Evaluating the Molecular Properties and Function of ANKHD1, and Its Role in Cancer
Abstract
:1. Structure and Function of ANKHD1
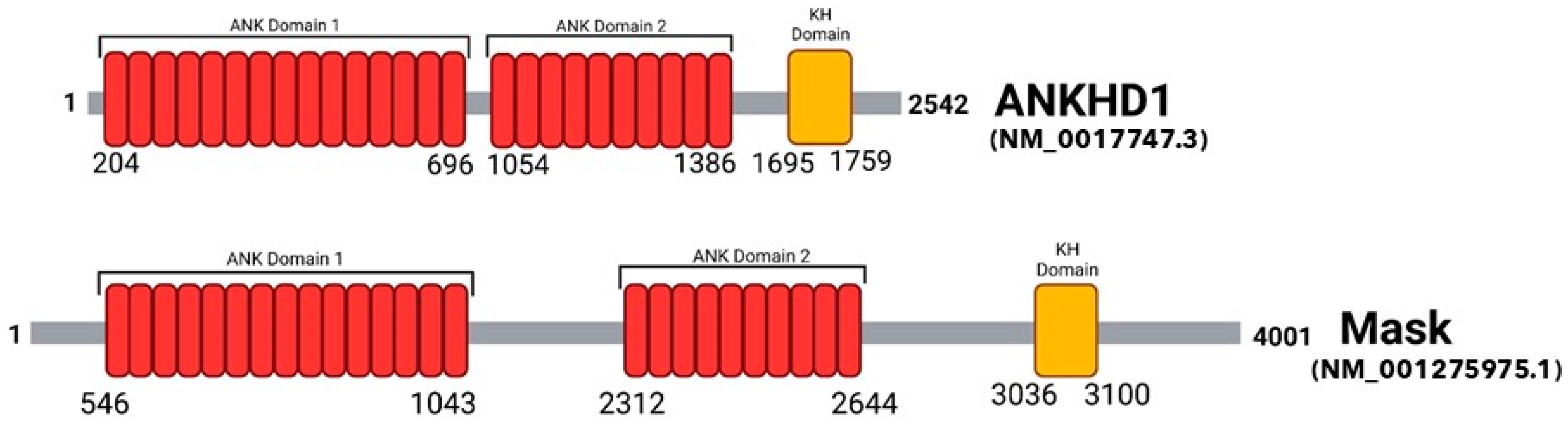
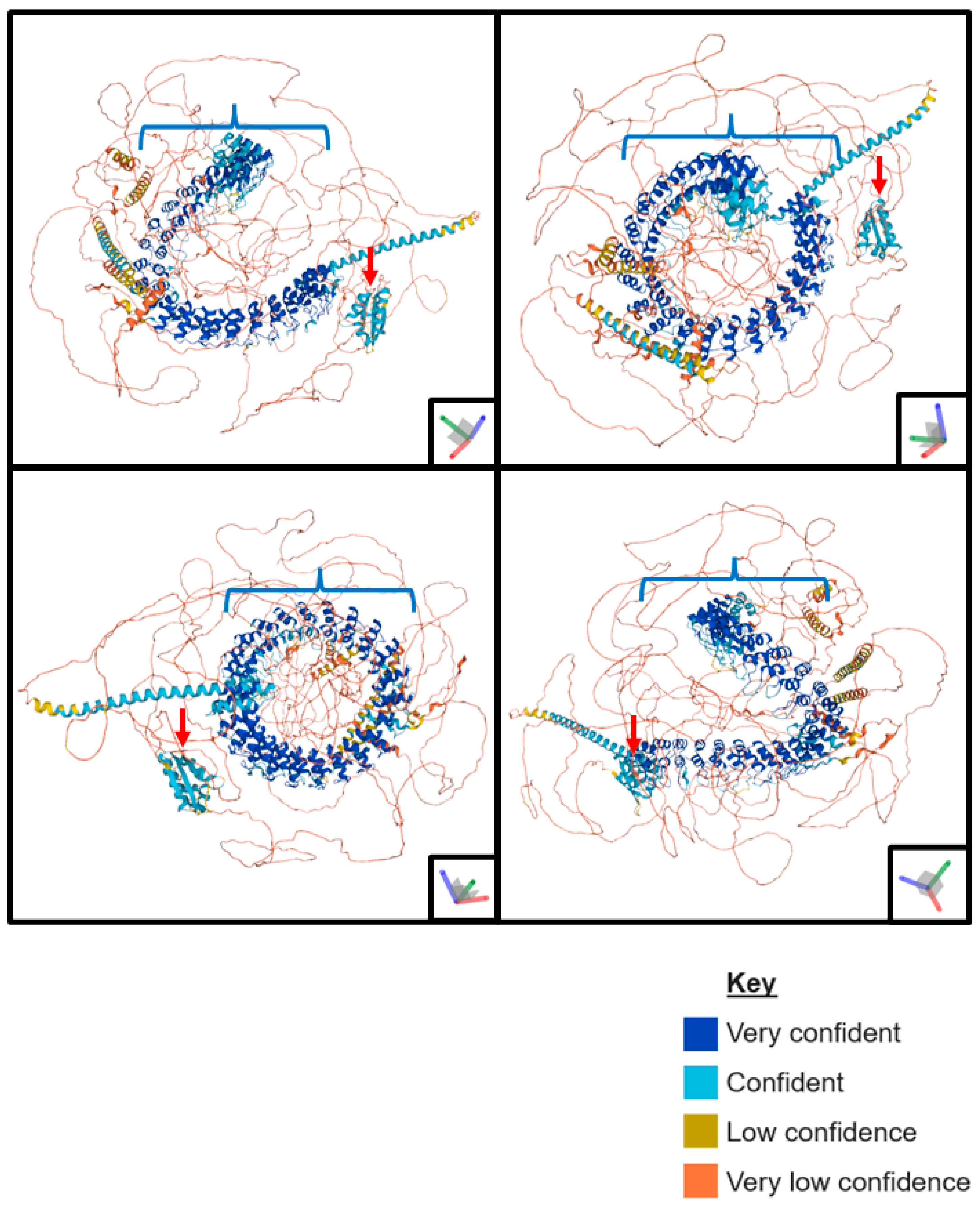
2. The Protein Domains of ANKHD1
3. Isoforms of ANKHD1
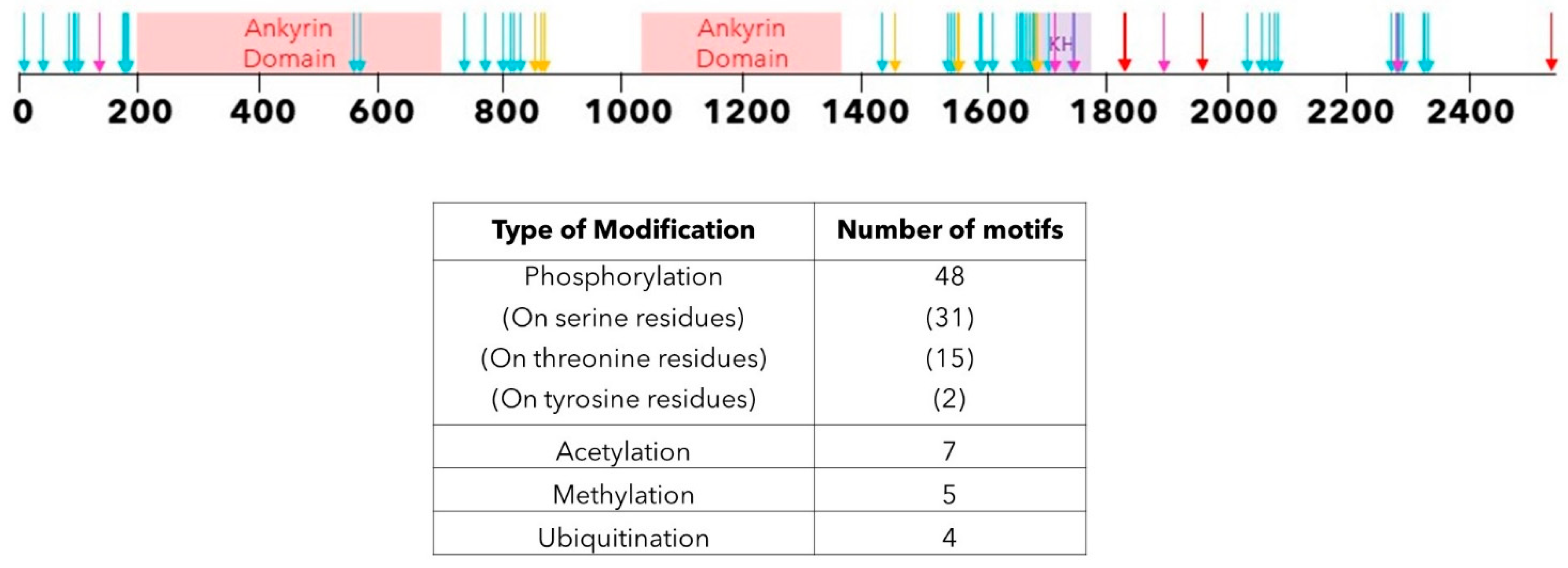
4. Post-Translational Modifications of ANKHD1
5. Subcellular Localisation of ANKHD1
6. Role of ANKHD1 in Signalling Pathways
6.1. SHP2 Signalling
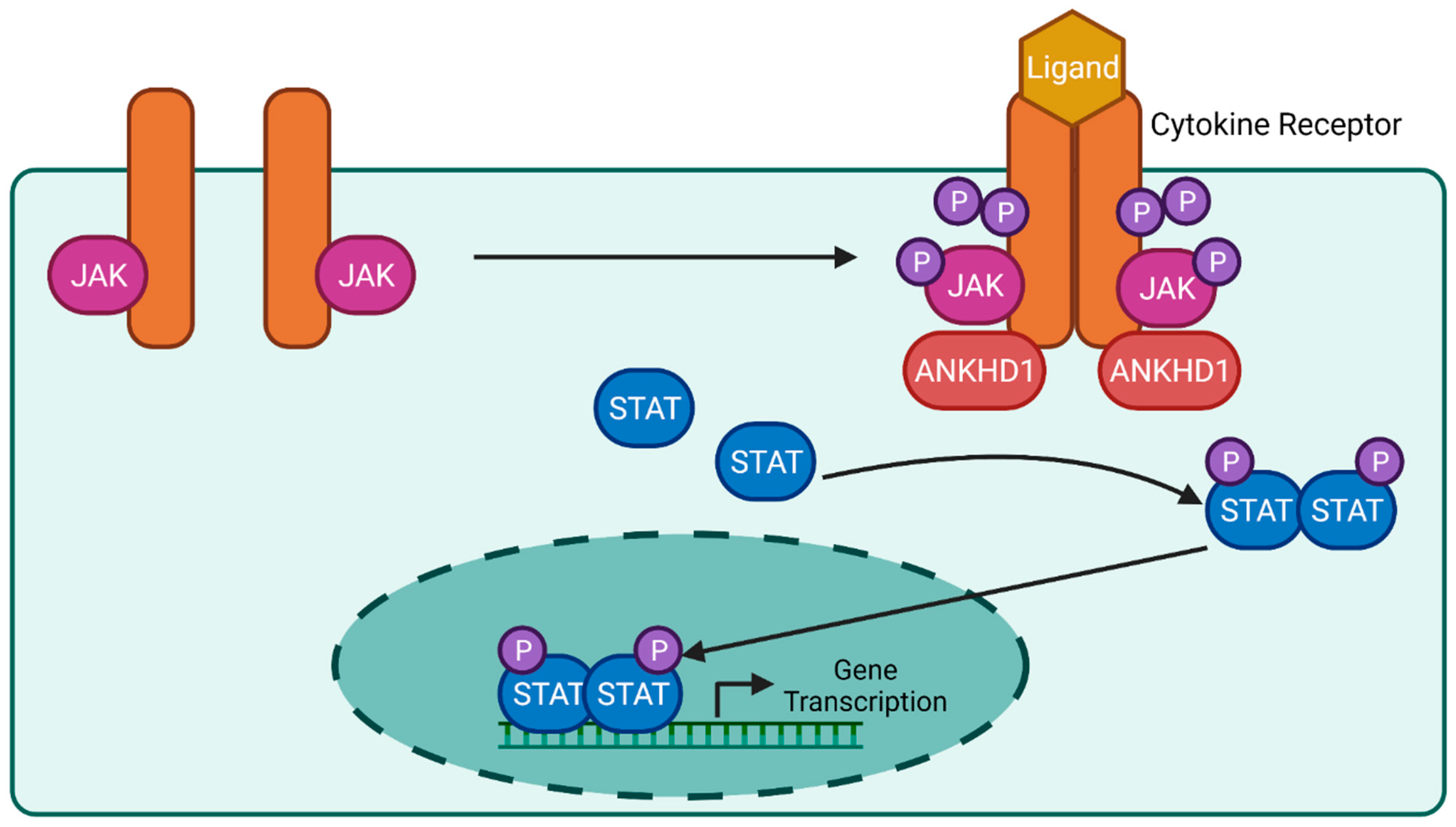
6.2. JAK/STAT Signalling Pathway
6.3. Hippo Signalling Pathway
6.4. p21 Signalling Pathway
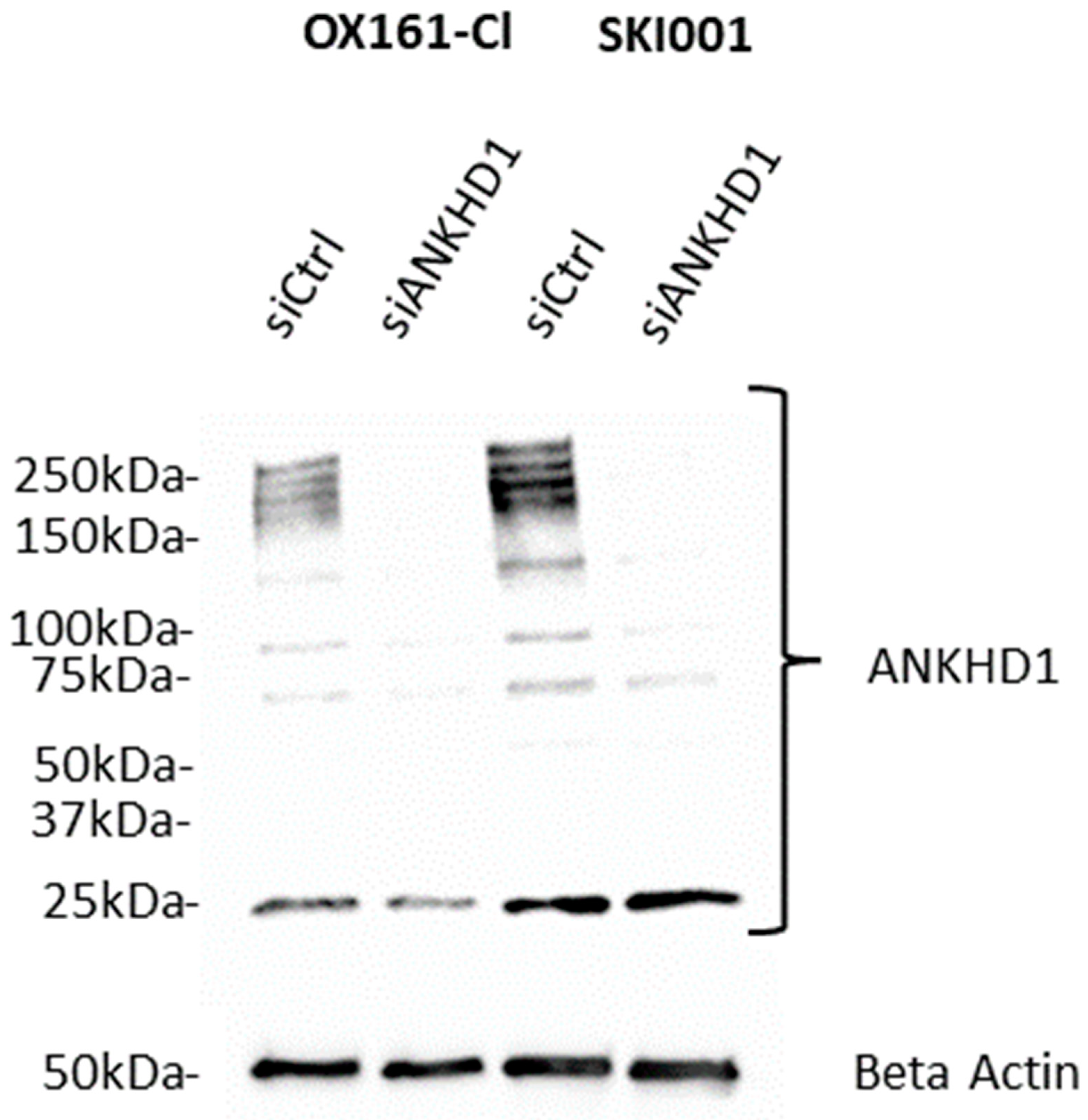
7. Role of ANKHD1 in Cancer
| Type of Cancer | References | Cellular Function of ANKHD1 | Clinical Features Associated with Enhanced ANKHD1 |
|---|---|---|---|
| Breast Cancer | [10,38] | Increases the viability, clonogenicity and migration of aggressive breast cancer cell lines, possibly due to the positive regulation of the YAP/Yki pathway. | Increased levels of ANKHD1 were correlated with a reduction in relapse-free survival, but the extent differs based on the breast cancer subtype. |
| Colorectal Cancer | [54] | Increases cell proliferation, migration, and invasion. It also dysregulates the epithelial organisation of tissues by increasing epithelial-to-mesenchymal transition. | ANKHD1 expression was correlated with greater tumour infiltration. It was demonstrated to promote metastasis and growth in tumorous nodules. |
| Glioma | [17] | Promotes angiogenesis. | An increase in ANKHD1 is detected in glioma-associated endothelial cells, but there are no data to show how this may affect prognosis. |
| Hepatocellular Cancer | [61] | Enhances cellular migration and invasion. | Higher levels of ANKHD1 are correlated with larger tumours, more nodes, poorer differentiation of tumour boundaries, more advanced metastasis, shorter time between recurrence and overall reduced survival rates. |
| Leukaemia | [13,33] | Increases cellular and clonal proliferation and migration. It enhances tumorigenicity in xenografts. | Higher levels of ANKHD1 (protein and mRNA) are observed in leukaemia cell lines and (mRNA only) are observed in primary acute leukaemia samples. |
| Non-Small-Cell Lung Cancer | [53,62] | Stimulates proliferation, invasion, and colony formation. May have a role in promoting DDP-chemoresistance. | ANKHD1 expression was correlated with greater growth, metastasis, overall reduced survival rates and poorer prognosis. ANKHD1 is required for SMYD3-induced DDP chemoresistance. |
| Multiple Myeloma | [34,36,57] | Increases proliferation and cell-cycle progression. It enhances tumorigenicity in xenografts. It also promotes growth, division, and migration of cell colonies. | ANKHD1 is highly expressed in neoplastic plasma cells. |
| Pancreatic Ductal Adenocarcinoma | [64,65] | Inferred control of cell growth by controlling the cell cycle inhibitor, p21. | Premature termination of the ANKHD1 protein due to a nonsense mutation (E2410*) was correlated with Pancreatic Ductal Adenocarcinoma. |
| Prostate Cancer | [51] | Increases cell growth, cell cycle progression during S phase and enhances tumorigenicity in xenografts. | No data. |
| Renal Cell Carcinoma | [16] | Increases proliferation by upregulating proliferative genes and drives cellular division via mitosis. | ANKHD1 expression (protein and mRNA) is upregulated early in the kidneys of patients with renal cell carcinoma. |
| Uterine Corpus Endometrial Carcinoma | [37] | In murine models, transcript variant ANKHD1-BP3 was demonstrated to promote cell proliferation whilst inhibiting metastasis. | Transcript variant ANKHD1-BP3 was highly expressed in primary tissue samples. In clinical samples, it was demonstrated to promote proliferation but inhibit metastasis, invasion, apoptosis, and necrosis of cells. |

8. Summary and Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Expression Atlas. EBI. Available online: https://www.ebi.ac.uk/gxa/experiments/E-MTAB-3358/Results (accessed on 23 January 2023).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Smith, R.K.; Carroll, P.M.; Allard, J.D.; Simon, M.A. MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development 2002, 129, 71–82. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Sigrist, C.J.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef]
- Breeden, L.; Nasmyth, K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature 1987, 329, 651–654. [Google Scholar] [CrossRef]
- Li, J.; Mahajan, A.; Tsai, M.D. Ankyrin repeat: A unique motif mediating protein-protein interactions. Biochemistry 2006, 45, 15168–15178. [Google Scholar] [CrossRef]
- RefSeq. Available online: http://genome.cse.ucsc.edu/cgi-bin/hgc?hgsid=1171240211_sPUEWAVI3sxH7RAAT8w39UaBm9OX&db=hg19&c=chr5&l=112173878&r=180837595&o=139781417&t=139852062&g=refGene&i=NM_017978 (accessed on 23 January 2023).
- Muller, P.; Kuttenkeuler, D.; Gesellchen, V.; Zeidler, M.P.; Boutros, M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature 2005, 436, 871–875. [Google Scholar] [CrossRef]
- Sansores-Garcia, L.; Atkins, M.; Moya, I.M.; Shahmoradgoli, M.; Tao, C.; Mills, G.B.; Halder, G. Mask is required for the activity of the Hippo pathway effector Yki/YAP. Curr. Biol. 2013, 23, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Sidor, C.M.; Brain, R.; Thompson, B.J. Mask proteins are cofactors of Yorkie/YAP in the Hippo pathway. Curr. Biol. 2013, 23, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Li, X.; Tian, X.; Wu, C. Mask loss-of-function rescues mitochondrial impairment and muscle degeneration of Drosophila pink1 and parkin mutants. Hum. Mol. Genet. 2015, 24, 3272–3285. [Google Scholar] [CrossRef]
- Machado-Neto, J.A.; Lazarini, M.; Favaro, P.; de Melo Campos, P.; Scopim-Ribeiro, R.; Franchi Junior, G.C.; Nowill, A.E.; Lima, P.R.; Costa, F.F.; Benichou, S.; et al. ANKHD1 silencing inhibits Stathmin 1 activity, cell proliferation and migration of leukemia cells. Biochim. Biophys. Acta 2015, 1853, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.; Edwards, L.; Regan, L. Structure and function of KH domains. FEBS J. 2008, 275, 2712–2726. [Google Scholar] [CrossRef] [PubMed]
- Hollingworth, D.; Candel, A.M.; Nicastro, G.; Martin, S.R.; Briata, P.; Gherzi, R.; Ramos, A. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 2012, 40, 6873–6886. [Google Scholar] [CrossRef] [PubMed]
- Fragiadaki, M.; Zeidler, M.P. Ankyrin repeat and single KH domain 1 (ANKHD1) drives renal cancer cell proliferation via binding to and altering a subset of miRNAs. J. Biol. Chem. 2018, 293, 9570–9579. [Google Scholar] [CrossRef]
- Yang, C.; Zheng, J.; Liu, X.; Xue, Y.; He, Q.; Dong, Y.; Wang, D.; Li, Z.; Liu, L.; Ma, J.; et al. Role of ANKHD1/LINC00346/ZNF655 Feedback Loop in Regulating the Glioma Angiogenesis via Staufen1-Mediated mRNA Decay. Mol. Ther. Nucleic Acids 2020, 20, 866–878. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Poulin, F.; Brueschke, A.; Sonenberg, N. Gene fusion and overlapping reading frames in the mammalian genes for 4E-BP3 and MASK. J. Biol. Chem. 2003, 278, 52290–52297. [Google Scholar] [CrossRef]
- National Cancer Institute. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/relapse-free-survival (accessed on 23 January 2023).
- Ensembl. Available online: https://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000131503;r=5:140401814-140539856 (accessed on 23 January 2023).
- Miles, M.C.; Janket, M.L.; Wheeler, E.D.; Chattopadhyay, A.; Majumder, B.; Dericco, J.; Schafer, E.A.; Ayyavoo, V. Molecular and functional characterization of a novel splice variant of ANKHD1 that lacks the KH domain and its role in cell survival and apoptosis. FEBS J. 2005, 272, 4091–4102. [Google Scholar] [CrossRef]
- Poulin, F.; Gingras, A.C.; Olsen, H.; Chevalier, S.; Sonenberg, N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 1998, 273, 14002–14007. [Google Scholar] [CrossRef]
- Pasek, S.; Risler, J.L.; Brezellec, P. Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins. Bioinformatics 2006, 22, 1418–1423. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 23 January 2023).
- Wilke, C.O. Transcriptional robustness complements nonsense-mediated decay in humans. PLoS Genet. 2011, 7, e1002296. [Google Scholar] [CrossRef] [PubMed]
- Parker, E.; Newby, L.J.; Sharpe, C.C.; Rossetti, S.; Streets, A.J.; Harris, P.C.; O'Hare, M.J.; Ong, A.C. Hyperproliferation of PKD1 cystic cells is induced by insulin-like growth factor-1 activation of the Ras/Raf signalling system. Kidney Int. 2007, 72, 157–165. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 23 January 2023).
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Kitamata, M.; Hanawa-Suetsugu, K.; Maruyama, K.; Suetsugu, S. Membrane-Deformation Ability of ANKHD1 Is Involved in the Early Endosome Enlargement. iScience 2019, 17, 101–118. [Google Scholar] [CrossRef]
- Traina, F.; Favaro, P.M.; Medina Sde, S.; Duarte Ada, S.; Winnischofer, S.M.; Costa, F.F.; Saad, S.T. ANKHD1, ankyrin repeat and KH domain containing 1, is overexpressed in acute leukemias and is associated with SHP2 in K562 cells. Biochim. Biophys. Acta 2006, 1762, 828–834. [Google Scholar] [CrossRef]
- Dhyani, A.; Duarte, A.S.; Machado-Neto, J.A.; Favaro, P.; Ortega, M.M.; Olalla Saad, S.T. ANKHD1 regulates cell cycle progression and proliferation in multiple myeloma cells. FEBS Lett. 2012, 586, 4311–4318. [Google Scholar] [CrossRef]
- Fisher, K.H.; Fragiadaki, M.; Pugazhendhi, D.; Bausek, N.; Arredondo, M.A.; Thomas, S.J.; Brown, S.; Zeidler, M.P. A genome-wide RNAi screen identifies MASK as a positive regulator of cytokine receptor stability. J. Cell Sci. 2018, 131, jcs209551. [Google Scholar] [CrossRef]
- Dhyani, A.; Machado-Neto, J.A.; Favaro, P.; Saad, S.T. ANKHD1 represses p21 (WAF1/CIP1) promoter and promotes multiple myeloma cell growth. Eur. J. Cancer 2015, 51, 252–259. [Google Scholar] [CrossRef]
- Sun, W.; Huang, R.; Li, Z.; Zhu, Y.; Bai, Y.; Wu, S.; Wang, J.; Xiao, Y.; Xian, S.; Tong, X.; et al. Alternative ANKHD1 transcript promotes proliferation and inhibits migration in uterine corpus endometrial carcinoma. NPJ Genom. Med. 2022, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, B.O.; de Almeida, L.C.; Costa-Lotufo, L.V.; Machado-Neto, J.A. ANKHD1 contributes to the malignant phenotype of triple-negative breast cancer cells. Cell Biol. Int. 2022, 46, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E., Jr. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, M.P.; Bach, E.A.; Perrimon, N. The roles of the Drosophila JAK/STAT pathway. Oncogene 2000, 19, 2598–2606. [Google Scholar] [CrossRef]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984–2009. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Darnell, J.E., Jr. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- Seif, F.; Khoshmirsafa, M.; Aazami, H.; Mohsenzadegan, M.; Sedighi, G.; Bahar, M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun. Signal 2017, 15, 23. [Google Scholar] [CrossRef]
- Banerjee, S.; Biehl, A.; Gadina, M.; Hasni, S.; Schwartz, D.M. JAK-STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs 2017, 77, 521–546. [Google Scholar] [CrossRef]
- Vainchenker, W.; Constantinescu, S.N. JAK/STAT signaling in hematological malignancies. Oncogene 2013, 32, 2601–2613. [Google Scholar] [CrossRef]
- Boopathy, G.T.K.; Hong, W. Role of Hippo Pathway-YAP/TAZ Signaling in Angiogenesis. Front. Cell Dev. Biol. 2019, 7, 49. [Google Scholar] [CrossRef]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef] [PubMed]
- Werneburg, N.; Gores, G.J.; Smoot, R.L. The Hippo Pathway and YAP Signaling: Emerging Concepts in Regulation, Signaling, and Experimental Targeting Strategies With Implications for Hepatobiliary Malignancies. Gene Expr. 2020, 20, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Zygulska, A.L.; Krzemieniecki, K.; Pierzchalski, P. Hippo pathway—Brief overview of its relevance in cancer. J. Physiol. Pharmacol. 2017, 68, 311–335. [Google Scholar] [PubMed]
- Zhang, X.; Zhao, H.; Li, Y.; Xia, D.; Yang, L.; Ma, Y.; Li, H. The role of YAP/TAZ activity in cancer metabolic reprogramming. Mol. Cancer 2018, 17, 134. [Google Scholar] [CrossRef]
- Machado-Neto, J.A.; Lazarini, M.; Favaro, P.; Franchi, G.C., Jr.; Nowill, A.E.; Saad, S.T.; Traina, F. ANKHD1, a novel component of the Hippo signaling pathway, promotes YAP1 activation and cell cycle progression in prostate cancer cells. Exp. Cell Res. 2014, 324, 137–145. [Google Scholar] [CrossRef]
- Sidor, C.; Borreguero-Munoz, N.; Fletcher, G.C.; Elbediwy, A.; Guillermin, O.; Thompson, B.J. Mask family proteins ANKHD1 and ANKRD17 regulate YAP nuclear import and stability. Elife 2019, 8, e48601. [Google Scholar] [CrossRef]
- Liu, X.F.; Han, Q.; Rong, X.Z.; Yang, M.; Han, Y.C.; Yu, J.H.; Lin, X.Y. ANKHD1 promotes proliferation and invasion of nonsmallcell lung cancer cells via regulating YAP oncoprotein expression and inactivating the Hippo pathway. Int. J. Oncol. 2020, 56, 1175–1185. [Google Scholar] [CrossRef]
- Yao, P.; Li, Y.; Shen, W.; Xu, X.; Zhu, W.; Yang, X.; Cao, J.; Xing, C. ANKHD1 silencing suppresses the proliferation, migration and invasion of CRC cells by inhibiting YAP1-induced activation of EMT. Am. J. Cancer Res. 2018, 8, 2311–2324. [Google Scholar]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair. 2016, 42, 63–71. [Google Scholar] [CrossRef]
- Dhyani, A.; Favaro, P.; Olalla Saad, S.T. ANKHD1 is an S phase protein required for histone synthesis and DNA repair in multiple myeloma cells. Blood Cells Mol. Dis. 2020, 84, 102460. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Seeger-Nukpezah, T.; Geynisman, D.M.; Nikonova, A.S.; Benzing, T.; Golemis, E.A. The hallmarks of cancer: Relevance to the pathogenesis of polycystic kidney disease. Nat. Rev. Nephrol. 2015, 11, 515–534. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yu, Y.; Zheng, S.; Zhao, X.; Dong, Q.; He, Z.; Liang, Y.; Lu, Q.; Fang, Y.; Gan, X.; et al. Overexpression of Shp2 tyrosine phosphatase is implicated in leukemogenesis in adult human leukemia. Blood 2005, 106, 3142–3149. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, H.; Tu, K.; Yu, W.; Zhang, J.; Hu, Z.; Zhang, H.; Hao, D.; Huang, P.; Wang, J.; et al. ANKHD1 is required for SMYD3 to promote tumor metastasis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 18. [Google Scholar] [CrossRef]
- Lv, H.W.; Xing, W.Q.; Ba, Y.F.; Li, H.M.; Wang, H.R.; Li, Y. SMYD3 confers cisplatin chemoresistance of NSCLC cells in an ANKHD1-dependent manner. Transl. Oncol. 2021, 14, 101075. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Ren, D.; Sun, Y.; Li, D.; Wu, H.; Jin, X. USP22-mediated deubiquitination of PTEN inhibits pancreatic cancer progression by inducing p21 expression. Mol. Oncol. 2021, 16, 1200–1217. [Google Scholar] [CrossRef]
- Werner, J.; Bernhard, P.; Cosenza-Contreras, M.; Pinter, N.; Fahrner, M.; Pallavi, P.; Eberhard, J.; Bronsert, P.; Ruckert, F.; Schilling, O. Targeted and explorative profiling of kallikrein proteases and global proteome biology of pancreatic ductal adenocarcinoma, chronic pancreatitis, and normal pancreas highlights disease-specific proteome remodelling. Neoplasia 2023, 36, 100871. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mullenger, J.L.; Zeidler, M.P.; Fragiadaki, M. Evaluating the Molecular Properties and Function of ANKHD1, and Its Role in Cancer. Int. J. Mol. Sci. 2023, 24, 12834. https://doi.org/10.3390/ijms241612834
Mullenger JL, Zeidler MP, Fragiadaki M. Evaluating the Molecular Properties and Function of ANKHD1, and Its Role in Cancer. International Journal of Molecular Sciences. 2023; 24(16):12834. https://doi.org/10.3390/ijms241612834
Chicago/Turabian StyleMullenger, Jordan L., Martin P. Zeidler, and Maria Fragiadaki. 2023. "Evaluating the Molecular Properties and Function of ANKHD1, and Its Role in Cancer" International Journal of Molecular Sciences 24, no. 16: 12834. https://doi.org/10.3390/ijms241612834







