Analysis of the RNA Editing Sites and Orthologous Gene Function of Transcriptome and Chloroplast Genomes in the Evolution of Five Deutzia Species
Abstract
:1. Introduction
2. Results
2.1. Chloroplast Genomes’ Features
2.2. Inverted Repeat Boundary Analysis
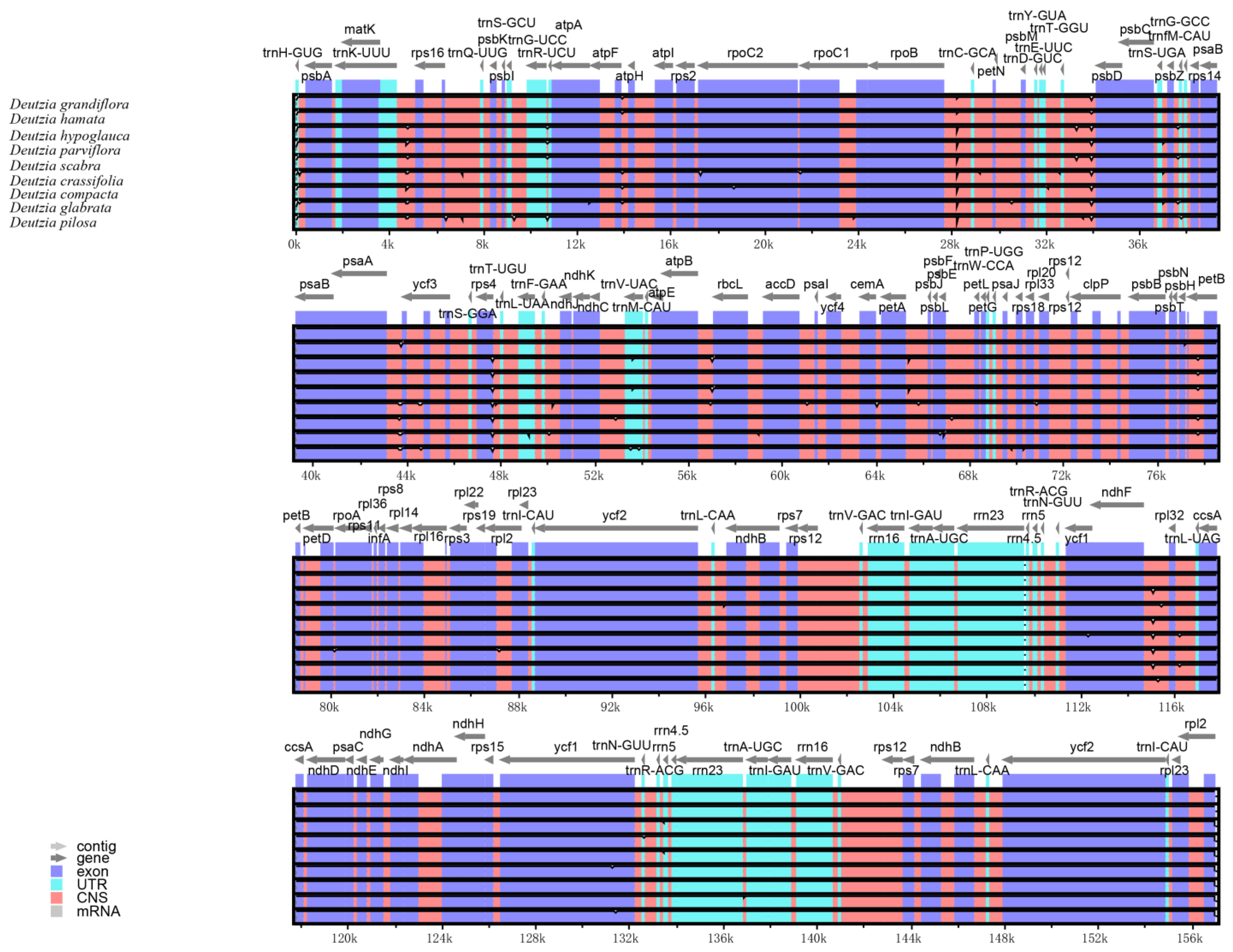
2.3. Comparative Genomic and Pi Analysis
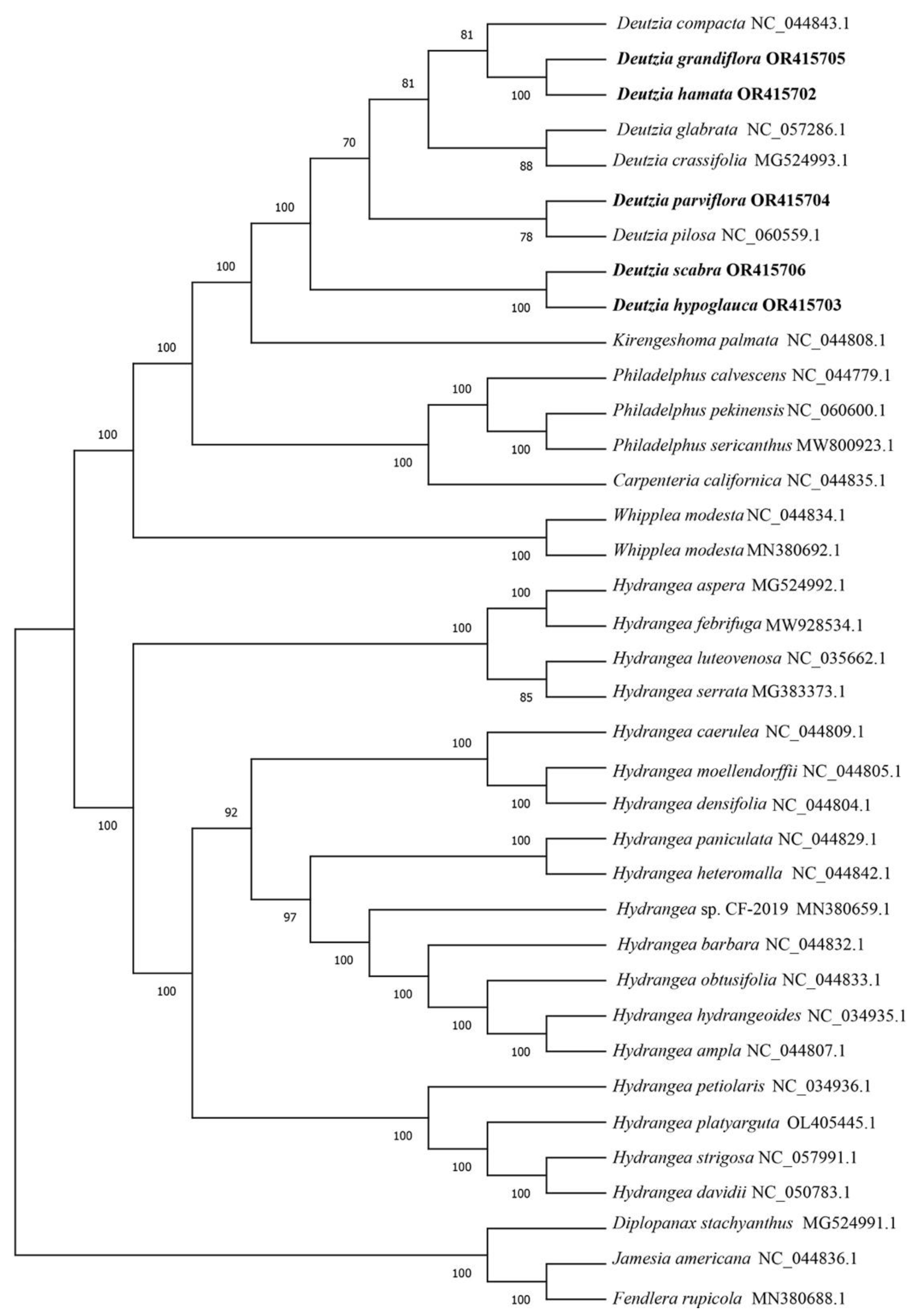
2.4. Phylogenetic Analysis
2.5. KaKs Analysis
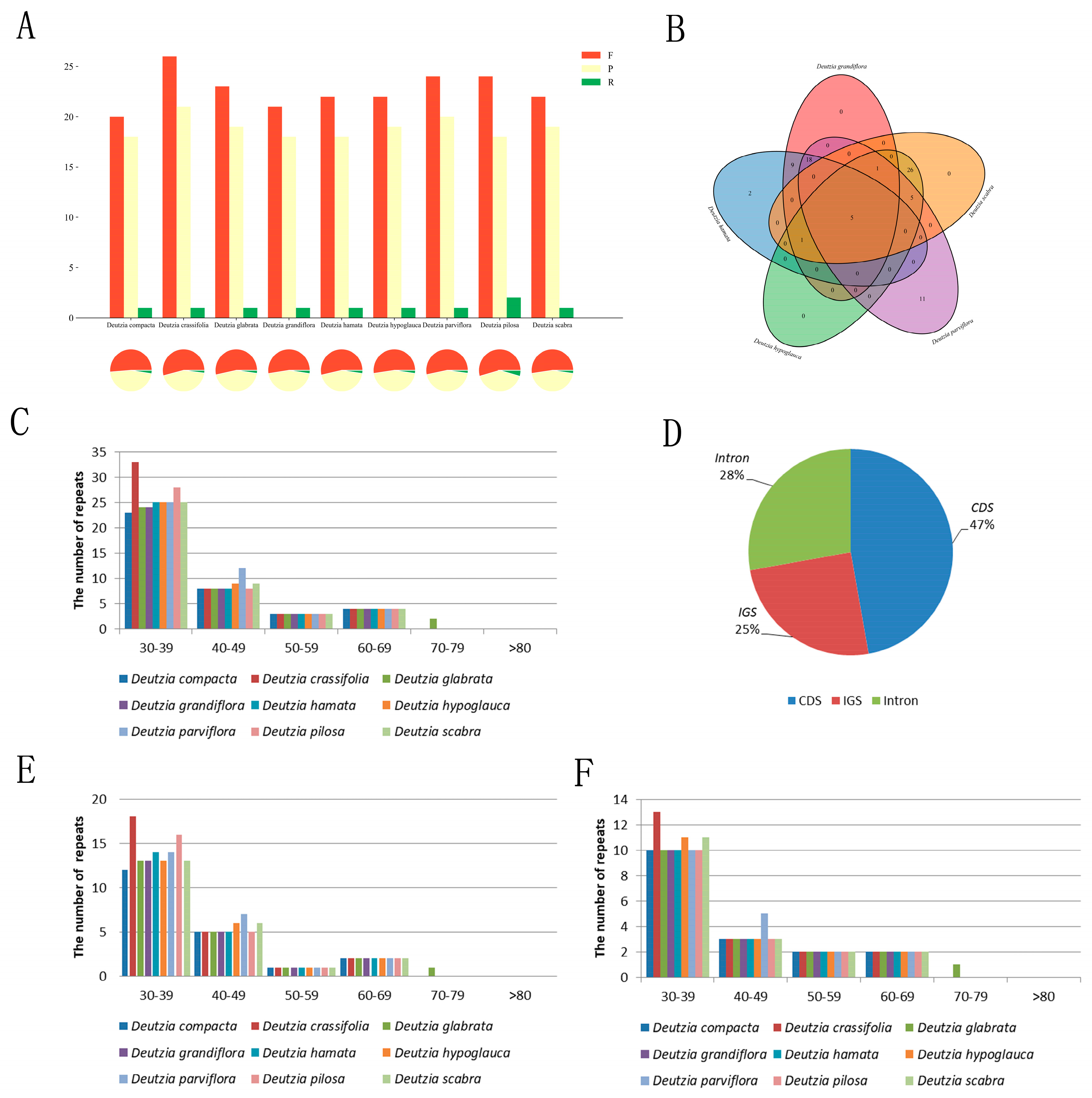
2.6. Repeat Analysis
2.6.1. Scattered Repetition Analysis
2.6.2. SSR Analysis
2.7. Chloroplast Genome RNA Editing Analysis
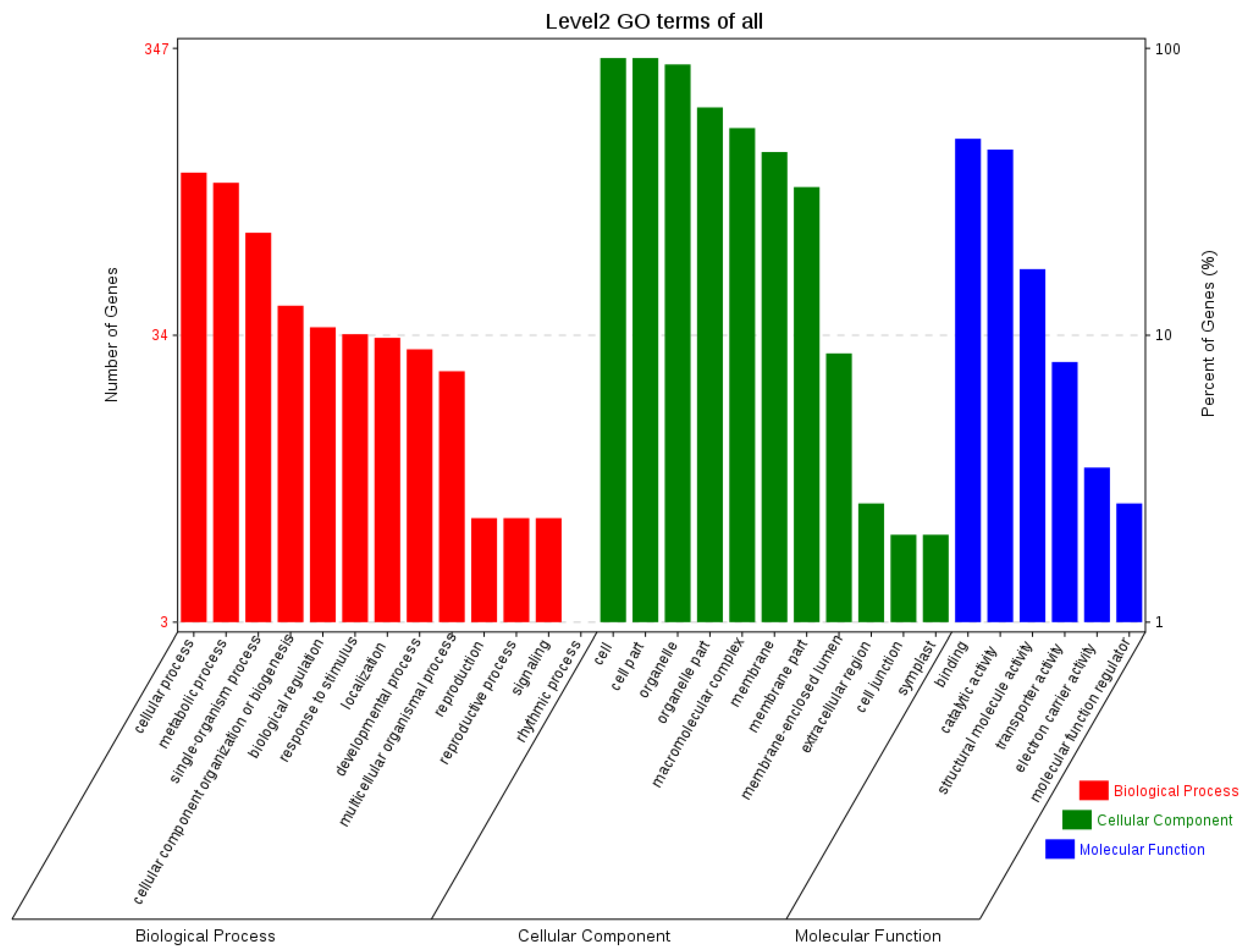
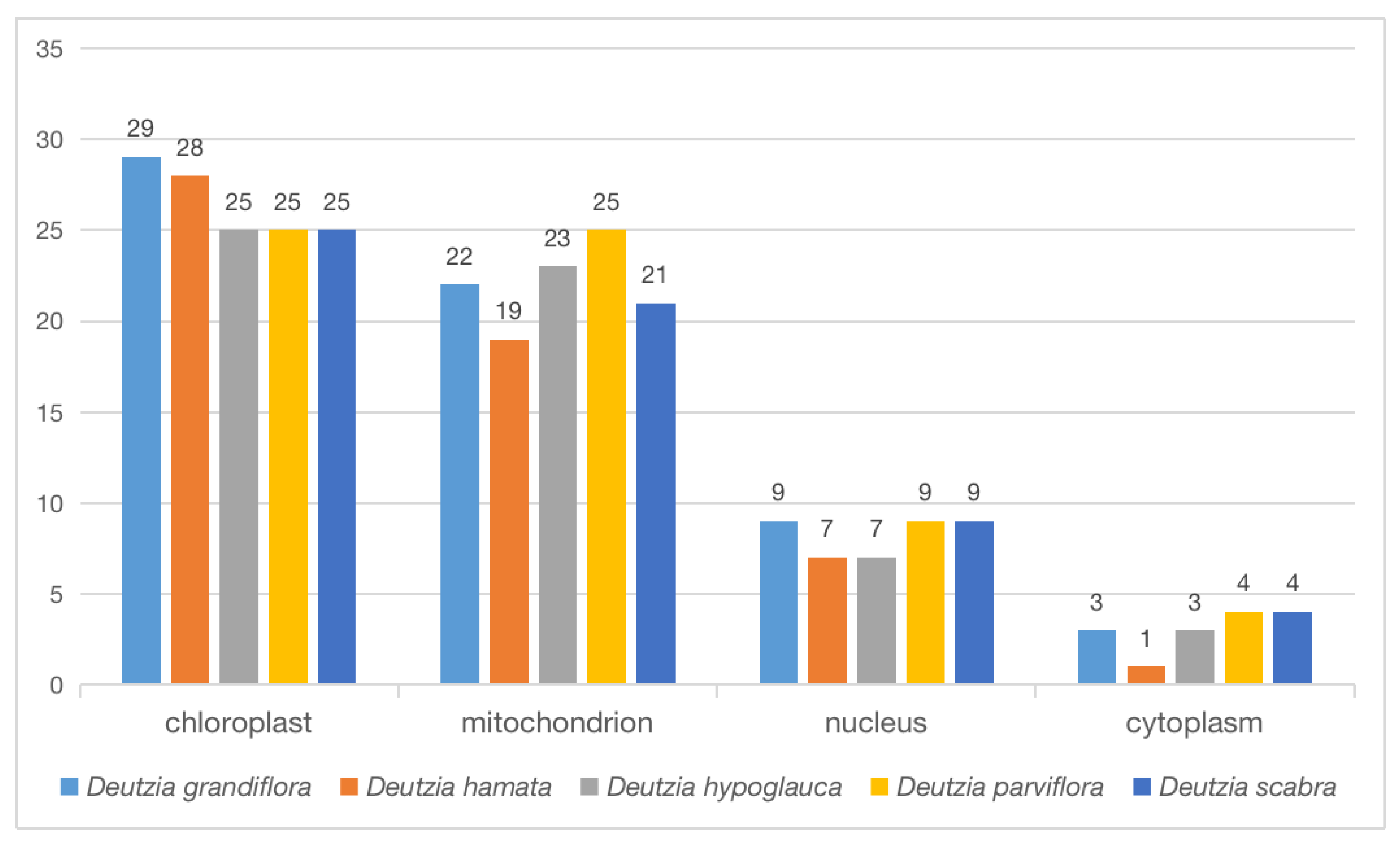
2.8. Orthologous Gene Analysis of Transcriptome and Chloroplast Genome
2.9. Discovery of Genes Potentially Encoding Medicinal Ingredients
3. Discussion
3.1. Relatively Conserved Chloroplast Genome of Deutzia Plants
3.2. Evolution Analysis
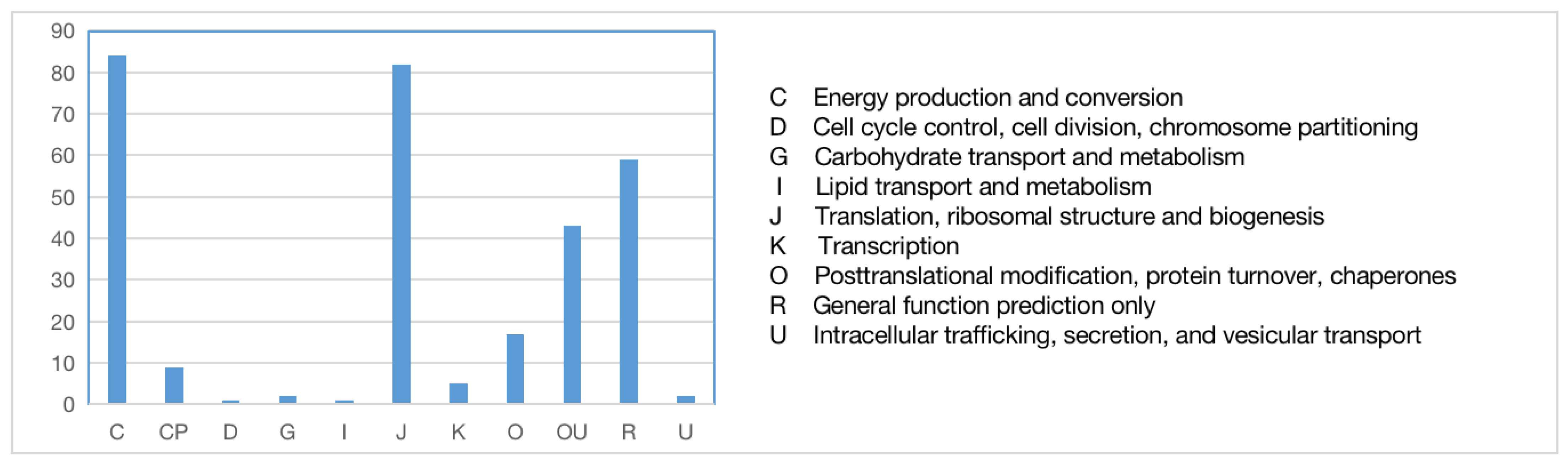
3.3. Functional Annotation of Chloroplast and Transcriptome Orthologous Genes and Genes Related to Major Active Ingredients
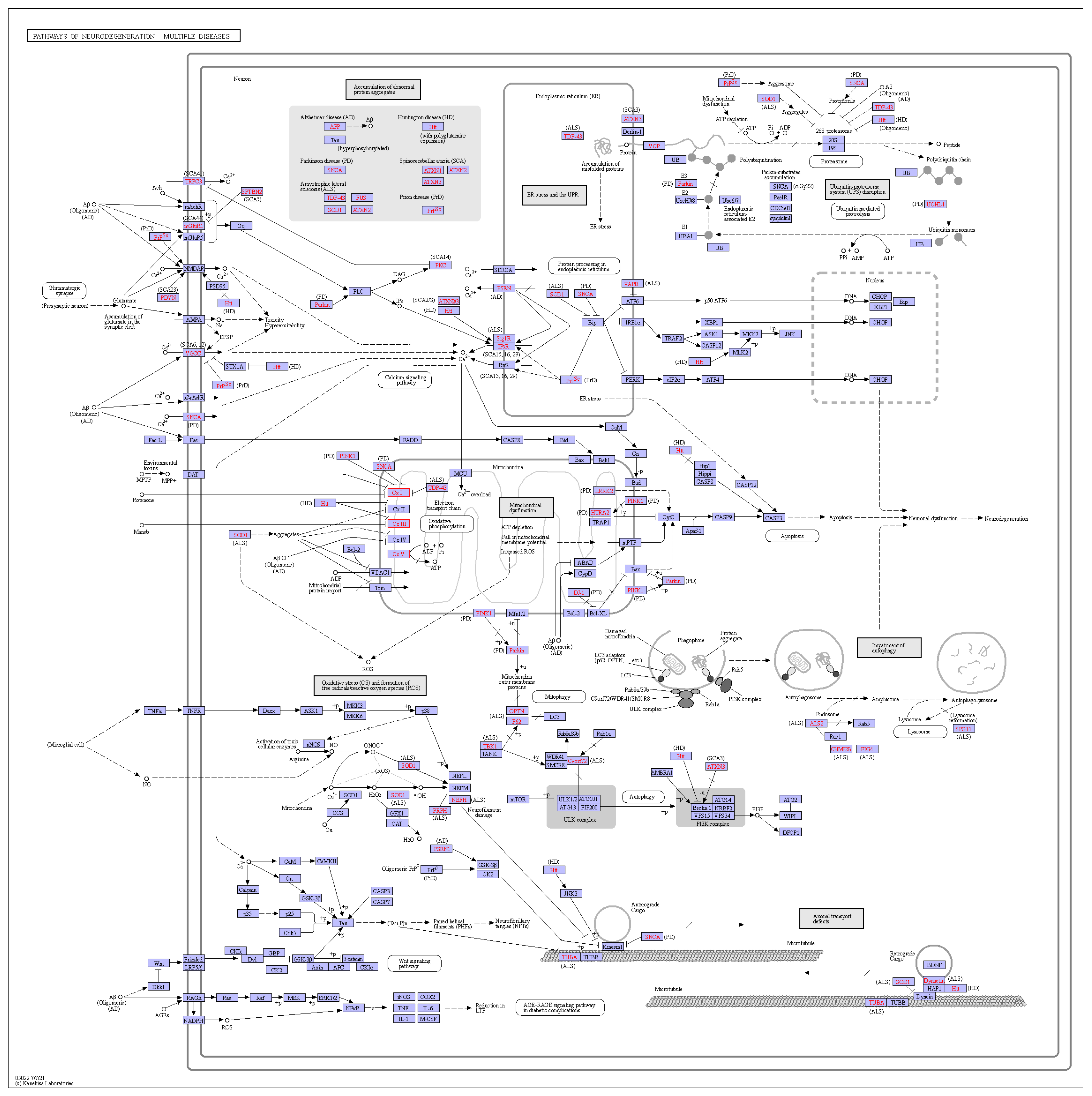
3.4. Genes Related to Major Active Ingredients
4. Materials and Methods
4.1. Plant Material and Chloroplast Genome Sequences Collection
4.2. Chloroplast Genome Sequencing, Assembly, and Annotation of Five Deutzia Species
4.3. Transcriptome Sequencing, Assembly, and Annotation
4.4. Analysis of Repeat Structures of Transcriptome and Chloroplast Genome
4.5. KaKs Analysis
4.6. Chloroplast Genomes Comparative Analysis
4.7. Evolution
4.8. RNA Editing Site Identification
4.9. Identification and Analysis of Orthologous Genes of Chloroplast Genome and Transcriptome
4.10. Discovery of Genes Potentially Encoding Medicinal Ingredients
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Styer, C.; Stern, W. Comparative anatomy and systematics of woody Saxifragaceae. Deutzia. Bot. J. Linn. Soc. 1979, 79, 291–319. [Google Scholar] [CrossRef]
- Shu-mei, H. The classification and distribution of genus Deutzia in China. J. Syst. Evol. 1993, 31, 105. [Google Scholar]
- Lu, L.; Huang, S. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1995. [Google Scholar]
- Yang, S. The Divine Farmer’s Materia Medica: A Translation of the Shen Nong Ben Cao Jing; Blue Poppy Enterprises, Inc.: Portland, OR, USA, 1998. [Google Scholar]
- Gu, X.; Hao, D.; Xiao, P. Research progress of Chinese herbal medicine compounds and their bioactivities: Fruitful 2020. Chin. Herb. Med. 2022, 14, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Zaikonnikova, T.I. Deutzias Ornamental Shrubs. In A Monographs of Genus Deutzia; Nauka: Moscow, Russia, 1966. [Google Scholar]
- Junjie, L.; Ying, R.; Xiurong, X.; Tingting, L.; Dekui, Z. Genetic Diversity Analysis of Deutzia baroniana Based on SRAP Marker. Shandong Agric. Sci. 2014, 46, 18–21+25. [Google Scholar]
- Junjie, L.; Yan, M.; Ying, R.; Dekui, Z. Genetic diversity analysis of Deutzia glabrata in Shandong Province based on SRAP Markers. China For. Sci. Technol. 2015, 29, 13–16. [Google Scholar]
- Kim, C.; Deng, T.; Wen, J.; Nie, Z.-L.; Sun, H. Systematics, biogeography, and character evolution of Deutzia (Hydrangeaceae) inferred from nuclear and chloroplast DNA sequences. Mol. Phylogenetics Evol. 2015, 87, 91–104. [Google Scholar] [CrossRef]
- Chan, C.X.; Bhattacharya, D. The origin of plastids. Nat. Educ. 2010, 3, 84-1–84-5. [Google Scholar]
- Howe, C.J.; Barbrook, A.C.; Koumandou, V.L.; Nisbet, R.E.R.; Symington, H.A.; Wightman, T.F. Evolution of the chloroplast genome. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2003, 358, 99–107. [Google Scholar] [CrossRef]
- Martin, W.; Stoebe, B.; Goremykin, V.; Hansmann, S.; Hasegawa, M.; Kowallik, K.V. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 1998, 393, 162–165. [Google Scholar] [CrossRef]
- Xing, S. Progress in chloroplast genome analysis. Prog. Biochem. Biophys. 2006, 12, wpr-590708. [Google Scholar]
- Wolfe, K.H.; Morden, C.W.; Ems, S.C.; Palmer, J.D. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: Loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J. Mol. Evol. 1992, 35, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, T.; Tsudzuki, J.; Ito, S.; Nakashima, K.; Tsudzuki, T.; Sugiura, M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 1994, 91, 9794–9798. [Google Scholar] [CrossRef] [PubMed]
- Ayliffe, M.A.; Timmis, J.N. Plastid DNA sequence homologies in the tobacco nuclear genome. Mol. Gen. Genet. MGG 1992, 236, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Hill, J.; Hsiao, J.; Moffat, K.; Ouyang, S.; Cheng, Z.; Jiang, J.; Buell, C.R. Genome sequencing of a 239-kb region of rice chromosome 10 L reveals a high frequency of gene duplication and a large chloroplast DNA insertion. Mol. Genet. Genom. 2002, 267, 713–720. [Google Scholar] [CrossRef]
- Maul, J.E.; Lilly, J.W.; Cui, L.Y.; dePamphilis, C.W.; Miller, W.; Harris, E.H.; Stern, D.B. The Chlamydomonas reinhardtti plastid chromosome: Islands of genes in a sea of repeats. Plant Cell 2002, 14, 2659–2679. [Google Scholar] [CrossRef]
- Huang, C.Y.; Ayliffe, M.A.; Timmis, J.N. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 2003, 422, 72–76. [Google Scholar] [CrossRef]
- Sasaki, Y.; Kozaki, A.; Ohmori, A.; Iguchi, H.; Nagano, Y. Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J. Biol. Chem. 2001, 276, 3937–3940. [Google Scholar] [CrossRef]
- Petriccione, M.; Di Cecco, I.; Arena, S.; Scaloni, A.; Scortichini, M. Proteomic changes in Actinidia chinensis shoot during systemic infection with a pandemic Pseudomonas syringae pv. actinidiae strain. J. Proteom. 2013, 78, 461–476. [Google Scholar] [CrossRef]
- Maier, R.M.; Neckermann, K.; Igloi, G.L.; Kossel, H. Complete Sequence of the Maize Chloroplast Genome-Gene Content, Hotspots of Divergence and Fine-Tuning of Genetic Information by Transcript Editing. J. Mol. Biol. 1995, 251, 614–628. [Google Scholar] [CrossRef]
- Wakasugi, T.; Hirose, T.; Horihata, M.; Tsudzuki, T.; Kossel, H.; Sugiura, M. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: The pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl. Acad. Sci. USA 1996, 93, 8766–8770. [Google Scholar] [CrossRef]
- Tangphatsornruang, S.; Uthaipaisanwong, P.; Sangsrakru, D.; Chanprasert, J.; Yoocha, T.; Jomchai, N.; Tragoonrung, S. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene 2011, 475, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.M.; Hoch, B.; Zeltz, P.; Kossel, H. Internal editing of the maize chloroplast ndhA transcript restores codons for conserved amino acids. Plant Cell 1992, 4, 609–616. [Google Scholar]
- Freyer, R.; KieferMeyer, M.C.; Kossel, H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. USA 1997, 94, 6285–6290. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Cao, S.-K.; Yang, H.; Liu, R.; Sun, F.; Wang, L.; Wang, M.; Tan, B.-C. DEK48 Is Required for RNA Editing at Multiple Mitochondrial Sites and Seed Development in Maize. Int. J. Mol. Sci. 2022, 23, 3064. [Google Scholar] [CrossRef]
- Lu, Y. RNA editing of plastid-encoded genes. Photosynthetica 2018, 56, 48–61. [Google Scholar] [CrossRef]
- Chu, D.; Wei, L. The chloroplast and mitochondrial C-to-U RNA editing in Arabidopsis thaliana shows signals of adaptation. Plant Direct 2019, 3, e00169. [Google Scholar] [CrossRef]
- Zhao, Y.; Gao, R.; Zhao, Z.; Hu, S.; Han, R.; Jeyaraj, A.; Arkorful, E.; Li, X.; Chen, X. Genome-wide identification of RNA editing sites in chloroplast transcripts and multiple organellar RNA editing factors in tea plant (Camellia sinensis L.): Insights into the albinism mechanism of tea leaves. Gene 2023, 848, 146898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cui, X.; Wang, Y.; Wu, J.; Gu, X.; Lu, T. The RNA Editing Factor WSP1 Is Essential for Chloroplast Development in Rice. Mol. Plant 2017, 10, 86–98. [Google Scholar] [CrossRef]
- Chen, C.-Z.; Wang, Y.-L.; He, M.-X.; Li, Z.-W.; Lan, S.; Qing, L.; Re, D.-Y.; Jiang, H.; Li, Z.; Zhang, G.-H. OsPPR9 encodes a DYW-type PPR protein that affects editing efficiency of multiple RNA editing sites and is essential for chloroplast development. J. Integr. Agric. 2022, 22, 972–980. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Haertel, B.; Brennicke, A. RNA Editing in Plants and Its Evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef]
- Cui, L.Y.; Leebens-Mack, J.; Wang, L.S.; Tang, J.J.; Rymarquis, L.; Stern, D.B.; dePamphilis, C.W. Adaptive evolution of chloroplast genome structure inferred using a parametric bootstrap approach. BMC Evol. Biol. 2006, 6, 13. [Google Scholar] [CrossRef]
- Xinbing, Y. The Study of Introduction and Propagation of Wild Deutzia. Master’s Thesis, Hebei Agriculture University, Baoding, China, 2004. [Google Scholar]
- Diroma, M.A.; Ciaccia, L.; Pesole, G.; Picardi, E. Elucidating the editome: Bioinformatics approaches for RNA editing detection. Brief. Bioinform. 2019, 20, 436–447. [Google Scholar] [CrossRef]
- Li, C.-S.; Yu, H.-W.; Chen, X.-Z.; Wu, X.-Q.; Fang, D.-M.; Li, G.-Y. Chemical Constituents of Deutzia setchuenensis. Nat. Prod. Res. Dev. 2012, 24, 465–468. [Google Scholar]
- Esposito, P.; Iavarone, C.; Sen, A.; Trogolo, C. Scabrosidol, a new highly oxygenated iridoid glucoside from Deutzia scabra. J. Nat. Prod. 1983, 46, 614–618. [Google Scholar] [CrossRef]
- Esposito, P.; Iavarone, C.; Sen, A.; Trogolo, C. Iridoid aglycones from Deutzia scabra. Phytochemistry 1984, 23, 803–807. [Google Scholar] [CrossRef]
- Lin, S.; Liu, J.; He, X.; Wang, J.; Wang, Z.; Zhang, X.; Bao, M.; Fu, X. Comprehensive Comparative Analysis and Development of Molecular Markers for Dianthus Species Based on Complete Chloroplast Genome Sequences. Int. J. Mol. Sci. 2022, 23, 12567. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Schmidt, A.M. Use of an intron region of a chloroplast tRNA gene (trnL) as a target for PCR identification of specific food crops including sources of potential allergens. Food Res. Int. 2004, 37, 395–402. [Google Scholar]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Shi, W.; Song, W.; Liu, J.; Shi, C.; Wang, S. Comparative chloroplast genome analysis of Citrus (Rutaceae) species: Insights into genomic characterization, phylogenetic relationships, and discrimination of subgenera. Sci. Hortic. 2023, 313, 111909. [Google Scholar]
- Danderson, C.A.; Downie, S.R.; Hermann, M. Rampant polyphyly in the Arracacia Glade (Apiaceae) and an assessment of the phylogenetic utility of 20 noncoding plastid loci. Mol. Phylogenetics Evol. 2018, 118, 286–305. [Google Scholar]
- Lin, P.; Yin, H.; Wang, K.; Gao, H.; Liu, L.; Yao, X. Comparative Genomic Analysis Uncovers the Chloroplast Genome Variation and Phylogenetic Relationships of Camellia Species. Biomolecules 2022, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, C.; Guo, X.; Liu, Q.; Wang, K. Complete chloroplast genome of Camellia japonica genome structures, comparative and phylogenetic analysis. PLoS ONE 2019, 14, e0216645. [Google Scholar]
- Wang, Y.; Zhang, C.-F.; Odago, W.O.; Jiang, H.; Yang, J.-X.; Hu, G.-W.; Wang, Q.-F. Evolution of 101 Apocynaceae plastomes and phylogenetic implications. Mol. Phylogenetics Evol. 2023, 180, 107688. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.F.; Borsch, T.; Hilu, K.W. Phylogenetic utility of rapidly evolving DNA at high taxonomical levels: Contrasting matK, trnT-F, and rbcL in basal angiosperms. Mol. Phylogenetics Evol. 2006, 41, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Pan, J.; Xue, Q.; Zhu, S.; Liu, W.; Ding, X. Plastome-wide comparison reveals new SNV resources for the authentication of Dendrobium huoshanense and its corresponding medicinal slice (Huoshan Fengdou). Acta Pharm. Sin. B 2018, 8, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Xiang, Q.Y.; Hufford, L. Relationships and Evolution of Hydrangeaceae Based on Rbcl Sequence DATA. Am. J. Bot. 1995, 82, 504–514. [Google Scholar] [CrossRef]
- Fan, C.Z.; Xiang, Q.Y. Phylogenetic analyses of Cornales based on 26S rRNA and combined 26S rDNA-matK-rbcL sequence data. Am. J. Bot. 2003, 90, 1357–1372. [Google Scholar]
- Acosta, M.C.; Premoli, A.C. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol. Phylogenetics Evol. 2010, 54, 235–242. [Google Scholar] [CrossRef]
- Hufford, L. A phylogenetic analysis of Hydrangeaceae based on morphological data. Int. J. Plant Sci. 1997, 158, 652–672. [Google Scholar] [CrossRef]
- Hufford, L. Hydrangeaceae. In Flowering Plants. Dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales; Kubitzki, K., Ed.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2004; pp. 202–215. [Google Scholar]
- Yang, Q.; Xin, C. Characterization of the complete chloroplast genome sequence of Deutzia glabrata (Saxifragaceae). Mitochondrial DNA Part B-Resour. 2020, 5, 764–765. [Google Scholar]
- Adams, K.L.; Daley, D.O.; Qiu, Y.L.; Whelan, J.; Palmer, J.D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 2000, 408, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Compilation of China’s Medicinal Herb; People’s Medical Publishing House: Beijing, China, 1975. [Google Scholar]
- Tao, H. Notes on Herbal Classics; People’s Medical Publishing House: Beijing, China, 1994. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Son, P.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Hill, K.; Krugmann, S.; Andrews, S.R.; Coadwell, W.J.; Finan, P.; Welch, H.C.E.; Hawkins, P.T.; Stephens, L.R. Regulation of P-Rex1 by phosphatidylinositol (3,4,5)-trisphosphate and G beta gamma subunits. J. Biol. Chem. 2005, 280, 4166–4173. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Wang, M.; Liu, H.; Ge, L.; Xing, G.; Wang, M.; Weining, S.; Nie, X. Identification and analysis of RNA editing sites in the chloroplast transcripts of Aegilops tauschii L. Genes 2016, 8, 13. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Mangalam, A.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef]



| Region | Pi | Total Number of Mutations | Region Length (bp) |
|---|---|---|---|
| SSC.gene4.trnL-UAG | 0.00625 | 1 | 80 |
| LSC.gene21.petN | 0.00556 | 1 | 90 |
| LSC.gene82.rps3 | 0.00431 | 5 | 657 |
| LSC.gene53.cemA | 0.00266 | 3 | 690 |
| LSC.gene17.rpoC2 | 0.00253 | 20 | 4158 |
| LSC.gene5.rps16 | 0.00253 | 1 | 264 |
| SSC.gene5.ccsA | 0.00242 | 4 | 966 |
| LSC.gene78.infA | 0.00214 | 1 | 234 |
| LSC.gene49.rbcL | 0.0021 | 5 | 1428 |
| LSC.gene4.matK | 0.00209 | 6 | 1518 |
| Region | Pi | Total Number of Mutations | Region Length (bp) |
|---|---|---|---|
| SSC.gene4.ndhD-psaC | 0.01122 | 3 | 132 |
| LSC.gene59.petD-CDS2-rpoA | 0.01099 | 5 | 187 |
| SSC.gene3.ccsA-ndhD | 0.00889 | 6 | 258 |
| LSC.gene57.petB-CDS2-petD-CDS1 | 0.00802 | 7 | 196 |
| LSC.gene10.atpF-CDS1-atpH | 0.00734 | 8 | 355 |
| LSC.gene13.rps2-rpoC2 | 0.00704 | 5 | 225 |
| SSC.gene2.rpl32-ccsA | 0.00669 | 21 | 1148 |
| LSC.gene14.rpoC2-rpoC1-CDS2 | 0.00654 | 5 | 174 |
| LSC.gene46.rps18-rpl20 | 0.00591 | 4 | 298 |
| SSC.gene4.ndhD-psaC | 0.01122 | 3 | 132 |
| Deutzia grandiflora | Deutzia hamata | Deutzia hypoglauca | Deutzia scabra | |
|---|---|---|---|---|
| vs | vs | vs | vs | |
| ccsA | Deutzia pilosa | Deutzia pilosa | ||
| matK | Deutzia glabrata | Deutzia glabrata | Deutzia glabrata | Deutzia glabrata |
| rbcL | Deutzia pilosa | Deutzia pilosa | Deutzia pilosa | Deutzia pilosa |
| ycf1 | Deutzia glabrata | Deutzia crassifolia Deutzia glabrata |
| Number | Species | Accession | Number | Species | Accession |
|---|---|---|---|---|---|
| 1 | Deutzia glabrata | NC_057286.1 | 17 | Hydrangea caerulea | NC_044809.1 |
| 2 | Deutzia crassifolia | MG524993.1 | 18 | Hydrangea serrata | MG383373.1 |
| 3 | Deutzia pilosa | NC_060559.1 | 19 | Hydrangea barbara | NC_044832.1 |
| 4 | Deutzia compacta | NC_044843.1 | 20 | Hydrangea petiolaris | NC_034936.1 |
| 5 | Philadelphus calvescens | NC_044779.1 | 21 | Hydrangea moellendorffii | NC_044805.1 |
| 6 | Philadelphus sericanthus | MW800923.1 | 22 | Hydrangea febrifuga | MW928534.1 |
| 7 | Philadelphus pekinensis | NC_060600.1 | 23 | Hydrangea aspera | MG524992.1 |
| 8 | Kirengeshoma palmata | NC_044808.1 | 24 | Hydrangea luteovenosa | NC_035662.1 |
| 9 | Carpenteria californica | NC_044835.1 | 25 | Hydrangea obtusifolia | NC_044833.1 |
| 10 | Whipplea modesta | MN380692.1 | 26 | Hydrangea hydrangeoides | NC_034935.1 |
| 11 | Hydrangea davidii | NC_050783.1 | 27 | Hydrangea ampla | NC_044807.1 |
| 12 | Hydrangea sp. CF-2019 | MN380659.1 | 28 | Hydrangea densifolia | NC_044804.1 |
| 13 | Hydrangea strigosa | NC_057991.1 | 29 | Jamesia americana | NC_044836.1 |
| 14 | Hydrangea heteromalla | NC_044842.1 | 30 | Diplopanax stachyanthus | MG524991.1 |
| 15 | Hydrangea paniculata | NC_044829.1 | 31 | Whipplea modesta | NC_044834.1 |
| 16 | Hydrangea platyarguta | OL405445.1 | 32 | Fendlera rupicola | MN380688.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, H.; Ren, Y.; Du, J.; Liu, L.; Long, L.; Yang, M. Analysis of the RNA Editing Sites and Orthologous Gene Function of Transcriptome and Chloroplast Genomes in the Evolution of Five Deutzia Species. Int. J. Mol. Sci. 2023, 24, 12954. https://doi.org/10.3390/ijms241612954
Cai H, Ren Y, Du J, Liu L, Long L, Yang M. Analysis of the RNA Editing Sites and Orthologous Gene Function of Transcriptome and Chloroplast Genomes in the Evolution of Five Deutzia Species. International Journal of Molecular Sciences. 2023; 24(16):12954. https://doi.org/10.3390/ijms241612954
Chicago/Turabian StyleCai, Hongyu, Yachao Ren, Juan Du, Lingyun Liu, Lianxiang Long, and Minsheng Yang. 2023. "Analysis of the RNA Editing Sites and Orthologous Gene Function of Transcriptome and Chloroplast Genomes in the Evolution of Five Deutzia Species" International Journal of Molecular Sciences 24, no. 16: 12954. https://doi.org/10.3390/ijms241612954
APA StyleCai, H., Ren, Y., Du, J., Liu, L., Long, L., & Yang, M. (2023). Analysis of the RNA Editing Sites and Orthologous Gene Function of Transcriptome and Chloroplast Genomes in the Evolution of Five Deutzia Species. International Journal of Molecular Sciences, 24(16), 12954. https://doi.org/10.3390/ijms241612954






