Unnatural Amino Acid Crosslinking for Increased Spatiotemporal Resolution of Chromatin Dynamics
Abstract
:Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Richmond, T.J. DNA binding within the nucleosome core. Curr. Opin. Struc. Biol. 1998, 8, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.K.; Cheung, P. A brief histone in time: Understanding the combinatorial functions of histone PTMs in the nucleosome context. Biochem. Cell Biol. 2016, 94, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-translational modifications of histones: Mechanisms, biological functions, and therapeutic targets. MedComm 2023, 4, e292. [Google Scholar] [CrossRef] [PubMed]
- Bowman, G.D.; Poirier, M.G. Post-Translational Modifications of Histones That Influence Nucleosome Dynamics. Chem. Rev. 2014, 115, 2274–2295. [Google Scholar] [CrossRef]
- Chan, J.C.; Maze, I. Nothing Is yet Set in (Hi)stone: Novel Post-Translational Modifications Regulating Chromatin Function. Trends Biochem. Sci. 2020, 45, 829–844. [Google Scholar] [CrossRef]
- Bartholomew, B. Regulating the Chromatin Landscape: Structural and Mechanistic Perspectives. Annu. Rev. Biochem. 2014, 83, 671–696. [Google Scholar] [CrossRef]
- Simone, C. SWI/SNF: The crossroads where extracellular signaling pathways meet chromatin. J. Cell. Physiol. 2006, 207, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Lans, H.; Marteijn, J.A.; Vermeulen, W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 2012, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Qiu, J.; Ratnakumar, K.; Laurent, B.C. RSC functions as an early double-strand-break sensor in the cell’s response to DNA damage. Curr. Biol. 2007, 17, 1432–1437. [Google Scholar] [CrossRef]
- Tsabar, M.; Haber, J.E. Chromatin modifications and chromatin remodeling during DNA repair in budding yeast. Curr. Opin. Genet. Dev. 2013, 23, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Czaja, W.; Mao, P.; Smerdon, M.J. Chromatin remodelling complex RSC promotes base excision repair in chromatin of Saccharomyces cerevisiae. DNA Repair 2014, 16, 35–43. [Google Scholar] [CrossRef]
- Brahma, S.; Henikoff, S. Epigenome Regulation by Dynamic Nucleosome Unwrapping. Trends Biochem. Sci. 2020, 45, 13–26. [Google Scholar] [CrossRef]
- Prajapati, H.K.; Ocampo, J.; Clark, D.J. Interplay among ATP-Dependent Chromatin Remodelers Determines Chromatin Organisation in Yeast. Biology 2020, 9, 190. [Google Scholar] [CrossRef]
- Längst, G.; Manelyte, L. Chromatin Remodelers: From Function to Dysfunction. Genes 2015, 6, 299–324. [Google Scholar] [CrossRef]
- Becker, P.B.; Workman, J.L. Nucleosome Remodeling and Epigenetics. Cold Spring Harb. Perspect. Biol. 2013, 5, a017905. [Google Scholar] [CrossRef]
- Tyagi, M.; Imam, N.; Verma, K.; Patel, A.K. Chromatin remodelers: We are the drivers!! Nucleus 2016, 7, 388–404. [Google Scholar] [CrossRef]
- Chatterjee, N.; Sinha, D.; Lemma-Dechassa, M.; Tan, S.; Shogren-Knaak, M.A.; Bartholomew, B. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 2011, 39, 8378–8391. [Google Scholar] [CrossRef] [PubMed]
- Dechassa, M.L.; Zhang, B.; Horowitz-Scherer, R.; Persinger, J.; Woodcock, C.L.; Peterson, C.L.; Bartholomew, B. Architecture of the SWI/SNF-nucleosome complex. Mol. Cell. Biol. 2008, 28, 6010–6021. [Google Scholar] [CrossRef] [PubMed]
- Ayala, R.; Willhoft, O.; Aramayo, R.J.; Wilkinson, M.; McCormack, E.A.; Ocloo, L.; Wigley, D.B.; Zhang, X. Structure and regulation of the human INO80–nucleosome complex. Nature 2018, 556, 391–395. [Google Scholar] [CrossRef]
- Eustermann, S.; Schall, K.; Kostrewa, D.; Lakomek, K.; Strauss, M.; Moldt, M.; Hopfner, K.-P. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 2018, 556, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wu, H.; Chen, K.; Clapier, C.R.; Verma, N.; Zhang, W.; Deng, H.; Cairns, B.R.; Gao, N.; Chen, Z. Structure of the RSC complex bound to the nucleosome. Science 2019, 366, 838–843. [Google Scholar] [CrossRef]
- Wagner, F.R.; Dienemann, C.; Wang, H.; Stützer, A.; Tegunov, D.; Urlaub, H.; Cramer, P. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 2020, 579, 448–451. [Google Scholar] [CrossRef]
- Han, Y.; Reyes, A.A.; Malik, S.; He, Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 2020, 579, 452–455. [Google Scholar] [CrossRef]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef]
- Chin, J.W.; Cropp, T.A.; Anderson, J.C.; Mukherji, M.; Zhang, Z.; Schultz, P.G. An expanded eukaryotic genetic code. Science 2003, 301, 964–967. [Google Scholar] [CrossRef]
- Young, T.S.; Schultz, P.G. Beyond the canonical 20 amino acids: Expanding the genetic lexicon. J. Biol. Chem. 2010, 285, 11039–11044. [Google Scholar] [CrossRef]
- Neumann, H.; Hancock, S.M.; Buning, R.; Routh, A.; Chapman, L.; Somers, J.; Owen-Hughes, T.; Noort, J.; van Rhodes, D.; Chin, J.W. A Method for Genetically Installing Site-Specific Acetylation in Recombinant Histones Defines the Effects of H3 K56 Acetylation. Mol. Cell 2009, 36, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Neumann, H.; Peak-Chew, S.Y.; Chin, J.W. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008, 4, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, B.J.; Hahn, L.E.; Heitmüller, S.; Frauendorf, H.; Valerius, O.; Braus, G.H.; Neumann, H. Genetically Encoding Lysine Modifications on Histone H4. ACS Chem. Biol. 2015, 10, 939–944. [Google Scholar] [CrossRef]
- Dorman, G.; Prestwich, G.D. Benzophenone photophores in biochemistry. Biochemistry 1994, 33, 5661–5673. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Mimasu, S.; Sato, A.; Hino, N.; Sakamoto, K.; Umehara, T.; Yokoyama, S. Crystallographic Study of a Site-Specifically Cross-Linked Protein Complex with a Genetically Incorporated Photoreactive Amino Acid. Biochemistry 2011, 50, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, B.J.; Rall, N.A.; Ostwal, Y.; Kruitwagen, T.; Hiragami-Hamada, K.; Winkler, M.; Barral, Y.; Fischle, W.; Neumann, H. A cascade of histone modifications induces chromatin condensation in mitosis. Science 2014, 343, 77–80. [Google Scholar] [CrossRef]
- Schalch, T.; Duda, S.; Sargent, D.F.; Richmond, T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 2005, 436, 138–141. [Google Scholar] [CrossRef]
- Dorigo, B.; Schalch, T.; Kulangara, A.; Duda, S.; Schroeder, R.R.; Richmond, T.J. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 2004, 306, 1571–1573. [Google Scholar] [CrossRef]
- Jain, N.; Janning, P.; Neumann, H. 14-3-3 protein Bmh1 triggers short-range compaction of mitotic chromosomes by recruiting sirtuin deacetylase Hst2. J. Biol. Chem. 2020, 296, 100078. [Google Scholar] [CrossRef]
- Kruitwagen, T.; Denoth-Lippuner, A.; Wilkins, B.J.; Neumann, H.; Barral, Y. Axial contraction and short-range compaction of chromatin synergistically promote mitotic chromosome condensation. eLife 2015, 4, 332. [Google Scholar] [CrossRef]
- Hiragami-Hamada, K.; Soeroes, S.; Nikolov, M.; Wilkins, B.; Kreuz, S.; Chen, C.; Rosa-Velazquez, I.A.D.L.; Zenn, H.M.; Kost, N.; Pohl, W.; et al. Dynamic and flexible H3K9me3 bridging via HP1β dimerization establishes a plastic state of condensed chromatin. Nat. Commun. 2016, 7, 11310. [Google Scholar] [CrossRef]
- Belotserkovskaya, R.; Oh, S.; Bondarenko, V.A.; Orphanides, G.; Studitsky, V.M.; Reinberg, D. FACT Facilitates Transcription-Dependent Nucleosome Alteration. Science 2003, 301, 1090–1093. [Google Scholar] [CrossRef]
- Hoffmann, C.; Neumann, H. In Vivo Mapping of FACT-Histone Interactions Identifies a Role of Pob3 C-terminus in H2A-H2B Binding. ACS Chem. Biol. 2015, 10, 2753–2763. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Tamborrini, D.; Evans, B.; Chaudhry, S.; Wilkins, B.J.; Neumann, H. Interaction of RSC Chromatin Remodeling Complex with Nucleosomes Is Modulated by H3 K14 Acetylation and H2B SUMOylation In Vivo. iScience 2020, 23, 101292. [Google Scholar] [CrossRef]
- Neumann, H.; Wilkins, B.J. Spanning the gap: Unraveling RSC dynamics in vivo. Curr. Genet. 2021, 67, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Parnell, T.J.; Huff, J.T.; Cairns, B.R. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008, 27, 100–110. [Google Scholar] [CrossRef]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending Healthy Life Span—From Yeast to Humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
- Sigismondo, G.; Papageorgiou, D.N.; Krijgsveld, J. Cracking chromatin with proteomics: From chromatome to histone modifications. Proteomics 2022, 22, 2100206. [Google Scholar] [CrossRef]
- Zheng, H.; Lin, S.; Chen, P.R. Genetically encoded protein labeling and crosslinking in living Pseudomonas aeruginosa. Bioorg. Med. Chem. 2020, 28, 115545. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, Z.; Xu, H.; Li, L.; Chen, S.; Li, J.; Hao, Z.; Chen, P.R. Site-Specific Incorporation of Photo-Cross-Linker and Bioorthogonal Amino Acids into Enteric Bacterial Pathogens. J. Am. Chem. Soc. 2011, 133, 20581–20587. [Google Scholar] [CrossRef]
- Takahashi, H.; Dohmae, N.; Kim, K.S.; Shimuta, K.; Ohnishi, M.; Yokoyama, S.; Yanagisawa, T. Genetic incorporation of non-canonical amino acid photocrosslinkers in Neisseria meningitidis: New method provides insights into the physiological function of the function-unknown NMB1345 protein. PLoS ONE 2020, 15, e0237883. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.H.; Poshtiban, A.; Klippenstein, V.; Ghisi, V.; Plested, A.J.R. Gating modules of the AMPA receptor pore domain revealed by unnatural amino acid mutagenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 13358–13367. [Google Scholar] [CrossRef]
- Neumann, H.; Wang, K.; Davis, L.; Garcia-Alai, M.; Chin, J.W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 2010, 464, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Aydin, Y.; Coin, I. Genetically encoded crosslinkers to address protein–protein interactions. Protein Sci. 2023, 32, e4637. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, B.J.; Daggett, K.A.; Cropp, T.A. Peptide mass fingerprinting using isotopically encoded photo-crosslinking amino acids. Mol. BioSystems 2008, 4, 934–936. [Google Scholar] [CrossRef]
- Johnson, D.S.; Mortazavi, A.; Myers, R.M.; Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007, 316, 1497–1502. [Google Scholar] [CrossRef]
- Orlando, V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000, 25, 99–104. [Google Scholar] [CrossRef]
- Lee, J.B.; Keung, A.J. Chromatin Immunoprecipitation in Human and Yeast Cells. Methods Mol. Biol. 2018, 1767, 257–269. [Google Scholar]
- Hsieh, T.-H.S.; Weiner, A.; Lajoie, B.; Dekker, J.; Friedman, N.; Rando, O.J. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell 2015, 162, 108–119. [Google Scholar] [CrossRef]
- Ramachandran, S.; Zentner, G.E.; Henikoff, S. Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res. 2015, 25, 381–390. [Google Scholar] [CrossRef]
- Brahma, S.; Henikoff, S. RSC-Associated Subnucleosomes Define MNase- Sensitive Promoters in Yeast. Mol. Cell 2019, 73, 238–249.e3. [Google Scholar] [CrossRef]
- Donovan, D.A.; Crandall, J.G.; Banks, O.G.B.; Jensvold, Z.D.; Truong, V.; Dinwiddie, D.; McKnight, L.E.; McKnight, J.N. Engineered Chromatin Remodeling Proteins for Precise Nucleosome Positioning. Cell Rep. 2019, 29, 2520–2535.e4. [Google Scholar] [CrossRef] [PubMed]
- Kubik, S.; O’Duibhir, E.; Jonge, W.J.; de Mattarocci, S.; Albert, B.; Falcone, J.-L.; Bruzzone, M.J.; Holstege, F.C.P.; Shore, D. Sequence-Directed Action of RSC Remodeler and General Regulatory Factors Modulates +1 Nucleosome Position to Facilitate Transcription. Mol Cell 2018, 71, 89–102.e5. [Google Scholar] [CrossRef] [PubMed]
- Skene, P.J.; Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 2017, 6, e21856. [Google Scholar] [CrossRef] [PubMed]
- Menoni, H.; Shukla, M.S.; Gerson, V.; Dimitrov, S.; Angelov, D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic Acids Res. 2012, 40, 692–700. [Google Scholar] [CrossRef]
- Chai, B.; Huang, J.; Cairns, B.R.; Laurent, B.C. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005, 19, 1656–1661. [Google Scholar] [CrossRef]
- Shim, E.Y.; Hong, S.J.; Oum, J.-H.; Yanez, Y.; Zhang, Y.; Lee, S.E. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 2007, 27, 1602–1613. [Google Scholar] [CrossRef]
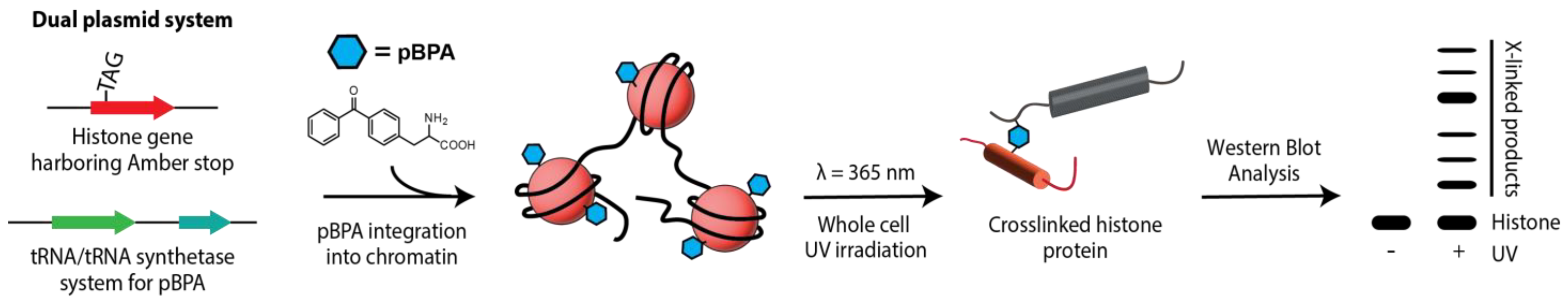
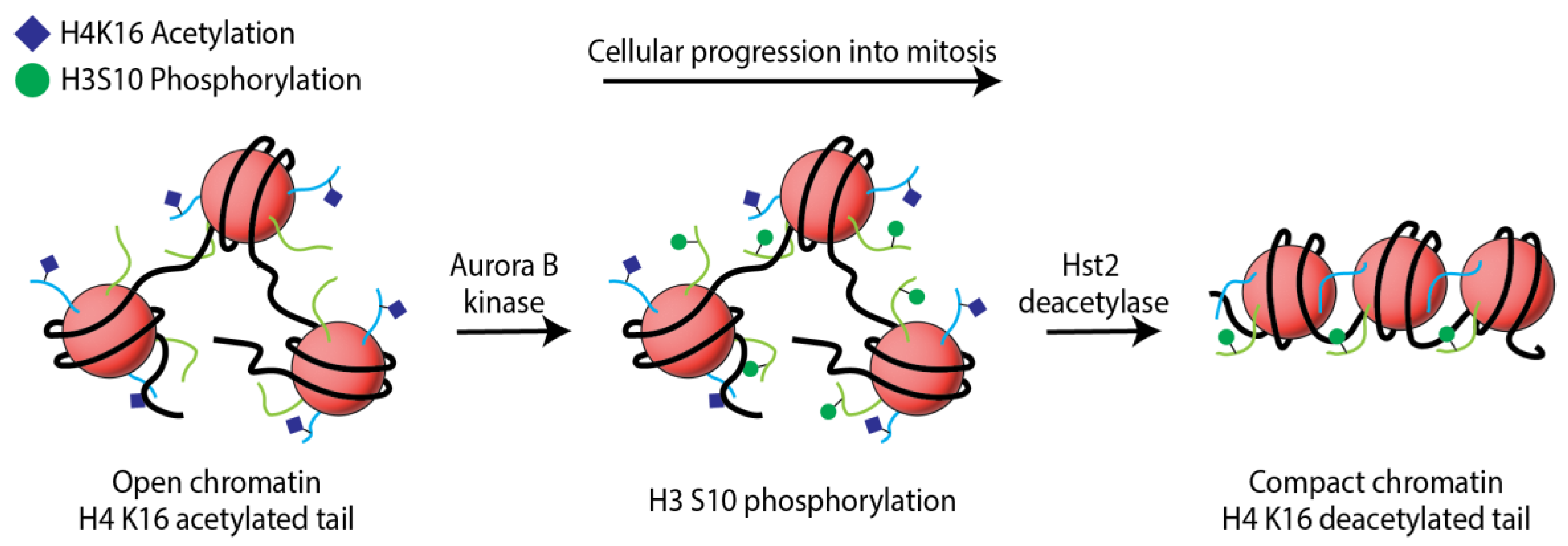

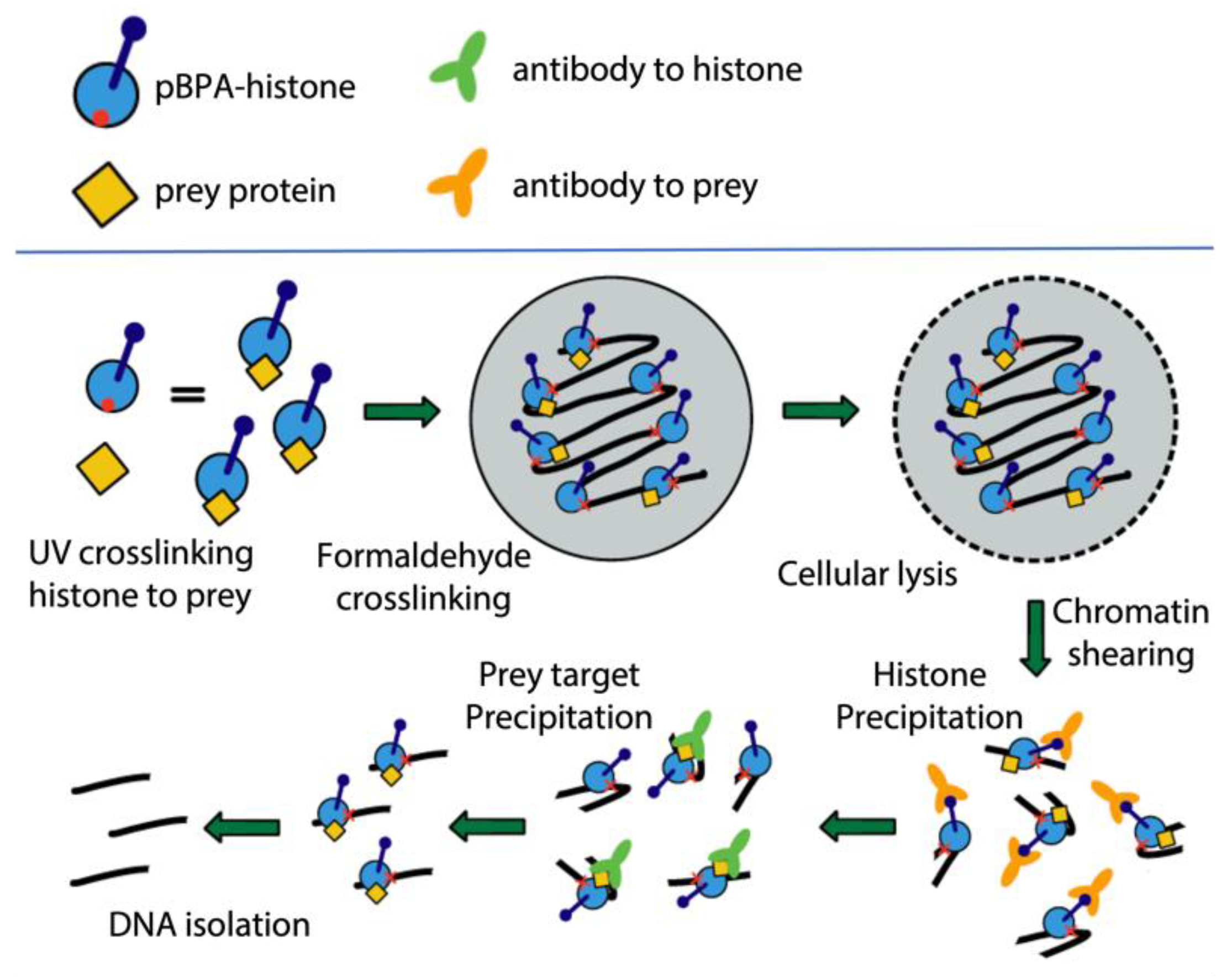
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moleri, P.; Wilkins, B.J. Unnatural Amino Acid Crosslinking for Increased Spatiotemporal Resolution of Chromatin Dynamics. Int. J. Mol. Sci. 2023, 24, 12879. https://doi.org/10.3390/ijms241612879
Moleri P, Wilkins BJ. Unnatural Amino Acid Crosslinking for Increased Spatiotemporal Resolution of Chromatin Dynamics. International Journal of Molecular Sciences. 2023; 24(16):12879. https://doi.org/10.3390/ijms241612879
Chicago/Turabian StyleMoleri, Pamela, and Bryan J. Wilkins. 2023. "Unnatural Amino Acid Crosslinking for Increased Spatiotemporal Resolution of Chromatin Dynamics" International Journal of Molecular Sciences 24, no. 16: 12879. https://doi.org/10.3390/ijms241612879
APA StyleMoleri, P., & Wilkins, B. J. (2023). Unnatural Amino Acid Crosslinking for Increased Spatiotemporal Resolution of Chromatin Dynamics. International Journal of Molecular Sciences, 24(16), 12879. https://doi.org/10.3390/ijms241612879



