Genome-Wide Identification of GmSPS Gene Family in Soybean and Expression Analysis in Response to Cold Stress
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of GmSPS Genes in Soybean
2.2. Phylogenetic Analysis of SPS Proteins
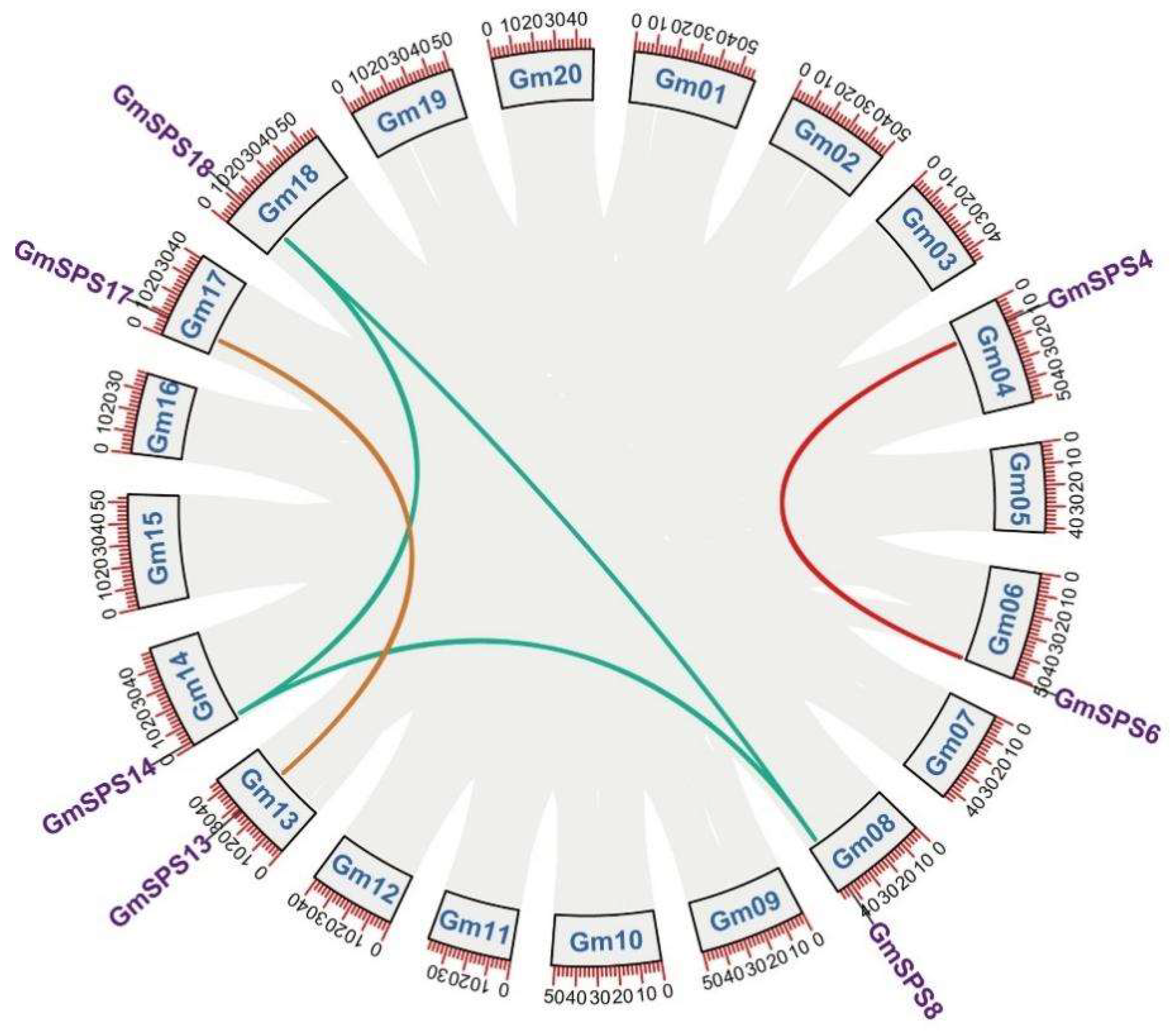
2.3. Collinearity Analysis of GmSPS Genes
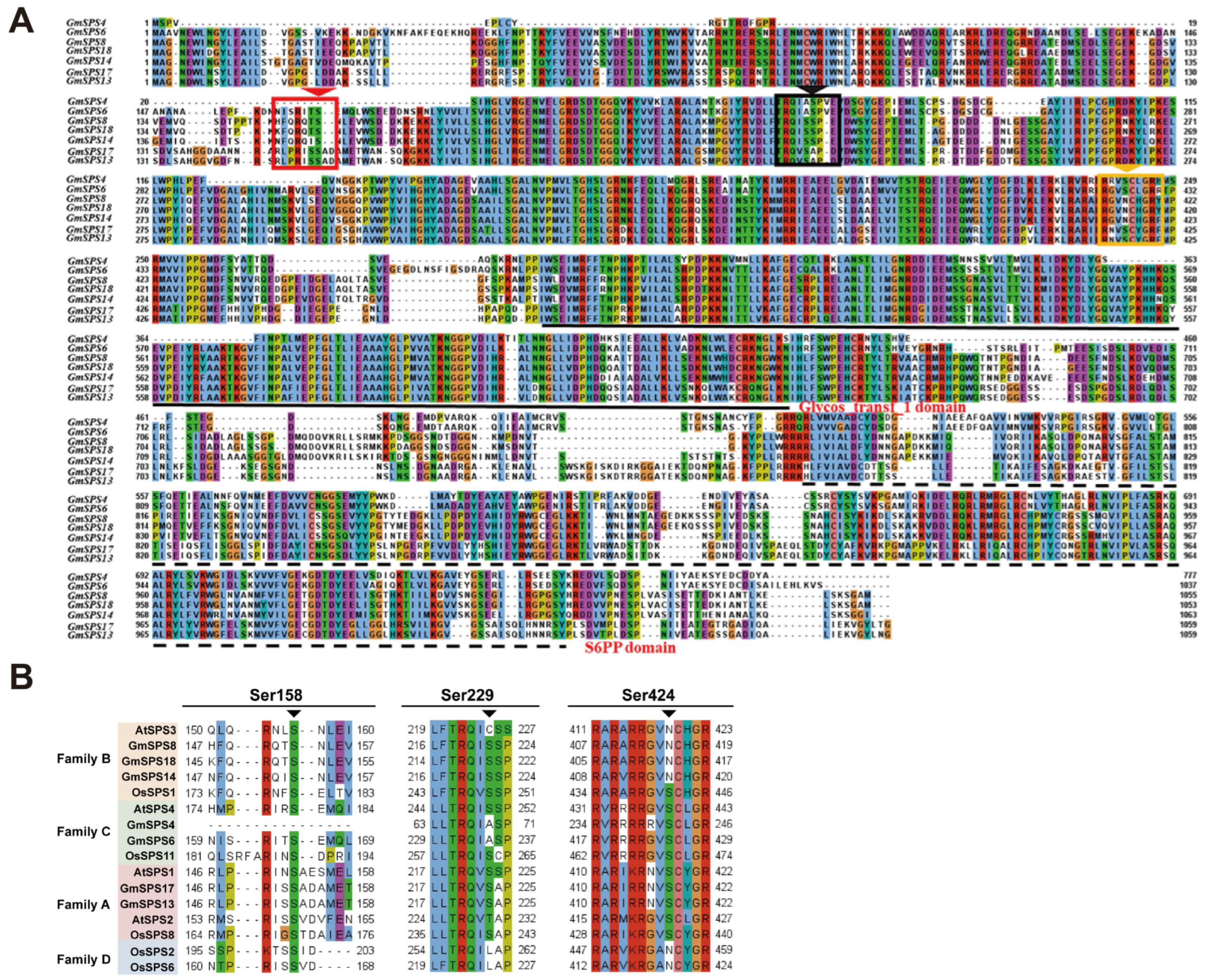
2.4. Gene Structure and Conserved Motif Analyses of GmSPSs
2.5. Analysis of Phosphorylation Sites in GmSPS Proteins
2.6. Promoter cis-Element Analysis of GmSPS
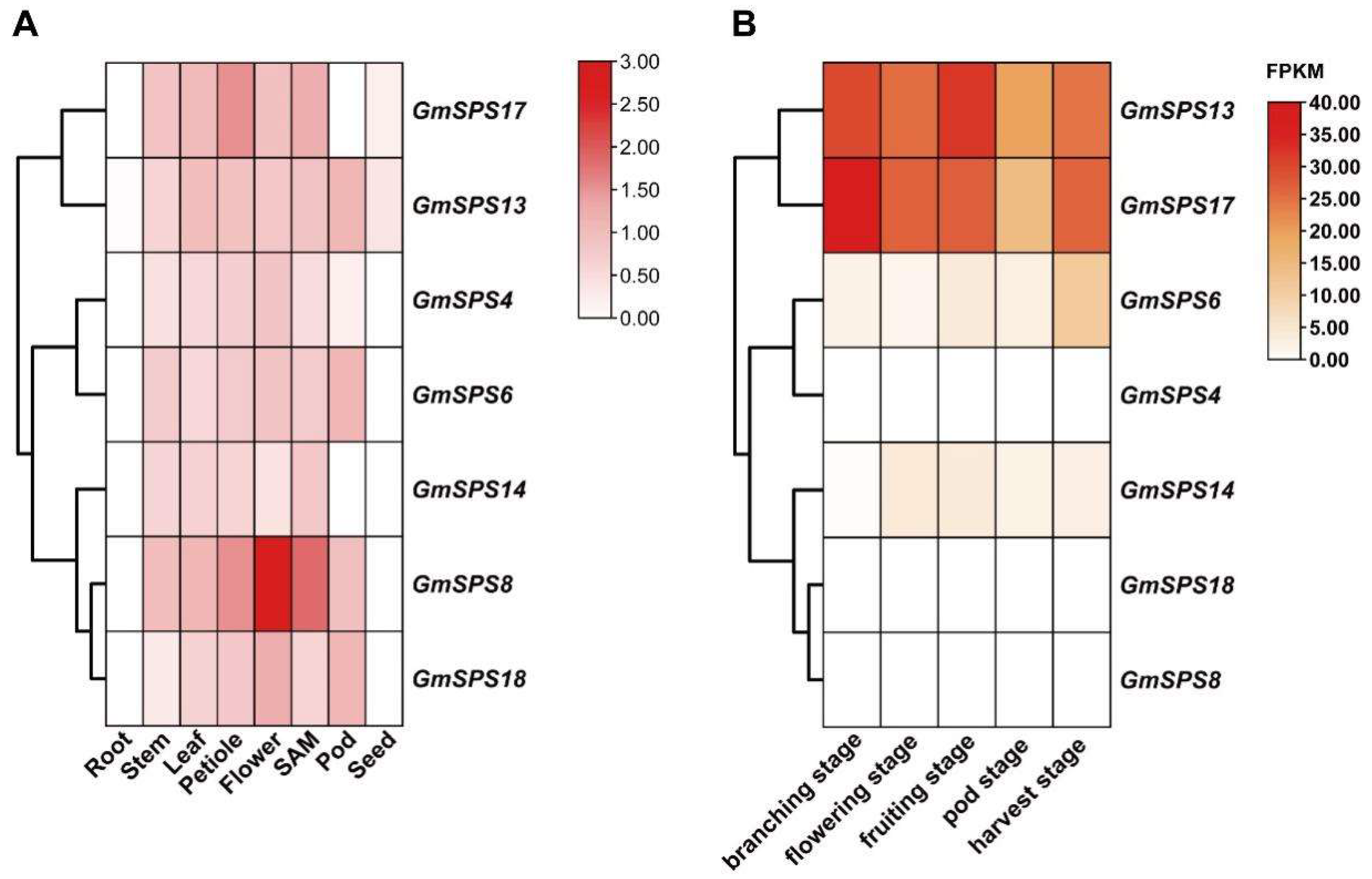
2.7. Tissue-Specific Expression Patterns of GmSPS Members in Soybean
2.8. Expression Analysis of GmSPS Genes in Response to Cold Stress
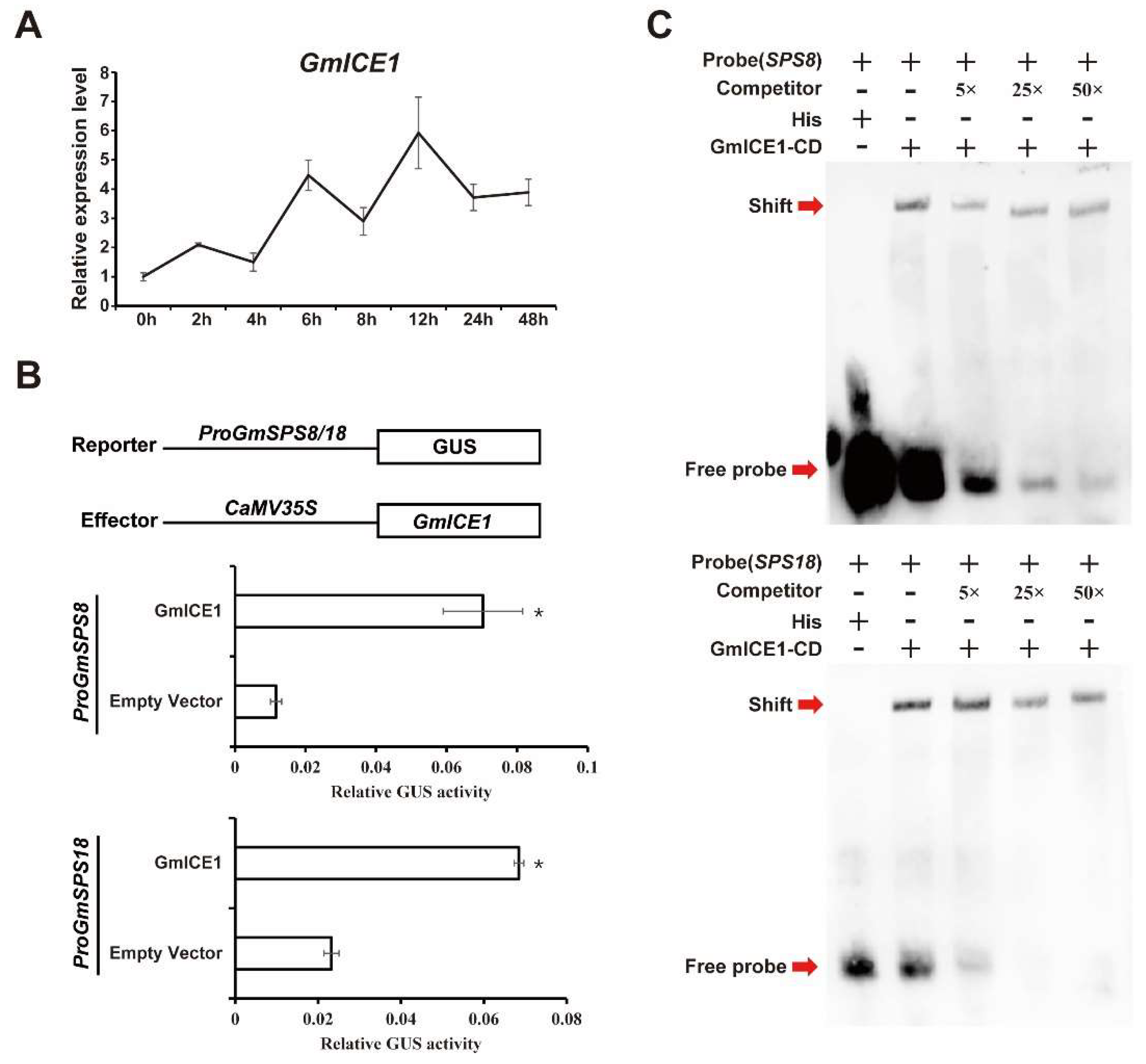
2.9. GmSPS8/18 Were Upregulated by GmICE1 Involved in Cold Stress
3. Discussion
4. Materials and Methods
4.1. Identification of GmSPS Genes in Soybean
4.2. Phylogenetic Analysis of SPS Proteins
4.3. Analysis of Gene Structure and Conserved Motifs
4.4. Phosphorylation Sites Analysis in GmSPSs
4.5. Chromosomal Location and Collinearity Analysis
4.6. cis-Regulatory Element in the Promoter Regions
4.7. Plant Materials and Cold Stress Treatment
4.8. RNA Extraction and qRT-PCR Analysis
4.9. GUS Activity Assay in Transiently Transformed N. benthamiana Leaves
4.10. Electrophoretic Mobility Shift Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Jia, Y.; Shi, Y.; Zhang, X.; Song, C.; Gong, Z.; Yang, S. OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 2018, 37, e98228. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Wilson, R.F. Soybean: Market Driven Research Needs; Springer: New York, NY, USA, 2008. [Google Scholar]
- Maruyama, K.; Takeda, M.; Kidokoro, S.; Yamada, K.; Sakuma, Y.; Urano, K.; Fujita, M.; Yoshiwara, K.; Matsukura, S.; Morishita, Y.; et al. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 2009, 150, 1972–1980. [Google Scholar] [CrossRef]
- Robison, J.D.; Yamasaki, Y.; Randall, S.K. The ethylene signaling pathway negatively impacts CBF/DREB-regulated cold re sponse in soybean (Glycine max). Front. Plant Sci. 2019, 10, 121. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, Y.; Randall, S.K. Functionality of soybean CBF/DREB1 transcription factors. Plant Sci. 2016, 246, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Reddy, V.R.; Quebedeaux, B. Growth and photosynthetic responses of soybean to short-term cold temperature. Environ. Exp. Bot. 1997, 37, 13–24. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, H.; Li, X.; Ji, H. Enhancement of plant cold tolerance by soybean RCC1 family gene GmTCF1a. BMC Plant Biol. 2021, 21, 369. [Google Scholar] [CrossRef]
- Ji, H.; Wang, Y.; Cloix, C.; Li, K.; Jenkins, G.I.; Wang, S.; Shang, Z.; Shi, Y.; Yang, S.; Li, X. The Arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar] [CrossRef]
- Rathinasabapathi, B. Metabolic engineering for stress tolerance: Installing osmoprotectant synthesis pathways. Ann. Bot. 2000, 86, 709–716. [Google Scholar] [CrossRef]
- Wanner, L.A.; Junttila, O. Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 1999, 120, 391–400. [Google Scholar] [CrossRef]
- Zúñiga-Feest, A.; Ort, D.R.; Gutiérrez, A.; Gidekel, M.; Bravo, L.A.; Corcuera, L.J. Light regulation of sucrose-phosphate synthase activity in the freezing-tolerant grass Deschampsia antarctica. Photosynth. Res. 2005, 83, 75–86. [Google Scholar] [CrossRef]
- Bauerfeind, M.A.; Winkelmann, T.; Franken, P.; Druege, U. Transcriptome, carbohydrate, and phytohormone analysis of Petunia hybrida reveals a complex disturbance of plant functional integrity under mild chilling stress. Front. Plant Sci. 2015, 6, 583. [Google Scholar] [CrossRef]
- Peng, T.; Zhu, X.; Duan, N.; Liu, J.H. PtrBAM1, a β-amylase-coding gene of Poncirus trifoliata, is a CBF regulon member with function in cold tolerance by modulating soluble sugar levels. Plant Cell Environ. 2014, 37, 2754–2767. [Google Scholar] [CrossRef]
- Dong, S.; Beckles, D.M. Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response. J. Plant Physiol. 2019, 234–235, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Rolland, F.; Moore, B.; Sheen, J. Sugar sensing and signaling in plants. Plant Cell 2002, 14 (Suppl. S1), 185–205. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Canam, T.; Kang, K.Y.; Ellis, D.D.; Mansfield, S.D. Over-expression of an Arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 2008, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Okamura, M.; Aoki, N.; Hirose, T.; Yonekura, M.; Ohto, C.; Ohsugi, R. Tissue specificity and diurnal change in gene expression of the sucrose phosphate synthase gene family in rice. Plant Sci. 2011, 181, 159–166. [Google Scholar] [CrossRef]
- Volkert, K.; Debast, S.; Voll, L.M.; Voll, H.; Schießl, I.; Hofmann, J.; Schneider, S.; Börnke, F. Loss of the two major leaf isoforms of sucrose-phosphate synthase in Arabidopsis thaliana limits sucrose synthesis and nocturnal starch degradation but does not alter carbon partitioning during photosynthesis. J. Exp. Bot. 2014, 65, 5217–5229. [Google Scholar] [CrossRef]
- Haigler, C.H.; Singh, B.; Zhang, D.; Hwang, S.; Wu, C.; Cai, W.X.; Hozain, M.; Kang, W.; Kiedaisch, B.; Strauss, R.E.; et al. Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol. Biol. 2007, 63, 815–832. [Google Scholar] [CrossRef]
- Miron, D.; Schaffer, A.A. Sucrose phosphate synthase, sucrose synthase, and invertase activities in developing fruit of Lycopersicon esculentum Mill. and the sucrose accumulating Lycopersicon hirsutum Humb. and Bonpl. Plant Physiol. 1991, 95, 623–627. [Google Scholar] [CrossRef]
- Dali, N.; Michaud, D.; Yelle, S. Evidence for the involvement of sucrose phosphate synthase in the pathway of sugar accumulation in sucrose-accumulating tomato fruits. Plant Physiol. 1992, 99, 434–438. [Google Scholar] [CrossRef]
- do Nascimento, J.R.; Cordenunsi, B.R.; Lajolo, F.M.; Alcocer, M.J. Banana sucrose-phosphate synthase gene expression during fruit ripening. Planta 1997, 203, 83–88. [Google Scholar] [CrossRef]
- Guy, C.L.; Huber, J.L.; Huber, S.C. Sucrose phosphate synthase and sucrose accumulation at low temperature. Plant Physiol. 1992, 100, 502–508. [Google Scholar] [CrossRef]
- Huang, T.; Luo, X.; Wei, M.; Shan, Z.; Zhu, Y.; Yang, Y.; Fan, Z. Molecular cloning and expression analysis of sucrose phosphate synthase genes in cassava (Manihot esculenta Crantz). Sci. Rep. 2020, 10, 20707. [Google Scholar] [CrossRef] [PubMed]
- Lutfiyya, L.L.; Xu, N.; D’Ordine, R.L.; Morrell, J.A.; Miller, P.W.; Duff, S.M. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J. Plant Physiol. 2007, 164, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Yang, L.; Zhu, H.; Zhou, J.; Sun, H.; Gong, H. Structure and expression analysis of sucrose phosphate synthase, sucrose synthase and invertase gene families in Solanum lycopersicum. Int. J. Mol. Sci. 2021, 22, 4698. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sreenivasulu, N.; Harshavardhan, V.T.; Seiler, C.; Sharma, S.; Khalil, Z.N.; Akhunov, E.; Sehgal, S.K.; Röder, M.S. Delineating the structural, functional and evolutionary relationships of sucrose phosphate synthase gene family II in wheat and related grasses. BMC Plant Biol. 2010, 10, 134. [Google Scholar] [CrossRef]
- Wang, D.; Zhao, J.; Hu, B.; Li, J.; Qin, Y.; Chen, L.; Qin, Y.; Hu, G. Identification and expression profile analysis of the sucrose phosphate synthase gene family in Litchi chinensis Sonn. PeerJ 2018, 6, e4379. [Google Scholar] [CrossRef]
- Castleden, C.K.; Aoki, N.; Gillespie, V.J.; MacRae, E.A.; Quick, W.P.; Buchner, P.; Foyer, C.H.; Furbank, R.T.; Lunn, J.E. Evolution and function of the sucrose-phosphate synthase gene families in wheat and other grasses. Plant Physiol. 2004, 135, 1753–1764. [Google Scholar] [CrossRef]
- Huber, S.C. Biochemical basis for effects of k-deficiency on assimilate export rate and accumulation of soluble sugars in soybean leaves. Plant Physiol. 1984, 76, 424–430. [Google Scholar] [CrossRef]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, H.; Wu, J.; Xie, F. Effect of drought stress during soybean R2-R6 growth stages on sucrose metabolism in leaf and seed. Int. J. Mol. Sci. 2020, 21, 618. [Google Scholar] [CrossRef]
- Aleman, L.; Ortega, J.L.; Martinez-Grimes, M.; Seger, M.; Holguin, F.O.; Uribe, D.J.; Garcia-Ibilcieta, D.; Sengupta-Gopalan, C. Nodule-enhanced expression of a sucrose phosphate synthase gene member (MsSPSA) has a role in carbon and nitrogen metabolism in the nodules of alfalfa (Medicago sativa L.). Planta 2010, 231, 233–244. [Google Scholar] [CrossRef]
- Li, A.; Liu, A.; Du, X.; Chen, J.Y.; Yin, M.; Hu, H.Y.; Shrestha, N.; Wu, S.D.; Wang, H.Q.; Dou, Q.W.; et al. A chromosome-scale genome assembly of a diploid alfalfa, the progenitor of autotetraploid alfalfa. Hortic. Res. 2020, 7, 194. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Cheng, Y.; Zhang, C.; You, Q.; Shen, X.; Guo, W.; Jiao, Y. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 2017, 7, 5636. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Cao, D.; Yang, H.; Guo, W.; Ouyang, W.; Chen, H.; Shan, Z.; Yang, Z.; Chen, S.; Li, X.; et al. Genome-wide identification and characterization of soybean GmLOR gene family and expression analysis in response to abiotic stresses. Int. J. Mol. Sci. 2021, 22, 12515. [Google Scholar] [CrossRef] [PubMed]
- Salih, H.; Gong, W.; He, S.; Sun, G.; Sun, J.; Du, X. Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 2016, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef]
- Yuan, S.L.; Li, R.; Chen, H.F.; Zhang, C.J.; Chen, L.M.; Hao, Q.N.; Chen, S.L.; Shan, Z.H.; Yang, Z.L.; Zhang, X.J.; et al. RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113-2. Sci. Rep. 2017, 7, 42248. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, M.; Wang, M.; Zhang, W.; Qiao, C.; Luo, Q.; Lu, X. Freshwater Cyanobacterium Synechococcus elongatus PCC 7942 adapts to an environment with salt stress via ion-induced enzymatic balance of dompatible solutes. Appl. Environ. Microbiol. 2020, 86, e02904-19. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Wang, Z.; Zhu, Q. Activities of starch hydrolytic enzymes and sucrose-phosphate synthase in the stems of rice subjected to water stress during grain filling. J. Exp. Bot. 2001, 52, 2169–2179. [Google Scholar] [CrossRef]
- Solís-Guzmán, M.G.; Argüello-Astorga, G.; López-Bucio, J.; Ruiz-Herrera, L.F.; López-Meza, J.E.; Sánchez-Calderón, L.; Carreón-Abud, Y.; Martínez-Trujillo, M. Arabidopsis thaliana sucrose phosphate synthase (sps) genes are expressed differentially in organs and tissues, and their transcription is regulated by osmotic stress. Gene Expr. Patterns 2017, 25–26, 92–101. [Google Scholar] [CrossRef]
- Padhi, S.; Grimes, M.M.; Muro-Villanueva, F.; Ortega, J.L.; Sengupta-Gopalan, C. Distinct nodule and leaf functions of two different sucrose phosphate synthases in alfalfa. Planta 2019, 250, 1743–1755. [Google Scholar] [CrossRef]
- Bilska-Kos, A.; Mytych, J.; Suski, S.; Magoń, J.; Ochodzki, P.; Zebrowski, J. Sucrose phosphate synthase (SPS), sucrose synthase (SUS) and their products in the leaves of Miscanthus × giganteus and Zea mays at low temperature. Planta 2020, 252, 23. [Google Scholar] [CrossRef]
- Bertrand, A.; Bipfubusa, M.; Claessens, A.; Rocher, S.; Castonguay, Y. Effect of photoperiod prior to cold acclimation on freezing tolerance and carbohydrate metabolism in alfalfa (Medicago sativa L.). Plant Sci. 2017, 264, 122–128. [Google Scholar] [CrossRef]
- Zeller, G.; Henz, S.R.; Widmer, C.K.; Sachsenberg, T.; Rätsch, G.; Weigel, D.; Laubinger, S. Stress-induced changes in the Ara bidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J. 2009, 58, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Lee, M.; Lee, J.H.; Lee, H.J.; Park, C.M. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 2015, 89, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhao, L.; Ren, Y.; Yang, S.; Zhu, J.K.; Zhao, C. The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 2020, 62, 258–263. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, J.; Sun, T.; Wang, X.; Zhu, C.; Ai, Y.; Gu, H. The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant Biol. 2017, 59, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Langenkämper, G.; Fung, R.W.; Newcomb, R.D.; Atkinson, R.G.; Gardner, R.C.; MacRae, E.A. Sucrose phosphate synthase genes in plants belong to three different families. J. Mol. Evol. 2002, 54, 322–332. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and appli cations. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interac tive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phos phorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. KaKs_calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chow, C.N.; Lee, T.Y.; Hung, Y.C.; Li, G.Z.; Tseng, K.C.; Liu, Y.H.; Kuo, P.L.; Zheng, H.Q.; Chang, W.C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, 1155–1163. [Google Scholar] [CrossRef]
- Xia, W.; Mason, A.S.; Xiao, Y.; Liu, Z.; Yang, Y.; Lei, X.; Wu, X.; Ma, Z.; Peng, M. Analysis of multiple transcriptomes of the African oil palm (Elaeis guineensis) to identify reference genes for RT-qPCR. J. Biotechnol. 2014, 184, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, W.; Li, J.; Yue, P.; Bu, H.; Jiang, J.; Liu, W.; Xu, Y.; Yuan, H.; Li, T.; et al. Histone acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in pear fruit. Plant Physiol. 2020, 182, 2035–2046. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, H.; He, D.; Damaris, R.N.; Yang, P. A stress-associated protein OsSAP8 modulates gibberellic acid biosynthesis by reducing the promotive effect of transcription factor OsbZIP58 on OsKO2. J. Exp. Bot. 2022, 73, 2420–2433. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Gene ID | Genome Location | Transcript Numbers | Protein Size (aa) | MW (kDa) | pI | Subcellular Location |
|---|---|---|---|---|---|---|---|
| GmSPS4 | Glyma.04g110200 | Chr04: 11369193-11375676 | 1 | 778 | 87.58 | 5.99 | Cytoplasm |
| GmSPS6 | Glyma.06g323700 | Gm06: 50675372-50684602 | 1 | 1038 | 117.09 | 6.31 | Nucleus |
| GmSPS8 | Glyma.08g308600 | Gm08: 42128704-42135717 | 1 | 1056 | 118.80 | 6.18 | Cytoplasm |
| GmSPS13 | Glyma.13g161600 | Gm13: 27135652-27142180 | 5 | 1060 | 118.01 | 6.04 | Nucleus |
| GmSPS14 | Glyma.14g029100 | Gm14: 2121804-2128414 | 2 | 1064 | 119.33 | 5.94 | Cytoplasm |
| GmSPS17 | Glyma.17g109700 | Gm17: 8599983-8606617 | 4 | 1060 | 118.08 | 6.09 | Nucleus |
| GmSPS18 | Glyma.18g108100 | Gm18: 12235657-12242570 | 1 | 1054 | 118.39 | 6.1 | Nucleus |
| Segmental Duplicated Genes | Method | Ka | Ks | Ka/Ks | T (Mya) |
|---|---|---|---|---|---|
| GmSPS17-GmSPS13 | MA | 0.012484 | 0.110769 | 0.112704 | 9.079426 |
| GmSPS18-GmSPS14 | MA | 0.074892 | 0.634249 | 0.118079 | 51.98762 |
| GmSPS6-GmSPS4 | MA | 0.060117 | 0.099571 | 0.60376 | 8.161582 |
| GmSPS8-GmSPS14 | MA | 0.068167 | 0.640031 | 0.106505 | 52.46156 |
| GmSPS8-GmSPS18 | MA | 0.014738 | 0.118258 | 0.124628 | 9.693279 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, J.; Xu, Y.; Yuan, S.; Jin, F.; Huang, Y.; Chen, H.; Shan, Z.; Yang, Z.; Chen, S.; Zhou, X.; et al. Genome-Wide Identification of GmSPS Gene Family in Soybean and Expression Analysis in Response to Cold Stress. Int. J. Mol. Sci. 2023, 24, 12878. https://doi.org/10.3390/ijms241612878
Shen J, Xu Y, Yuan S, Jin F, Huang Y, Chen H, Shan Z, Yang Z, Chen S, Zhou X, et al. Genome-Wide Identification of GmSPS Gene Family in Soybean and Expression Analysis in Response to Cold Stress. International Journal of Molecular Sciences. 2023; 24(16):12878. https://doi.org/10.3390/ijms241612878
Chicago/Turabian StyleShen, Jiafang, Yiran Xu, Songli Yuan, Fuxiao Jin, Yi Huang, Haifeng Chen, Zhihui Shan, Zhonglu Yang, Shuilian Chen, Xinan Zhou, and et al. 2023. "Genome-Wide Identification of GmSPS Gene Family in Soybean and Expression Analysis in Response to Cold Stress" International Journal of Molecular Sciences 24, no. 16: 12878. https://doi.org/10.3390/ijms241612878
APA StyleShen, J., Xu, Y., Yuan, S., Jin, F., Huang, Y., Chen, H., Shan, Z., Yang, Z., Chen, S., Zhou, X., & Zhang, C. (2023). Genome-Wide Identification of GmSPS Gene Family in Soybean and Expression Analysis in Response to Cold Stress. International Journal of Molecular Sciences, 24(16), 12878. https://doi.org/10.3390/ijms241612878






