 495–504. [Google Scholar] [CrossRef]
- Liu, Y.; Lear, T.; Iannone, O.; Shiva, S.; Corey, C.; Rajbhandari, S.; Jerome, J.; Chen, B.B.; Mallampalli, R.K. The Proapoptotic F-box Protein Fbxl7 Regulates Mitochondrial Function by Mediating the Ubiquitylation and Proteasomal Degradation of Survivin. J. Biol. Chem. 2015, 290, 11843–11852. [Google Scholar] [CrossRef]
- Arora, V.; Cheung, H.H.; Plenchette, S.; Micali, O.C.; Liston, P.; Korneluk, R.G. Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J. Biol. Chem. 2007, 282, 26202–26209. [Google Scholar] [CrossRef]
- Li, Z.; Pei, X.H.; Yan, J.; Yan, F.; Cappell, K.M.; Whitehurst, A.W.; Xiong, Y. CUL9 mediates the functions of the 3M complex and ubiquitylates survivin to maintain genome integrity. Mol. Cell 2014, 54, 805–819. [Google Scholar] [CrossRef]
- Woo, S.M.; Kim, S.; Seo, S.U.; Kim, S.; Park, J.W.; Kim, G.; Choi, Y.R.; Hur, K.; Kwon, T.K. Inhibition of USP1 enhances anticancer drugs-induced cancer cell death through downregulation of survivin and miR-216a-5p-mediated upregulation of DR5. Cell Death Dis. 2022, 13, 821. [Google Scholar] [CrossRef]
- Chandrasekaran, A.P.; Tyagi, A.; Poondla, N.; Sarodaya, N.; Karapurkar, J.K.; Kaushal, K.; Park, C.H.; Hong, S.H.; Kim, K.S.; Ramakrishna, S. Dual role of deubiquitinating enzyme USP19 regulates mitotic progression and tumorigenesis by stabilizing survivin. Mol. Ther. 2022, 30, 3414–3429. [Google Scholar] [CrossRef]
- Wang, W.; Lin, H.; Zheng, E.; Hou, Z.; Liu, Y.; Huang, W.; Chen, D.; Feng, J.; Li, J.; Li, L. Regulation of survivin protein stability by USP35 is evolutionarily conserved. Biochem. Biophys. Res. Commun. 2021, 574, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.M.; Seo, S.U.; Kubatka, P.; Min, K.J.; Kwon, T.K. Honokiol Enhances TRAIL-Mediated Apoptosis through STAMBPL1-Induced Survivin and c-FLIP Degradation. Biomolecules 2019, 9, 838. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.U.; Min, K.J.; Woo, S.M.; Kwon, T.K. Z-FL-COCHO, a cathepsin S inhibitor, enhances oxaliplatin-mediated apoptosis through the induction of endoplasmic reticulum stress. Exp. Mol. Med. 2018, 50, 107. [Google Scholar] [CrossRef]
- Park, Y.S.; Kwon, Y.J.; Chun, Y.J. CYP1B1 Activates Wnt/beta-Catenin Signaling through Suppression of Herc5-Mediated ISGylation for Protein Degradation on beta-Catenin in HeLa Cells. Toxicol. Res. 2017, 33, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.M.; Seo, S.U.; Min, K.J.; Kwon, T.K. BIX-01294 sensitizes renal cancer Caki cells to TRAIL-induced apoptosis through downregulation of survivin expression and upregulation of DR5 expression. Cell Death Discov. 2018, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.U.; Woo, S.M.; Kim, M.W.; Lee, H.S.; Kim, S.H.; Kang, S.C.; Lee, E.W.; Min, K.J.; Kwon, T.K. Cathepsin K inhibition-induced mitochondrial ROS enhances sensitivity of cancer cells to anti-cancer drugs through USP27x-mediated Bim protein stabilization. Redox Biol. 2020, 30, 101422. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, T.G.; Woo, S.M.; Seo, S.U.; Kim, S.; Park, J.-W.; Chang, Y.-C.; Kwon, T.K. Inhibition of USP2 Enhances TRAIL-Mediated Cancer Cell Death through Downregulation of Survivin. Int. J. Mol. Sci. 2023, 24, 12816. https://doi.org/10.3390/ijms241612816
Lee TG, Woo SM, Seo SU, Kim S, Park J-W, Chang Y-C, Kwon TK. Inhibition of USP2 Enhances TRAIL-Mediated Cancer Cell Death through Downregulation of Survivin. International Journal of Molecular Sciences. 2023; 24(16):12816. https://doi.org/10.3390/ijms241612816
Chicago/Turabian StyleLee, Tak Gyeom, Seon Min Woo, Seung Un Seo, Shin Kim, Jong-Wook Park, Young-Chae Chang, and Taeg Kyu Kwon. 2023. "Inhibition of USP2 Enhances TRAIL-Mediated Cancer Cell Death through Downregulation of Survivin" International Journal of Molecular Sciences 24, no. 16: 12816. https://doi.org/10.3390/ijms241612816
APA StyleLee, T. G., Woo, S. M., Seo, S. U., Kim, S., Park, J.-W., Chang, Y.-C., & Kwon, T. K. (2023). Inhibition of USP2 Enhances TRAIL-Mediated Cancer Cell Death through Downregulation of Survivin. International Journal of Molecular Sciences, 24(16), 12816. https://doi.org/10.3390/ijms241612816






