The Light- and Jasmonic Acid-Induced AaMYB108-like Positive Regulates the Initiation of Glandular Secretory Trichome in Artemisia annua L.
Abstract
:1. Introduction
2. Results
2.1. Identification of the AaMYB108-like Gene from A. annua
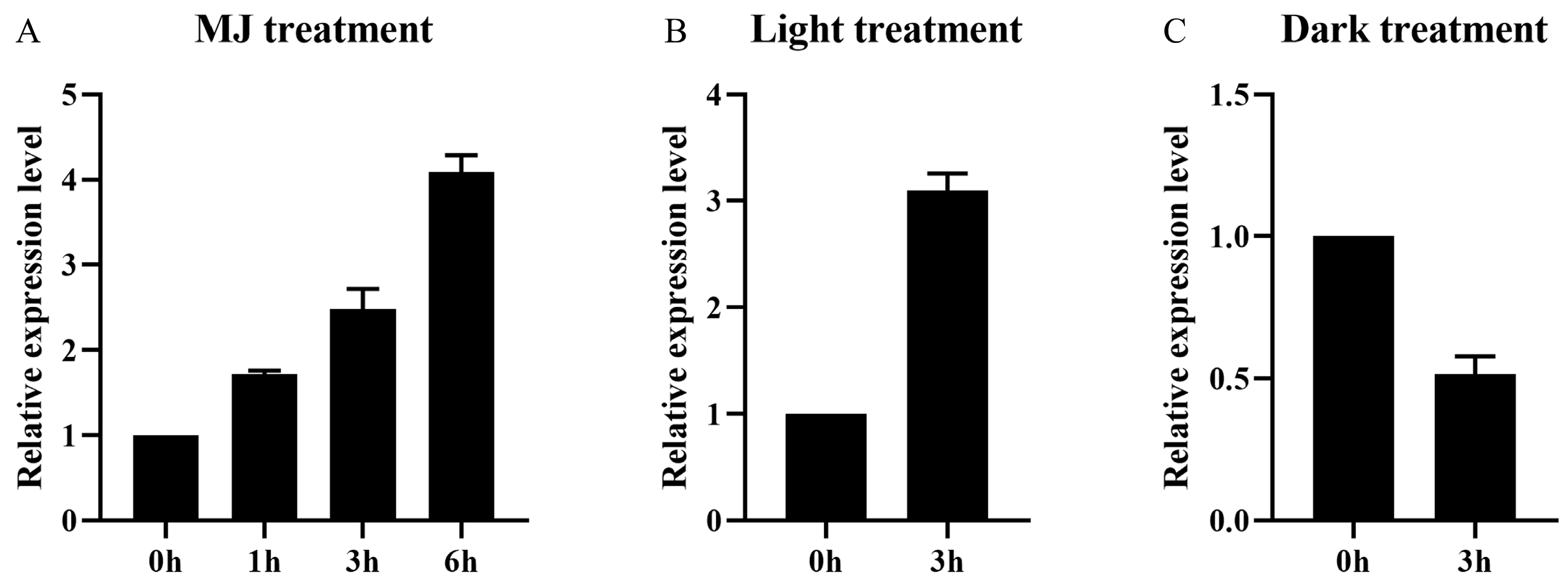
2.2. Co-Induction of AaMYB108-like Gene Expression by Light and Jasmonic Acid
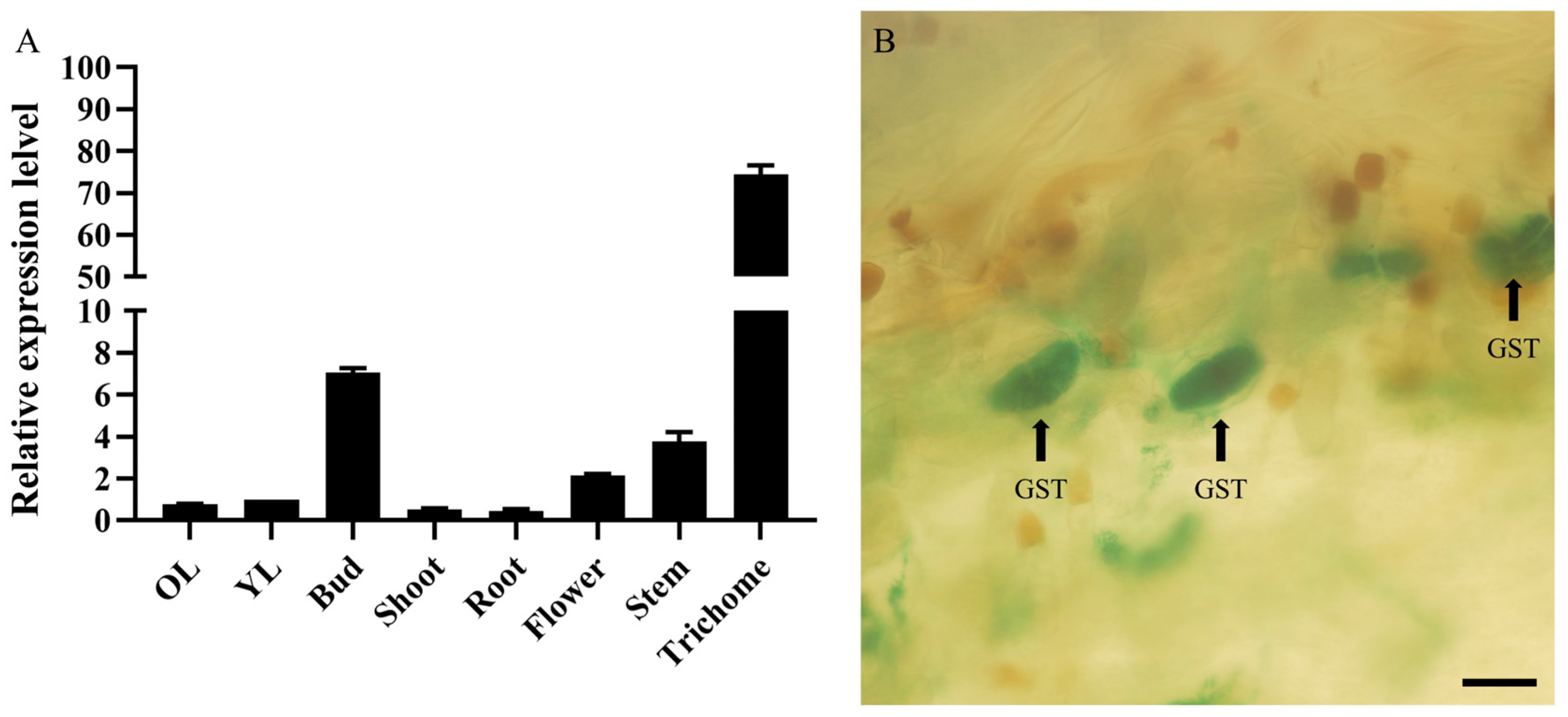
2.3. Expression Pattern of the AaMYB108-like Gene
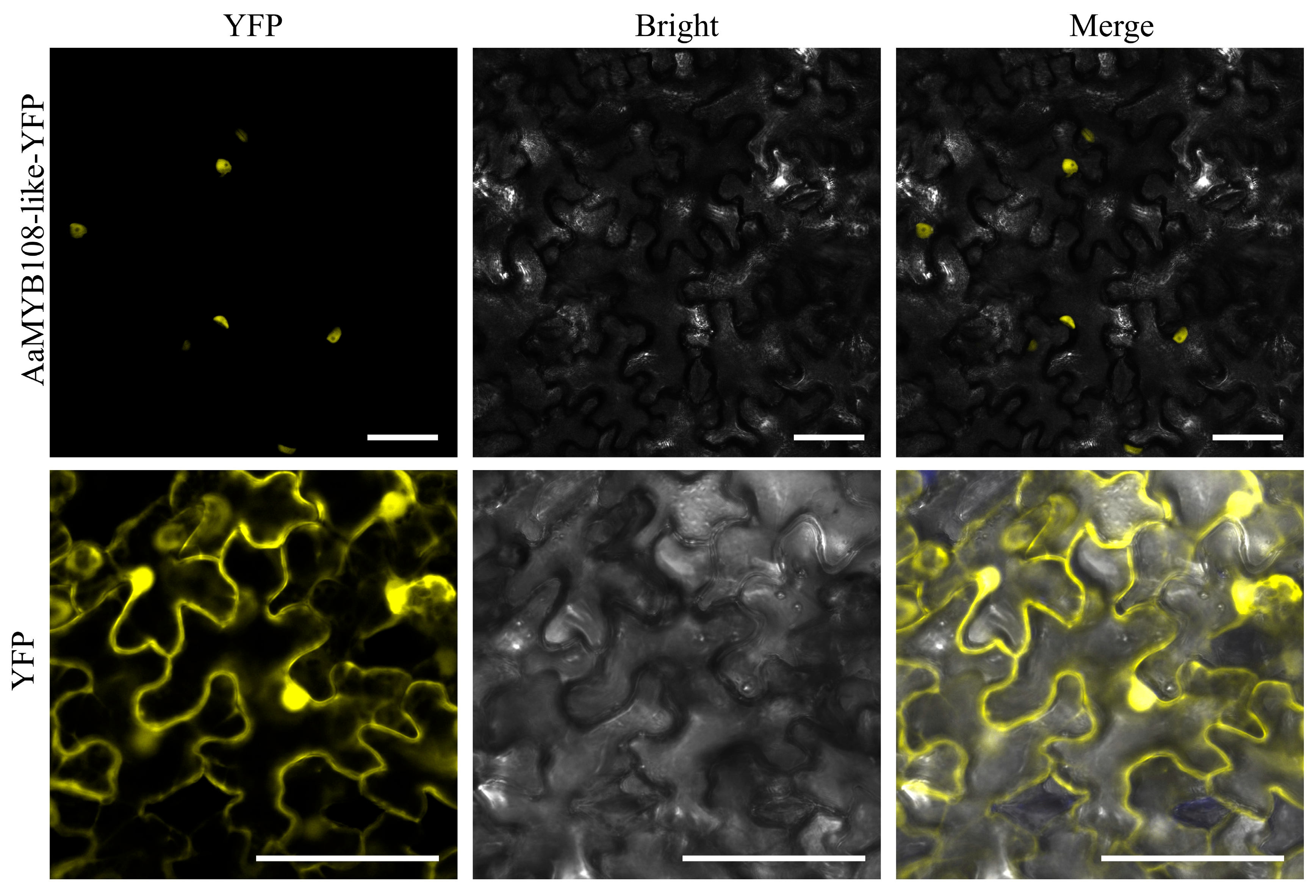
2.4. Subcellular Localization of the AaMYB108-like Gene
2.5. Overexpressing AaMYB108-like Gene Increased GST Density in A. annua
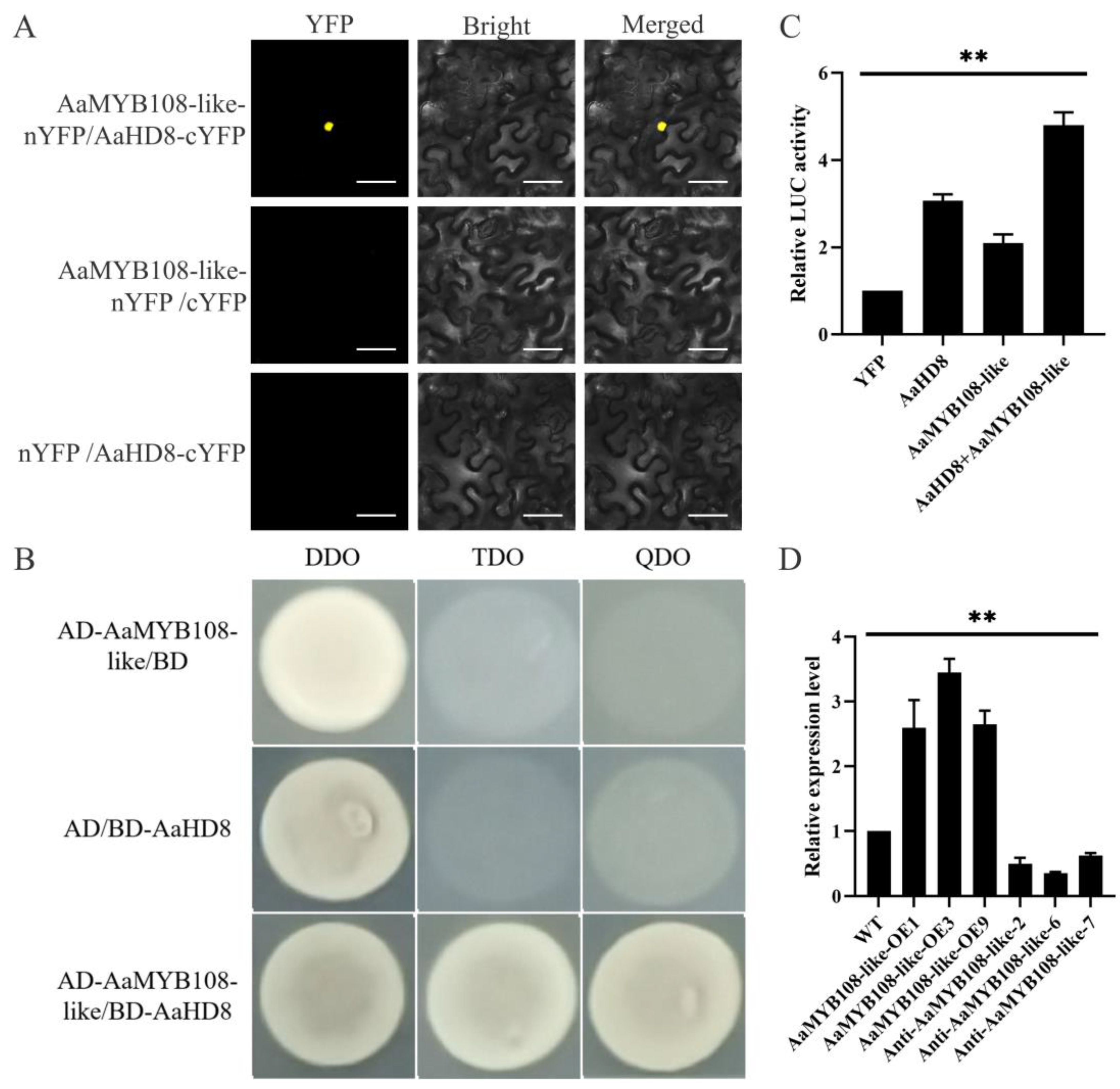
2.6. The AaMYB108-like Gene Interacts with AaHD8 to Promote AaHD1 Expression
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Treatment of Light, Dark and MJ
4.3. Isolation of Glandular Trichomes
4.4. RNA Extraction and RT-qPCR
4.5. Subcellular Localization
4.6. Transformation of Artemisia annua
4.7. GUS Expression in 1391Z-proMYB108-like-GUS Transgenic A. annua Plants
4.8. Artemisinin Content Measurement
4.9. Bimolecular Fluorescence Complementation Assays
4.10. Yeast Two-Hybrid (Y2H) Assays
4.11. Dual-LUC (Dual-Luciferase Reporter) Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garcia, L.S. Malaria. Clin. Lab. Med. 2010, 30, 93–129. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Zhang, Z.; Liao, F.; Jiang, T.; Tu, Y. The Birth of Artemisinin. Pharmacol. Ther. 2020, 216, 107658. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Fu, X.; Shao, J.; Tang, Y.; Yu, M.; Li, L.; Huang, L.; Tang, K. Transcriptional Regulatory Network of High-Value Active Ingredients in Medicinal Plants. Trends Plant Sci. 2023, 28, 429–446. [Google Scholar] [CrossRef]
- Qin, W.; Xie, L.; Li, Y.; Liu, H.; Fu, X.; Chen, T.; Hassani, D.; Li, L.; Sun, X.; Tang, K. An R2R3-MYB Transcription Factor Positively Regulates the Glandular Secretory Trichome Initiation in Artemisia annua L. Front. Plant Sci. 2021, 12, 657156. [Google Scholar] [CrossRef] [PubMed]
- Hassani, D.; Taheri, A.; Fu, X.; Qin, W.; Hang, L.; Ma, Y.; Tang, K. Elevation of Artemisinin Content by Co-Transformation of Artemisinin Biosynthetic Pathway Genes and Trichome-Specific Transcription Factors in Artemisia annua. Front. Plant Sci. 2023, 14, 1118082. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Yan, T.; Fu, X.; Tang, K. Transcriptional Regulation of Artemisinin Biosynthesis in Artemisia annua L. Sci. Bull. 2016, 61, 18–25. [Google Scholar] [CrossRef]
- Yu, Z.-X.; Li, J.-X.; Yang, C.-Q.; Hu, W.-L.; Wang, L.-J.; Chen, X.-Y. The Jasmonate-Responsive AP2/ERF Transcription Factors AaERF1 and AaERF2 Positively Regulate Artemisinin Biosynthesis in Artemisia annua L. Mol. Plant 2012, 5, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, L.; Zhang, F.; Jiang, W.; Shen, Q.; Zhang, L.; Lv, Z.; Wang, G.; Tang, K. AaORA, a Trichome-Specific AP2/ERF Transcription Factor of Artemisia annua, Is a Positive Regulator in the Artemisinin Biosynthetic Pathway and in Disease Resistance to Botrytis Cinerea. New Phytol. 2013, 198, 1191–1202. [Google Scholar] [CrossRef]
- Tan, H.; Xiao, L.; Gao, S.; Li, Q.; Chen, J.; Xiao, Y.; Ji, Q.; Chen, R.; Chen, W.; Zhang, L. TRICHOME AND ARTEMISININ REGULATOR 1 Is Required for Trichome Development and Artemisinin Biosynthesis in Artemisia annua. Mol. Plant 2015, 8, 1396–1411. [Google Scholar] [CrossRef]
- Zhang, F.; Fu, X.; Lv, Z.; Lu, X.; Shen, Q.; Zhang, L.; Zhu, M.; Wang, G.; Sun, X.; Liao, Z.; et al. A Basic Leucine Zipper Transcription Factor, AabZIP1, Connects Abscisic Acid Signaling with Artemisinin Biosynthesis in Artemisia annua. Mol. Plant 2015, 8, 163–175. [Google Scholar] [CrossRef]
- Chen, M.; Yan, T.; Shen, Q.; Lu, X.; Pan, Q.; Huang, Y.; Tang, Y.; Fu, X.; Liu, M.; Jiang, W.; et al. GLANDULAR TRICHOME-SPECIFIC WRKY 1 Promotes Artemisinin Biosynthesis in Artemisia annua. New Phytol. 2017, 214, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-N.; Xu, D.-B.; Yan, X.; Wu, Z.-K.; Kayani, S.I.; Shen, Q.; Fu, X.-Q.; Xie, L.-H.; Hao, X.-L.; Hassani, D.; et al. Jasmonate- and Abscisic Acid-Activated AaGSW1-AaTCP15/AaORA Transcriptional Cascade Promotes Artemisinin Biosynthesis in Artemisia annua. Plant Biotechnol. J. 2021, 19, 1412–1428. [Google Scholar] [CrossRef] [PubMed]
- Demarsy, E.; Fankhauser, C. Higher Plants Use LOV to Perceive Blue Light. Curr. Opin. Plant Biol. 2009, 12, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Nagatani, A. Phytochrome: Structural Basis for Its Functions. Curr. Opin. Plant Biol. 2010, 13, 565–570. [Google Scholar] [CrossRef]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.-O.; van der Horst, G.T.J.; Batschauer, A.; Ahmad, M. The Cryptochromes: Blue Light Photoreceptors in Plants and Animals. Annu. Rev. Plant Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef]
- Heijde, M.; Ulm, R. UV-B Photoreceptor-Mediated Signalling in Plants. Trends Plant Sci. 2012, 17, 230–237. [Google Scholar] [CrossRef]
- Fu, X.; Peng, B.; Hassani, D.; Xie, L.; Liu, H.; Li, Y.; Chen, T.; Liu, P.; Tang, Y.; Li, L.; et al. AaWRKY9 Contributes to Light- and Jasmonate-Mediated to Regulate the Biosynthesis of Artemisinin in Artemisia annua. New Phytol. 2021, 231, 1858–1874. [Google Scholar] [CrossRef]
- Liu, H.; Li, L.; Fu, X.; Li, Y.; Chen, T.; Qin, W.; Yan, X.; Wu, Z.; Xie, L.; Kayani, S.; et al. AaMYB108 Is the Core Factor Integrating Light and Jasmonic Acid Signaling to Regulate Artemisinin Biosynthesis in Artemisia annua. New Phytol. 2023, 237, 2224–2237. [Google Scholar] [CrossRef]
- Shi, P.; Fu, X.; Shen, Q.; Liu, M.; Pan, Q.; Tang, Y.; Jiang, W.; Lv, Z.; Yan, T.; Ma, Y.; et al. The Roles of AaMIXTA1 in Regulating the Initiation of Glandular Trichomes and Cuticle Biosynthesis in Artemisia annua. New Phytol. 2018, 217, 261–276. [Google Scholar] [CrossRef]
- Yan, T.; Chen, M.; Shen, Q.; Li, L.; Fu, X.; Pan, Q.; Tang, Y.; Shi, P.; Lv, Z.; Jiang, W.; et al. HOMEODOMAIN PROTEIN 1 Is Required for Jasmonate-Mediated Glandular Trichome Initiation in Artemisia annua. New Phytol. 2017, 213, 1145–1155. [Google Scholar] [CrossRef]
- Yan, T.; Li, L.; Xie, L.; Chen, M.; Shen, Q.; Pan, Q.; Fu, X.; Shi, P.; Tang, Y.; Huang, H.; et al. A Novel HD-ZIP IV/MIXTA Complex Promotes Glandular Trichome Initiation and Cuticle Development in Artemisia annua. New Phytol. 2018, 218, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Yan, T.; Li, L.; Chen, M.; Hassani, D.; Li, Y.; Qin, W.; Liu, H.; Chen, T.; Fu, X.; et al. An HD-ZIP-MYB Complex Regulates Glandular Secretory Trichome Initiation in Artemisia annua. New Phytol. 2021, 231, 2050–2064. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, T.; Li, Y.; Liu, H.; Qin, W.; Wu, Z.; Peng, B.; Wang, X.; Yan, X.; Fu, X.; et al. AaWIN1, an AP2/ERF Protein, Positively Regulates Glandular Secretory Trichome Initiation in Artemisia annua. Plant Sci. 2023, 329, 111602. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fu, X.; Li, L.; Sun, X.; Tang, K.; Zhao, J. AaSPL9 Affects Glandular Trichomes Initiation by Positively Regulating Expression of AaHD1 in Artemisia annua L. Plant Sci. 2022, 317, 111172. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, R.; Chai, Y.; Bonfill, M.; Moyano, E.; Oksman-Caldentey, K.-M.; Xu, T.; Pi, Y.; Wang, Z.; Zhang, H.; et al. Engineering Tropane Biosynthetic Pathway in Hyoscyamus Niger Hairy Root Cultures. Proc. Natl. Acad. Sci. USA 2004, 101, 6786–6791. [Google Scholar] [CrossRef]
- Pan, Q.; Chen, Y.; Wang, Q.; Yuan, F.; Xing, S.; Tian, Y.; Zhao, J.; Sun, X.; Tang, K. Effect of Plant Growth Regulators on the Biosynthesis of Vinblastine, Vindoline and Catharanthine in Catharanthus Roseus. Plant Growth Regul. 2010, 60, 133–141. [Google Scholar] [CrossRef]
- Lin, X.; Zhou, Y.; Zhang, J.; Lu, X.; Zhang, F.; Shen, Q.; Wu, S.; Chen, Y.; Wang, T.; Tang, K. Enhancement of Artemisinin Content in Tetraploid Artemisia annua Plants by Modulating the Expression of Genes in Artemisinin Biosynthetic Pathway. Biotechnol. Appl. Biochem. 2011, 58, 50–57. [Google Scholar] [CrossRef]
- Xing, S.-H.; Guo, X.-B.; Wang, Q.; Pan, Q.-F.; Tian, Y.-S.; Liu, P.; Zhao, J.-Y.; Wang, G.-F.; Sun, X.-F.; Tang, K.-X. Induction and Flow Cytometry Identification of Tetraploids from Seed-Derived Explants through Colchicine Treatments in Catharanthus roseus (L.) G. Don. BioMed Res. Int. 2011, 2011, 793198. [Google Scholar] [CrossRef]
- Wang, Q.; Xing, S.; Pan, Q.; Yuan, F.; Zhao, J.; Tian, Y.; Chen, Y.; Wang, G.; Tang, K. Development of Efficient Catharanthus Roseus Regeneration and Transformation System Using Agrobacterium Tumefaciens and Hypocotyls as Explants. BMC Biotechnol. 2012, 12, 34. [Google Scholar] [CrossRef]
- Tang, K.; Shen, Q.; Yan, T.; Fu, X. Transgenic Approach to Increase Artemisinin Content in Artemisia annua L. Plant Cell Rep. 2014, 33, 605–615. [Google Scholar] [CrossRef]
- Ochatt, S.; Alan, A.R.; Bhattacharya, A.; Hano, C.; Kiselev, K.V.; Marconi, P.L.; Otoni, W.C.; Park, S.Y.; Tang, K.X.; Weathers, P.J. Secondary Metabolites: A Boon from Plants, the Best Chemist in Nature: Preface from the Editors. Plant Cell Tissue Organ Cult. 2022, 149, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jing, F.; Li, F.; Li, M.; Wang, Y.; Wang, G.; Sun, X.; Tang, K. Development of Transgenic Artemisia annua (Chinese Wormwood) Plants with an Enhanced Content of Artemisinin, an Effective Anti-Malarial Drug, by Hairpin-RNA-Mediated Gene Silencing. Biotechnol. Appl. Biochem. 2009, 52, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lu, X.; Lv, Z.; Zhang, L.; Zhu, M.; Jiang, W.; Wang, G.; Sun, X.; Tang, K. Overexpression of the Artemisia Orthologue of ABA Receptor, AaPYL9, Enhances ABA Sensitivity and Improves Artemisinin Content in Artemisia annua L. PLoS ONE 2013, 8, e56697. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Chen, Y.; Wang, T.; Wu, S.; Lu, X.; Zhang, L.; Zhang, F.; Jiang, W.; Wang, G.; Tang, K. Overexpression of the Cytochrome P450 Monooxygenase (Cyp71av1) and Cytochrome P450 Reductase (Cpr) Genes Increased Artemisinin Content in Artemisia annua (Asteraceae). Genet. Mol. Res. 2012, 11, 3298–3309. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Zhang, F.; Pan, Q.; Fu, X.; Jiang, W.; Shen, Q.; Yan, T.; Shi, P.; Lu, X.; Sun, X.; et al. Branch Pathway Blocking in Artemisia annua Is a Useful Method for Obtaining High Yield Artemisinin. Plant Cell Physiol. 2016, 57, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wang, Q.; Yuan, F.; Xing, S.; Zhao, J.; Choi, Y.H.; Verpoorte, R.; Tian, Y.; Wang, G.; Tang, K. Overexpression of ORCA3 and G10H in Catharanthus Roseus Plants Regulated Alkaloid Biosynthesis and Metabolism Revealed by NMR-Metabolomics. PLoS ONE 2012, 7, e43038. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, M.; Yang, C.; Liu, X.; Zhang, L.; Lan, X.; Tang, K.; Liao, Z. Enhancing the Scopolamine Production in Transgenic Plants of Atropa Belladonna by Overexpressing Pmt and H6h Genes. Physiol. Plant. 2011, 143, 309–315. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, B.; Lu, B.; Kai, G.; Wang, Z.; Xia, Y.; Ding, R.; Zhang, H.; Sun, X.; Chen, W.; et al. Tropane Alkaloids Production in Transgenic Hyoscyamus Niger Hairy Root Cultures Over-Expressing Putrescine N-Methyltransferase Is Methyl Jasmonate-Dependent. Planta 2007, 225, 887–896. [Google Scholar] [CrossRef]
- Hao, X.; Zhong, Y.; Fu, X.; Lv, Z.; Shen, Q.; Yan, T.; Shi, P.; Ma, Y.; Chen, M.; Lv, X.; et al. Transcriptome Analysis of Genes Associated with the Artemisinin Biosynthesis by Jasmonic Acid Treatment under the Light in Artemisia annua. Front. Plant Sci. 2017, 8, 971. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, L.; Liao, Z.; Wang, S.; Yan, T.; Shi, P.; Liu, M.; Fu, X.; Pan, Q.; Wang, Y.; et al. The Genome of Artemisia annua Provides Insight into the Evolution of Asteraceae Family and Artemisinin Biosynthesis. Mol. Plant 2018, 11, 776–788. [Google Scholar] [CrossRef]
- Matías-Hernández, L.; Jiang, W.; Yang, K.; Tang, K.; Brodelius, P.E.; Pelaz, S. AaMYB1 and Its Orthologue AtMYB61 Affect Terpene Metabolism and Trichome Development in Artemisia annua and Arabidopsis Thaliana. Plant J. 2017, 90, 520–534. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wang, C.; Xiong, Z.; Wang, H.; Fu, X.; Shen, Q.; Peng, B.; Ma, Y.; Sun, X.; Tang, K. CrERF5, an AP2/ERF Transcription Factor, Positively Regulates the Biosynthesis of Bisindole Alkaloids and Their Precursors in Catharanthus Roseus. Front. Plant Sci. 2019, 10, 931. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Huang, H.; Xie, L.; Hao, X.; Kayani, S.-I.; Liu, H.; Qin, W.; Chen, T.; Pan, Q.; Liu, P.; et al. Basic Helix-Loop-Helix Transcription Factors AabHLH2 and AabHLH3 Function Antagonistically With AaMYC2 and Are Negative Regulators in Artemisinin Biosynthesis. Front. Plant Sci. 2022, 13, 885622. [Google Scholar] [CrossRef] [PubMed]
- Shu, G.; Tang, Y.; Yuan, M.; Wei, N.; Zhang, F.; Yang, C.; Lan, X.; Chen, M.; Tang, K.; Xiang, L.; et al. Molecular Insights into AabZIP1-Mediated Regulation on Artemisinin Biosynthesis and Drought Tolerance in Artemisia annua. Acta Pharm. Sin. B 2022, 12, 1500–1513. [Google Scholar] [CrossRef]
- Xie, L.; Yan, T.; Li, L.; Chen, M.; Ma, Y.; Hao, X.; Fu, X.; Shen, Q.; Huang, Y.; Qin, W.; et al. The WRKY Transcription Factor AaGSW2 Promotes Glandular Trichome Initiation in Artemisia annua. J. Exp. Bot. 2021, 72, 1691–1701. [Google Scholar] [CrossRef]
- Kayani, S.-I.; Shen, Q.; Ma, Y.; Fu, X.; Xie, L.; Zhong, Y.; Tiantian, C.; Pan, Q.; Li, L.; Rahman, S.; et al. The YABBY Family Transcription Factor AaYABBY5 Directly Targets Cytochrome P450 Monooxygenase (CYP71AV1) and Double-Bond Reductase 2 (DBR2) Involved in Artemisinin Biosynthesis in Artemisia annua. Front. Plant Sci. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Kayani, S.-I.; Shen, Q.; Rahman, S.; Fu, X.; Li, Y.; Wang, C.; Hassani, D.; Tang, K. Transcriptional Regulation of Flavonoid Biosynthesis in Artemisia annua by AaYABBY5. Hortic. Res. 2021, 8, 257. [Google Scholar] [CrossRef]
- Ma, Y.-N.; Xu, D.-B.; Li, L.; Zhang, F.; Fu, X.-Q.; Shen, Q.; Lyu, X.-Y.; Wu, Z.-K.; Pan, Q.-F.; Shi, P.; et al. Jasmonate Promotes Artemisinin Biosynthesis by Activating the TCP14-ORA Complex in Artemisia annua. Sci. Adv. 2018, 4, eaas9357. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, S.; Zhang, F.; Chen, L.; Hao, X.; Pan, Q.; Fu, X.; Li, L.; Sun, X.; Tang, K. Overexpression of a Novel NAC Domain-Containing Transcription Factor Gene (AaNAC1) Enhances the Content of Artemisinin and Increases Tolerance to Drought and Botrytis Cinerea in Artemisia annua. Plant Cell Physiol. 2016, 57, 1961–1971. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, Y.; Liu, Y.; Peng, B.; Zhang, L.; Tang, K.; Chen, W. The SPB-Box Transcription Factor AaSPL2 Positively Regulates Artemisinin Biosynthesis in Artemisia annua L. Front. Plant Sci. 2019, 10, 409. [Google Scholar] [CrossRef]
- Chen, T.-T.; Yao, X.-H.; Liu, H.; Li, Y.-P.; Qin, W.; Yan, X.; Wang, X.-Y.; Peng, B.-W.; Zhang, Y.-J.; Shao, J.; et al. MADS-Box Gene AaSEP4 Promotes Artemisinin Biosynthesis in Artemisia annua. Front. Plant Sci. 2022, 13, 1961–1971. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hao, X.; Liu, H.; Wang, W.; Fu, X.; Ma, Y.; Shen, Q.; Chen, M.; Tang, K. Jasmonic Acid-Responsive AabHLH1 Positively Regulates Artemisinin Biosynthesis in Artemisia annua. Biotechnol. Appl. Biochem. 2019, 66, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Jian, D.; Zhang, F.; Yang, C.; Bai, G.; Lan, X.; Chen, M.; Tang, K.; Liao, Z. The Cold-Induced Transcription Factor BHLH112 Promotes Artemisinin Biosynthesis Indirectly via ERF1 in Artemisia annua. J. Exp. Bot. 2019, 70, 4835–4848. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Zhong, Y.; Nützmann, H.-W.; Fu, X.; Yan, T.; Shen, Q.; Chen, M.; Ma, Y.; Zhao, J.; Osbourn, A.; et al. Light-Induced Artemisinin Biosynthesis Is Regulated by the BZIP Transcription Factor AaHY5 in Artemisia annua. Plant Cell Physiol. 2019, 60, 1747–1760. [Google Scholar] [CrossRef] [PubMed]
- Maes, L.; Van Nieuwerburgh, F.C.W.; Zhang, Y.; Reed, D.W.; Pollier, J.; Casteele, S.R.F.V.; Inzé, D.; Covello, P.S.; Deforce, D.L.D.; Goossens, A. Dissection of the Phytohormonal Regulation of Trichome Formation and Biosynthesis of the Antimalarial Compound Artemisinin in Artemisia annua Plants. New Phytol. 2011, 189, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-T.; Liu, H.; Li, Y.-P.; Yao, X.-H.; Qin, W.; Yan, X.; Wang, X.-Y.; Peng, B.-W.; Zhang, Y.-J.; Shao, J.; et al. AaSEPALLATA1 Integrates JA and Light-Regulated Glandular Secretory Trichome Initiation in Artemisia annua. Plant Physiol. 2023, 192, 1483–1497. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient Expression Vectors for Functional Genomics, Quantification of Promoter Activity and RNA Silencing in Plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; He, W.; Yao, X.; Yan, X.; Wang, X.; Peng, B.; Zhang, Y.; Shao, J.; Hu, X.; Miao, Q.; et al. The Light- and Jasmonic Acid-Induced AaMYB108-like Positive Regulates the Initiation of Glandular Secretory Trichome in Artemisia annua L. Int. J. Mol. Sci. 2023, 24, 12929. https://doi.org/10.3390/ijms241612929
Liu H, He W, Yao X, Yan X, Wang X, Peng B, Zhang Y, Shao J, Hu X, Miao Q, et al. The Light- and Jasmonic Acid-Induced AaMYB108-like Positive Regulates the Initiation of Glandular Secretory Trichome in Artemisia annua L. International Journal of Molecular Sciences. 2023; 24(16):12929. https://doi.org/10.3390/ijms241612929
Chicago/Turabian StyleLiu, Hang, Weizhi He, Xinghao Yao, Xin Yan, Xiuyun Wang, Bowen Peng, Yaojie Zhang, Jin Shao, Xinyi Hu, Qing Miao, and et al. 2023. "The Light- and Jasmonic Acid-Induced AaMYB108-like Positive Regulates the Initiation of Glandular Secretory Trichome in Artemisia annua L." International Journal of Molecular Sciences 24, no. 16: 12929. https://doi.org/10.3390/ijms241612929
APA StyleLiu, H., He, W., Yao, X., Yan, X., Wang, X., Peng, B., Zhang, Y., Shao, J., Hu, X., Miao, Q., Li, L., & Tang, K. (2023). The Light- and Jasmonic Acid-Induced AaMYB108-like Positive Regulates the Initiation of Glandular Secretory Trichome in Artemisia annua L. International Journal of Molecular Sciences, 24(16), 12929. https://doi.org/10.3390/ijms241612929





