Widely Targeted Metabolomic Profiling Combined with Transcriptome Analysis Provides New Insights into Lipid Biosynthesis in Seed Kernels of Pinus koraiensis
Abstract
:1. Introduction
2. Results
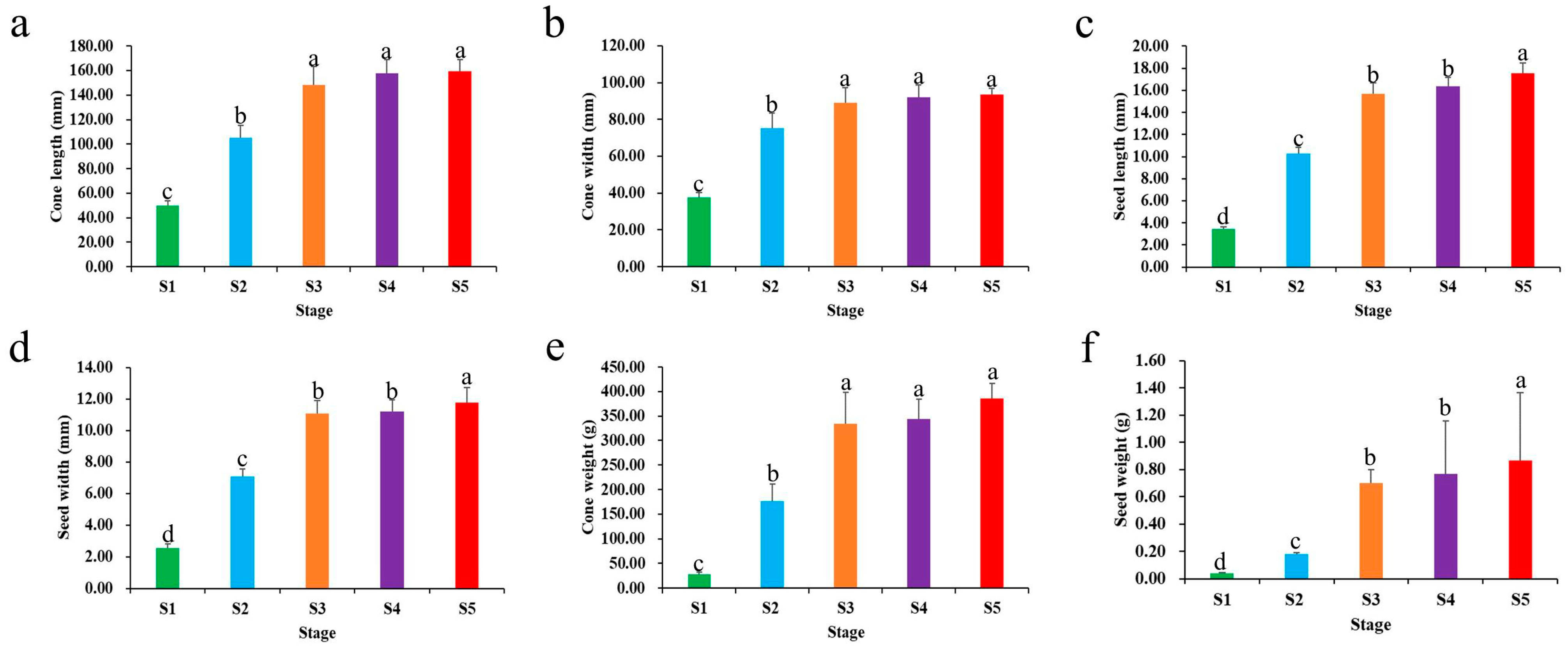
2.1. Observation and Characterization of Cone and Seed Development
2.2. Lipid Metabolite Profile for Seed Kernel Development
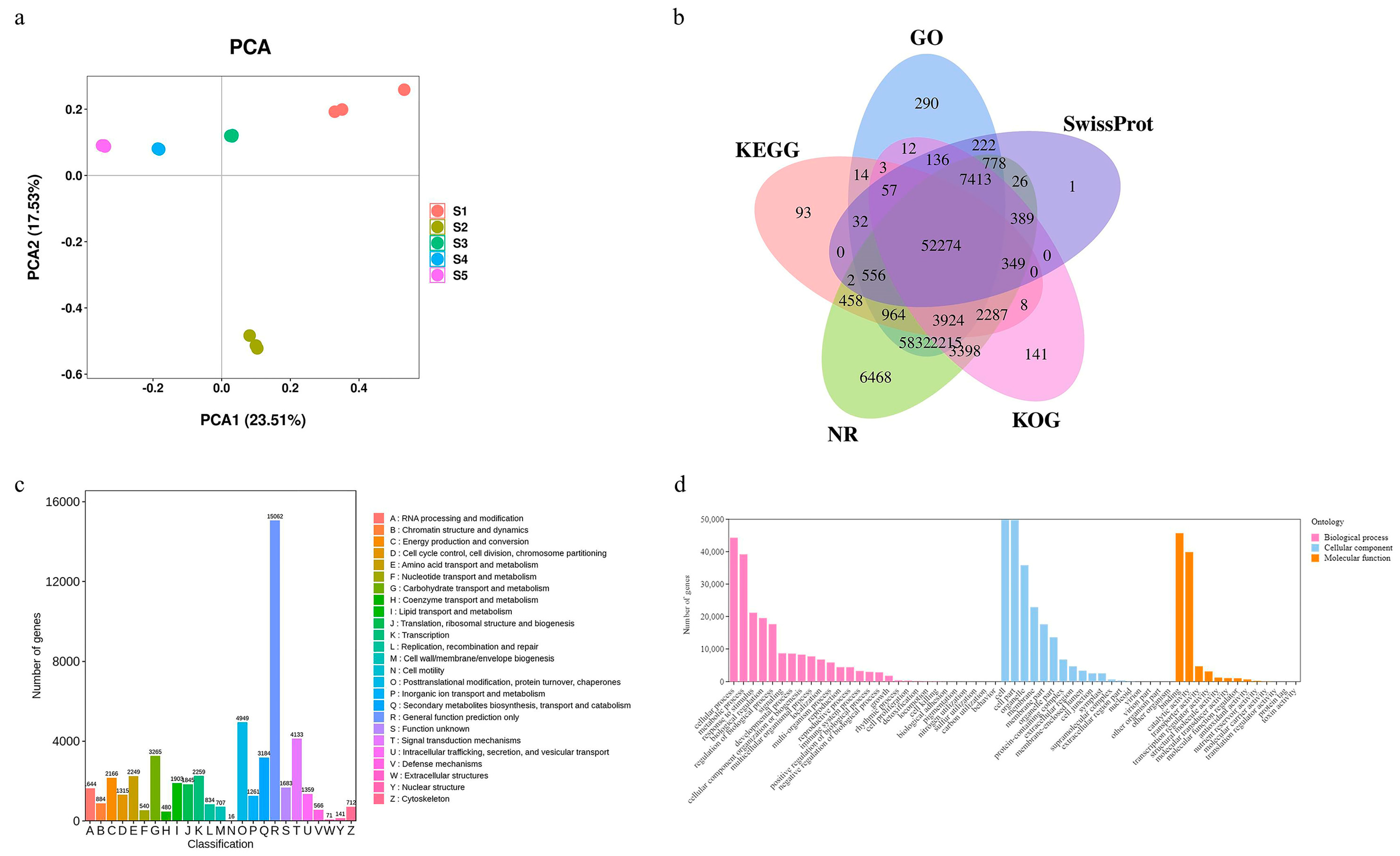
2.3. Statistical Analysis of Transcriptome Data and Gene Functional Annotation
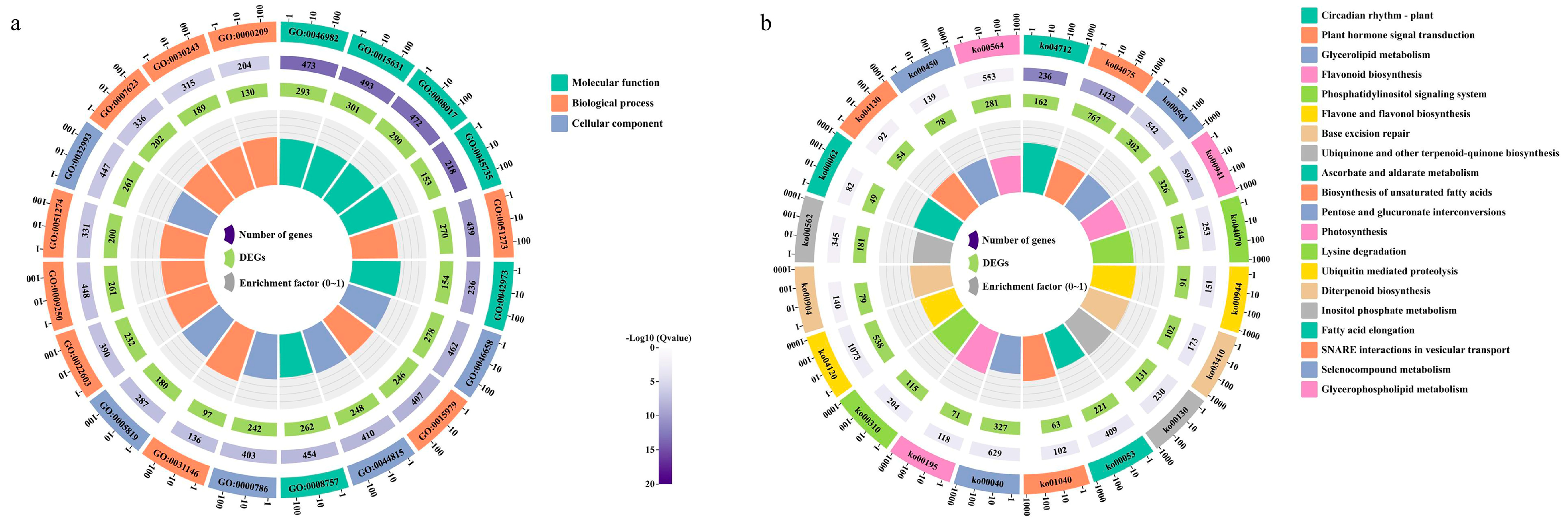
2.4. Differentially Expressed Gene Enrichment Analysis
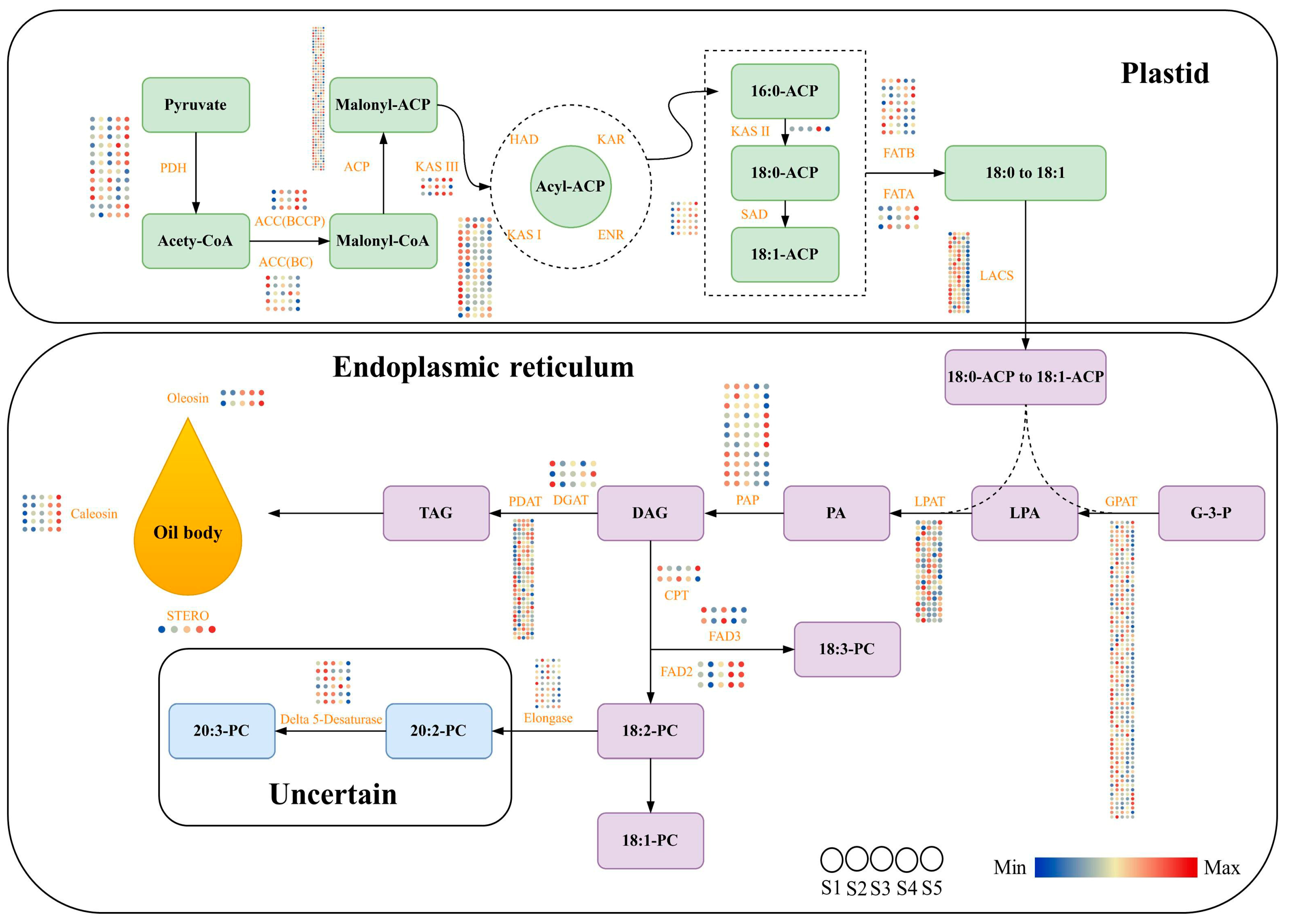
2.5. Identification and Expression Patterns of Potential Genes Related to Oil Body Biosynthesis in Kernels
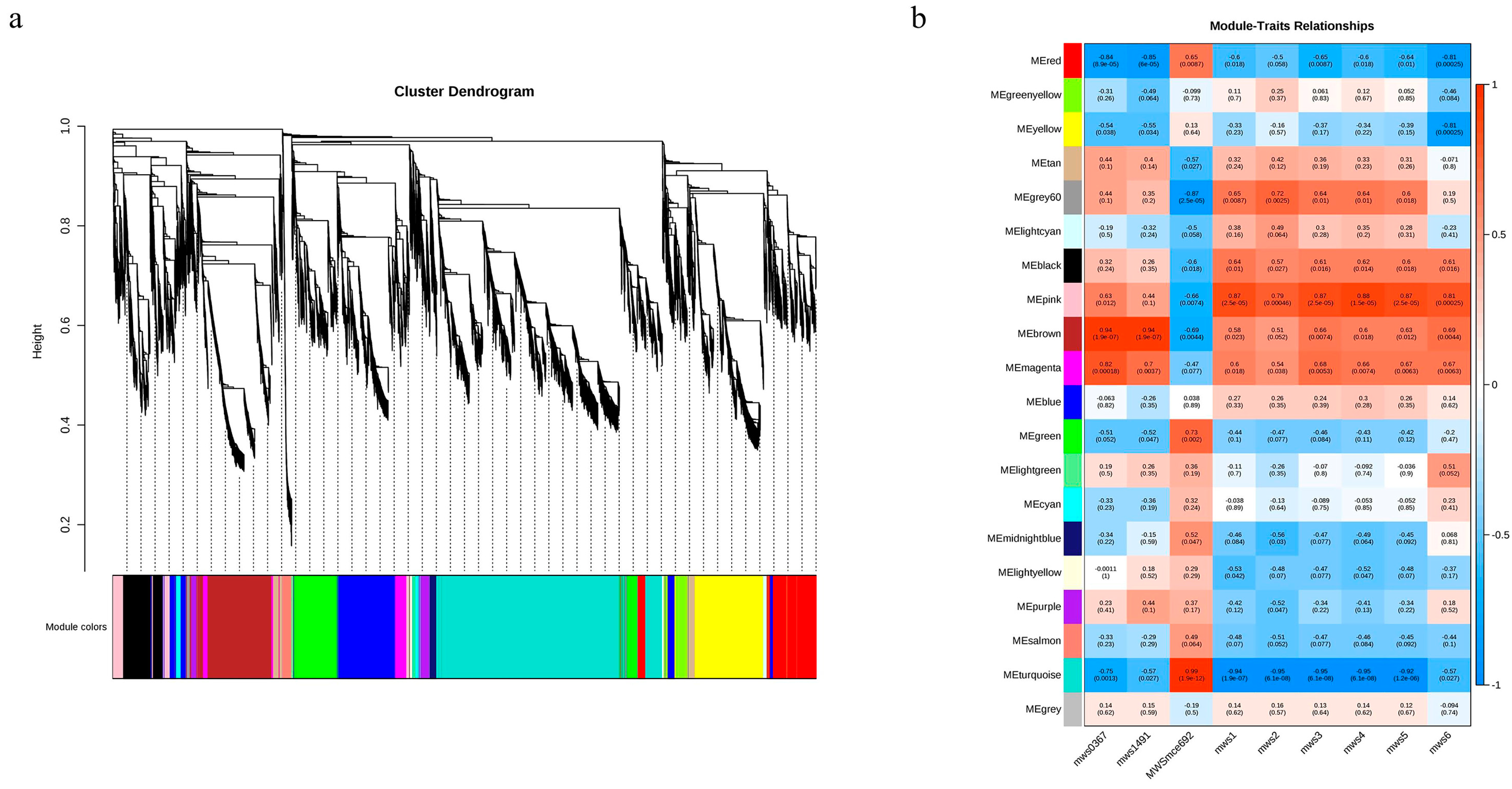
2.6. WGCNA
2.7. Gene Interaction Network Analysis
2.8. RT-qPCR Validation of Differentially Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Materials and Sampling
4.2. Metabolite Extraction and Analysis
4.3. RNA-Seq and Functional Annotation
4.4. Differentially Expressed Gene (DEG) and GO and KEGG Enrichment Analyses
4.5. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.6. Gene Interaction Network Analysis
4.7. Quantitative Real-Time PCR (qRT-PCR) Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, S.L.; Yan, S.Y.; Zhao, C.; Zhang, P.; Yang, L.; Wang, C.; Shen, H.L. Deep sequencing and analysis of transcriptomes of Pinus koraiensis Sieb. & Zucc. Forests 2020, 11, 350. [Google Scholar]
- Park, J.M.; Kwon, S.H.; Lee, H.J.; Na, S.J.; El-Kassaby, Y.A.; Kang, K.S. Integrating fecundity variation and genetic relatedness in estimating the gene diversity of seed crops: Pinus koraiensis seed orchard as an example. Can. J. For. Res. 2016, 47, 366–370. [Google Scholar] [CrossRef]
- Wang, X.J.; Liang, H.Y.; Guo, D.L.; Guo, L.L.; Duan, X.G.; Jia, Q.S.; Hou, X.G. Integrated analysis of transcriptomic and proteomic data from tree peony (P. ostii) seeds reveals key developmental stages and candidate genes related to oil biosynthesis and fatty acid metabolism. Hortic. Res. 2019, 6, 344–362. [Google Scholar] [CrossRef]
- Wei, J.T.; Li, X.; Xu, H.Z.; Wang, Y.L.; Zang, C.H.; Xu, J.W.; Pei, X.N.; Zhao, X.Y. Evaluation of the genetic diversity of Pinus koraiensis by EST-SSR and its management, utilization and protection. For. Ecol. Manag. 2021, 505, 119882. [Google Scholar] [CrossRef]
- Aizawa, M.; Kim, Z.S.; Yoshimaru, H. Phylogeography of the Korean pine (Pinus koraiensis) in northeast Asia: Inferences from organelle gene sequences. J. Plant Res. 2012, 125, 713–723. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, Q.H.; Tian, Y.G.; Yang, S.C.; Wang, H.W.; Wang, L.K.; Li, Y.L.; Zhang, P.; Zhao, X.Y. Comprehensive assessment of growth traits and wood properties in half-sib Pinus koraiensis families. Euphytica 2018, 214, 202. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.T.; Wei, J.T.; Li, Y.; Tigabu, M.; Zhao, X.Y. Genetic improvement of Pinus koraiensis in China: Current situation and future prospects. Forests 2020, 11, 148. [Google Scholar] [CrossRef]
- Tong, Y.W.; Lewis, B.J.; Zhou, W.M.; Mao, C.R.; Wang, Y.; Zhou, L.; Yu, D.P.; Dai, L.M.; Qi, L. Genetic diversity and population structure of natural Pinus koraiensis populations. Forests 2019, 11, 39. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Xin, C.; Qiu, J.Q.; Wang, Z.Y. Essential oil from Pinus Koraiensis pinecones inhibits gastric cancer cells via the HIPPO/YAP signaling pathway. Molecules 2019, 24, 3851. [Google Scholar] [CrossRef]
- Park, S.; Shin, S.; Lim, Y.; Shin, J.H.; Seong, J.K.; Han, S.N. Korean pine nut oil attenuated hepatic triacylglycerol accumulation in high-fat diet-induced obese mice. Nutrients 2015, 8, 59. [Google Scholar] [CrossRef]
- Lee, M.S.; Cho, S.M.; Lee, M.H.; Lee, E.O.; Kim, S.H.; Lee, H.J. Ethanol extract of Pinus koraiensis leaves containing lambertianic acid exerts antiobesity and hypolipidemic effects by activating adenosine monophosphateactivated protein kinase (AMPK). BMC Complement. Altern. Med. 2016, 16, 51. [Google Scholar]
- Kaviriri, D.K.; Liu, X.T.; Fan, Z.Y.; Wang, J.Y.; Wang, Q.; Wang, L.F.; Wang, L.X.; Khasa, D.; Zhao, X.Y. Genetic variation in growth and cone traits of Pinus koraiensis half-sib families in Northeast China. Phyton-Int. J. Exp. Bot. 2020, 89, 57–69. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.T.; Wei, J.T.; Li, Y.; Tigabu, M.; Zhao, X.Y. Development and transferability of EST-SSR markers for Pinus koraiensis from cold-Stressed transcriptome through illumine sequencing. Genes 2020, 11, 500. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, Y.C.; Zhang, H.; Dong, A.J.; Zhao, H.T.; Xu, D.C.; Ma, Y.; Wang, J. Diterpenoid acids from Pinus koraiensis. Chem. Nat. Compd. 2010, 46, 227–229. [Google Scholar] [CrossRef]
- Zhang, S.T.; Zhang, L.G.; Wang, L.; Zhao, Y.H. Total phenols, flavonoids, and procyanidins levels and total antioxidant activity of different Korean pine (Pinus koraiensis) varieties. J. For. Res. 2018, 30, 1743–1754. [Google Scholar] [CrossRef]
- Guo, Y. Extraction of Pine Nut Oil and Preparation of the Microcapsule. Master’s Thesis, Northeast Forestry University, Harbin, China, 2017. [Google Scholar]
- Shen, S.Y. Study on Isolation, Purification, Sturvture and Antioxidant Activity of Polysacchairides from Different Parts of Pinus koraiensis. Master’s Thesis, Harbin Institute of Technology, Harbin, China, 2014. [Google Scholar]
- Huang, Y.Y. The Separation, Isolation and Identification of Polyphenol Structures in Korean Pine (Pinus koraiensis) Bark and Evaluation of Its Antioxidant and Anticancer Activity. Ph.D. Thesis, Northeast Forestry University, Harbin, China, 2014. [Google Scholar]
- Wang, W.W. Study on the Extraction, Purification and Anti-Oxidation Activities of Polysaccharide from Pine Cone of Pinus koraiensis. Master’s Thesis, Qingdao University of Science & Technology, Qingdao, China, 2013. [Google Scholar]
- Miller, C.; Wells, R.; McKenzie, N.; Trick, M.; Ball, J.; Fatihi, A.; Dubreucq, B.; Chardot, T.; Lepiniec, L.; Bevan, M.W. Variation in expression of the HECT E3 ligase UPL3 modulates LEC2 levels, seed size, and crop yields in Brassica napus. Plant Cell 2019, 31, 2370–2385. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.Y.; Wallis, J.G.; Denolf, P.; Engelen, S.; Bengtsson, J.D.; Van, T.M.; Doerivkx, J.; Haesendonckx, B.; Browse, J. The biochemistry of headgroup exchange during triacylglycerol synthesis in canola. Plant J. 2020, 103, 83–94. [Google Scholar] [CrossRef]
- Shahid, M.; Cai, G.; Zu, F.; Zhao, Q.; Qasim, M.U.; Hong, Y.; Fan, C.; Zhou, Y. Comparative transcriptome analysis of developing seeds and silique wall reveals dynamic transcription networks for effective oil production in Brassica napus L. Int. J. Mol. Sci. 2019, 20, 1982. [Google Scholar] [CrossRef]
- Liao, P.; Woodfield, H.K.; Harwood, J.L.; Chye, M.L.; Scofield, S. Comparative transcriptomics analysis of Brassica napus L. during seed maturation reveals dynamic changes in gene expression between embryos and seed coats and distinct expression profiles of Acyl-CoA-binding proteins for lipid accumulation. Plant Cell Physiol. 2019, 61, 221. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Wang, Y.M.; Huang, Y.; Cui, Y.P.; Hua, J.P. Gene network of oil accumulation reveals expression profiles in developing embryos and fatty acid composition in Upland cotton. J. Plant Physiol. 2018, 288, 101–112. [Google Scholar] [CrossRef]
- Zhou, L.H.; Yan, T.; Chen, X.; Li, Z.L.; Wu, D.Z.; Hua, S.J.; Jiang, L.X. Effect of high night temperature on storage lipids and transcriptome changes in developing seeds of oilseed rape. J. Exp. Bot. 2017, 69, 1721–1733. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Kilaru, A.; Cao, X.; Durrett, T.P.; Fan, J.L.; Jensen, J.K.; Thrower, N.A.; Pauly, M.; Wilkerson, C.; Ohlrogge, J.B. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011, 68, 1014–1027. [Google Scholar] [CrossRef]
- Yeilaghi, H.; Arzani, A.; Shaderian, M.; Fotovat, R.; Feizi, M.; Pourdad, S.S. Effect of salinity on seed oil content and fatty acid composition of safflower (Carthamus tinctorius L.) genotypes. Food Chem. 2012, 130, 618–625. [Google Scholar] [CrossRef]
- Tan, H.L.; Yang, X.H.; Zhang, F.X.; Zheng, X.; Qu, C.M.; Mu, J.Y.; Fu, F.Y.; Li, J.A.; Guan, R.Z.; Zhang, H.S.; et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2015, 156, 1577–1588. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef]
- Andre, C.; Froehlich, J.E.; Moll, M.R.; Benning, C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2006–2022. [Google Scholar] [CrossRef] [PubMed]
- Stevanato, N.; Silva, C. Radish seed oil: Ultrasound-assisted extraction using ethanol as solvent and assessment of its potential for ester production. Ind. Crops Prod. 2019, 132, 283–291. [Google Scholar] [CrossRef]
- Ghasemlou, M.; Daver, F.; Ivanova, E.P.; Adhikari, B. Polyurethanes from seed oil-based polyols: A review of synthesis, mechanical and thermal properties. Ind. Crops Prod. 2019, 142, 111841. [Google Scholar] [CrossRef]
- Hua, W.; Li, R.J.; Zhan, G.M.; Liu, J.; Li, J.; Wang, X.F.; Liu, G.H.; Wang, H.Z. Maternal control of seed oil content in Brassica napus: The role of silique wall photosynthesis. Plant J. 2012, 69, 432–444. [Google Scholar] [CrossRef]
- Liu, N.A.; Liu, J.; Fan, S.H.; Liu, H.F.; Zhou, X.R.; Hua, W.; Zheng, M. An integrated omics analysis reveals the gene expression profiles of maize, castor bean, and rapeseed for seed oil biosynthesis. BMC Plant Biol. 2022, 22, 153. [Google Scholar] [CrossRef]
- Wan, H.F.; Cui, Y.X.; Ding, Y.J.; Mei, J.Q.; Dong, H.L.; Zhang, W.X.; Wu, S.Q.; Liang, Y.; Zhang, C.Y.; Li, J.N.; et al. Time-series analyses of transcriptomes and proteomes reveal molecular networks underlying oil accumulation in Canola. Front. Plant Sci. 2016, 7, 2007. [Google Scholar] [CrossRef]
- Deng, W.; Yan, F.; Zhang, X.L.; Tang, Y.W.; Yuan, Y.J. Transcriptional profiling of canola developing embryo and identification of the important roles of BnDof5.6 in embryo development and fatty acids synthesis. Plant Cell Physiol. 2015, 56, 1624–1640. [Google Scholar] [CrossRef]
- Chen, H.; Wang, F.W.; Dong, Y.Y.; Wang, N.; Sun, Y.P.; Li, X.Y.; Liu, L.; Fan, X.D.; Yin, H.L.; Jing, Y.Y.; et al. Sequence mining and transcript profiling to explore differentially expressed genes associated with lipid biosynthesis during soybean seed development. BMC Plant Biol. 2012, 12, 122. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.M.; Wang, Y.; Zhang, X.G.; Li, H.M.; Lu, X.; Zhang, J.S.; Zhang, W.K.; Chen, S.Y. De novo assembly of the peanut (Arachis hypogaea L.) seed transcriptome revealed candidate unigenes for oil accumulation pathways. PLoS ONE 2013, 8, e73767. [Google Scholar] [CrossRef]
- Dussert, S.; Guerin, C.; Andersson, M.; Joet, T.; Tranbarger, T.J.; Pizot, M.; Sarah, G.; Omore, A.; Durand-GAsselin, T.; Morcillo, F. Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol. 2013, 162, 1337–1358. [Google Scholar] [CrossRef]
- Wang, L.H.; Yu, S.; Tong, C.B.; Zhao, Y.Z.; Liu, Y.; Song, C.; Zhang, Y.X.; Zhang, X.D.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.B.; Yin, D.S.; Rodriguez-Calcerrada, J.; Zhang, J.Y.; Gil, L.; Zhang, P.; Shen, H.L. Cone-bearing effects on photosynthetic traits do not change with needle age in Pinus koraiensis trees. New For. 2022, 53, 607–626. [Google Scholar] [CrossRef]
- Kwon, S.H.; Kim, Y.G.; Kang, H.I.; Shim, D.; Kang, K.S. Morphology of strobili at different development positions and cone and seed characteristics of Pinus densiflora f. multicaulis. Dendrology 2021, 85, 51–59. [Google Scholar] [CrossRef]
- Niu, S.H.; Li, J.; Bo, W.H.; Yang, W.F.; Zuccolo, A.; Giacomello, S.; Chen, X.; Han, F.X.; Yang, J.H.; Song, Y.T.; et al. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell 2021, 185, 204. [Google Scholar] [CrossRef]
- Du, L.; Li, N.; Chen, L.L.; Xu, Y.X.; Li, Y.; Zhang, Y.Y.; Li, C.Y.; Li, Y.H. The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 2014, 26, 665–677. [Google Scholar] [CrossRef]
- Lau, S.; Jurgens, G.; Ge, S.I. The evolving complexity of the auxin pathway. Plant Cell 2018, 20, 1738–1746. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, X.J.; Kang, X.J.; Zhao, X.Y.; Zhang, X.S.; Ni, M. SHORT HYPOCOTYL UNDERBLUE1 associates with MINISEED3 and HAIKU2 promoters in vivoto regulate Arabidopsis seed development. Plant Cell 2009, 21, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.P.; Liang, J.A.; Wang, M.W.; Li, D.X.; Liu, Y.; Chen, T.H.H.; Yang, X.H. Genetic engineering of the biosynthesis of glycinebetaine enhances the fruit development and size of tomato. Plant Sci. 2019, 80, 355–366. [Google Scholar] [CrossRef]
- Guo, J.X.; Wang, S.F.; Yu, X.Y.; Dong, R.; Li, Y.Z.; Mei, X.R.; Shen, Y.Y. Polyamines regulate strawberry fruit ripening by abscisic 15 acid, auxin, and ethylene. Plant Physiol. 2018, 177, 339–351. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644-U130. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.L.; Ohlrogge, J.B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.T.; Jiang, C.; Ye, T.T.; Tan, D.X.; Reiter, R.J.; Zhang, H.; Liu, R.; Chan, Z.L. Comparative physiological, metabolomic, and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass [Cynodon dactylon (L). Pers.] by exogenous melatonin. J. Exp. Bot. 2015, 66, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.P.; Wu, J.N.; Xiong, H.X.; Zeb, A.; Yang, T.Z.; Su, X.M.; Su, L.J.; Liu, W.T. Impact of polystyrene nanoplastics (PSNPs) on seed germination and seedling growth of wheat (Triticum aestivum L.). J. Hazard. Mat. 2020, 385, 121620. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, B.J.W.; He, H.Z.; Hanson, J.; Willems, L.A.J.; Jamar, D.C.L.; Cueff, G.; Rajjou, L.; Hilhorst, H.W.M.; Bentsink, L. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 2016, 85, 451–465. [Google Scholar] [CrossRef]
- Roessner, U.; Luedemann, A.; Brust, D.; Fiehn, O.; Linke, T.; Willmitzer, L.; Fernie, A. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 2001, 13, 11–29. [Google Scholar] [CrossRef]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.C.; León, P.; Zhou, L.; Sheen, J. Hexokinase as a sugar sensor in higher plants. Plant Cell 1997, 9, 5–19. [Google Scholar]
- Lalonde, S.; Boles, E.; Hellmann, H.; Barker, L.; Patrick, J.W.; Frommer, W.B.; Ward, J.M. The dual function of sugar carriers: Transport and sugar sensing. Plant Cell 1999, 11, 707–726. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cai, K.W.; Zhang, Q.H.; Pei, X.N.; Chen, S.; Jiang, L.P.; Han, Z.M.; Zhao, M.H.; Li, Y.; Zhang, X.X.; et al. The Manchurian Walnut genome: Insights into Juglone and lipid biosynthesis. Gigascience 2021, 11, giac057. [Google Scholar] [CrossRef]
- Wang, F.; Liang, D.Y.; Pei, X.N.; Zhang, Q.H.; Zhang, P.; Zhang, J.Q.; Lu, Z.M.; Yang, Y.C.; Liu, G.F.; Zhao, X.Y. Study on the physiological indices of Pinus sibirica and Pinus koraiensis seedlings under cold stress. J. For. Res. 2018, 30, 1255–1265. [Google Scholar] [CrossRef]
- Gu, K.Y.; Yi, C.X.; Tian, D.S.; Sangha, J.S.; Hong, Y.; Yin, Z.C. Expression of fatty acid and lipid biosynthetic genes in developing endosperm of Jatropha curcas. Biotechnol. Biofuels 2012, 5, 47. [Google Scholar] [CrossRef]
- Ding, J.; Ruan, C.J.; Guan, Y.; Krishna, P. Identification of microRNAs involved in lipid biosynthesis and seed size in developing sea buckthorn seeds using high-throughput sequencing. Sci. Rep. 2018, 8, 4022. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.M.; Zhou, Y.; Zhang, J.P.; Ji, F.Y.; Jin, F.; Fan, W.; Pei, D. Transcriptome analysis of Walnut (Juglans regia L.) embryos reveals key developmental stages and genes involved in lipid biosynthesis and polyunsaturated fatty acid metabolism. J. Agric. Food Chem. 2021, 69, 377–396. [Google Scholar] [CrossRef]
- Liu, W.S.; Zhang, B.; Jiang, R.R. Improving acetyl-CoA biosynthesis in Saccharomyces cerevisiae via the overexpression of pantothenate kinase and PDH bypass. Biotechnol. Biofuels 2017, 10, 41. [Google Scholar] [CrossRef]
- Huang, R.M.; Huang, Y.J.; Sun, Z.C.; Huang, J.Q.; Wang, Z.J. Transcriptome analysis of genes involved in lipid biosynthesis in the developing embryo of pecan (Carya illinoinensis). J. Agric. Food Chem. 2017, 65, 4223–4236. [Google Scholar] [CrossRef]
- Beaudoin, F.; Lacey, D.J.; Napier, J.A. The biogenesis of the plant seed oil body: Oleosin protein is synthesised by ER-bound ribosomes. Plant Physiol. Biochem. 1999, 37, 481–490. [Google Scholar] [CrossRef]
- Frandsen, G.I.; Mundy, J.; Tzen, J.T.C. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plantarum 2001, 112, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Tai, S.S.K.; Chen, M.C.M.; Peng, C.C.; Tzen, J.T.C. Gene family of Oleosin isoforms and their structural stabilization in sesame seed oil bodies. Biosci. Biotech. Biochem. 2002, 66, 2146–2153. [Google Scholar] [CrossRef]
- Jiang, P.L.; Wang, C.S.; Hsu, C.M.; Jauh, G.Y.; Tzen, J.T.C. Stable oil bodies sheltered by a unique Oleosin in Lily pollen. Plant Cell Physiol. 2007, 48, 812–821. [Google Scholar] [CrossRef]
- Naested, H.; Frandsen, G.I.; Jauh, G.Y.; Hernandez-Pinzon, I.; Nielsen, H.B.; Murphy, D.J.; Rogers, J.C.; Mundy, D.J. Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol. Biol. 2000, 44, 463–476. [Google Scholar] [CrossRef]
- Lin, L.J.; Tai, S.S.; Peng, C.C.; Tzen, J.T. Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol. 2002, 128, 1200–1211. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Zhao, M.H.; Hu, Y.B.; Meng, F.J.; Song, X.S.; Togabu, M.; Chiang, V.L.; Sederoff, R.; Ma, W.J.; et al. Molecular and metabolic insights into anthocyanin biosynthesis for leaf color change in Chokecherry (Padus virginiana). Int. J. Mol. Sci. 2021, 22, 10697. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, X.X.; Li, Y.X.; Yan, H.L.; Cai, K.W.; Li, H.X.; Wu, Z.W.; Wu, J.G.; Yang, X.D.; Jiang, H.C.; Wang, Q.C.; et al. Integrated metabolomic and transcriptomic analyses reveal different metabolite biosynthesis profiles of Juglans mandshurica in shade. Front. Plant Sci. 2022, 13, 991874. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Q.; Zhang, C.H.; Huang, Z.J.; Lyu, L.F.; Li, W.L.; Wu, W.L. Integrative analysis of the metabolome and transcriptome provides insights into the mechanisms of flavonoid biosynthesis in blackberry. Food Res. Int. 2022, 153, 110948. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.T.; Hu, H.C.; Luo, Y.L.; Shi, Q.Q.; Wei, A.Z. Widely targeted metabolomic profiling combined with transcriptome analysis provides new insights into amino acid biosynthesis in green and red pepper fruits. Food Res. Int. 2022, 160, 111718. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cai, K.W.; Pei, X.N.; Li, Y.; Hu, Y.B.; Meng, F.J.; Song, X.S.; Tigabu, M.; Ding, C.J.; Zhao, X.Y. Genome-wide identification of NAC transcription factor family in Juglans mandshurica and their expression analysis during the fruit development and ripening. Int. J. Mol. Sci. 2021, 22, 12414. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative Toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, L.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Xu, Y.; Han, R.; Liu, L.; Pei, X.; Zhao, X. Widely Targeted Metabolomic Profiling Combined with Transcriptome Analysis Provides New Insights into Lipid Biosynthesis in Seed Kernels of Pinus koraiensis. Int. J. Mol. Sci. 2023, 24, 12887. https://doi.org/10.3390/ijms241612887
Li Y, Xu Y, Han R, Liu L, Pei X, Zhao X. Widely Targeted Metabolomic Profiling Combined with Transcriptome Analysis Provides New Insights into Lipid Biosynthesis in Seed Kernels of Pinus koraiensis. International Journal of Molecular Sciences. 2023; 24(16):12887. https://doi.org/10.3390/ijms241612887
Chicago/Turabian StyleLi, Yan, Yujin Xu, Rui Han, Lin Liu, Xiaona Pei, and Xiyang Zhao. 2023. "Widely Targeted Metabolomic Profiling Combined with Transcriptome Analysis Provides New Insights into Lipid Biosynthesis in Seed Kernels of Pinus koraiensis" International Journal of Molecular Sciences 24, no. 16: 12887. https://doi.org/10.3390/ijms241612887




