Differential Epigenetic Changes in the Dorsal Hippocampus of Male and Female SAMP8 Mice: A Preliminary Study
Abstract
:1. Introduction
2. Results
2.1. Markers of Neurodegeneration Are Altered in 9-Month-Old SAMP8 Mice
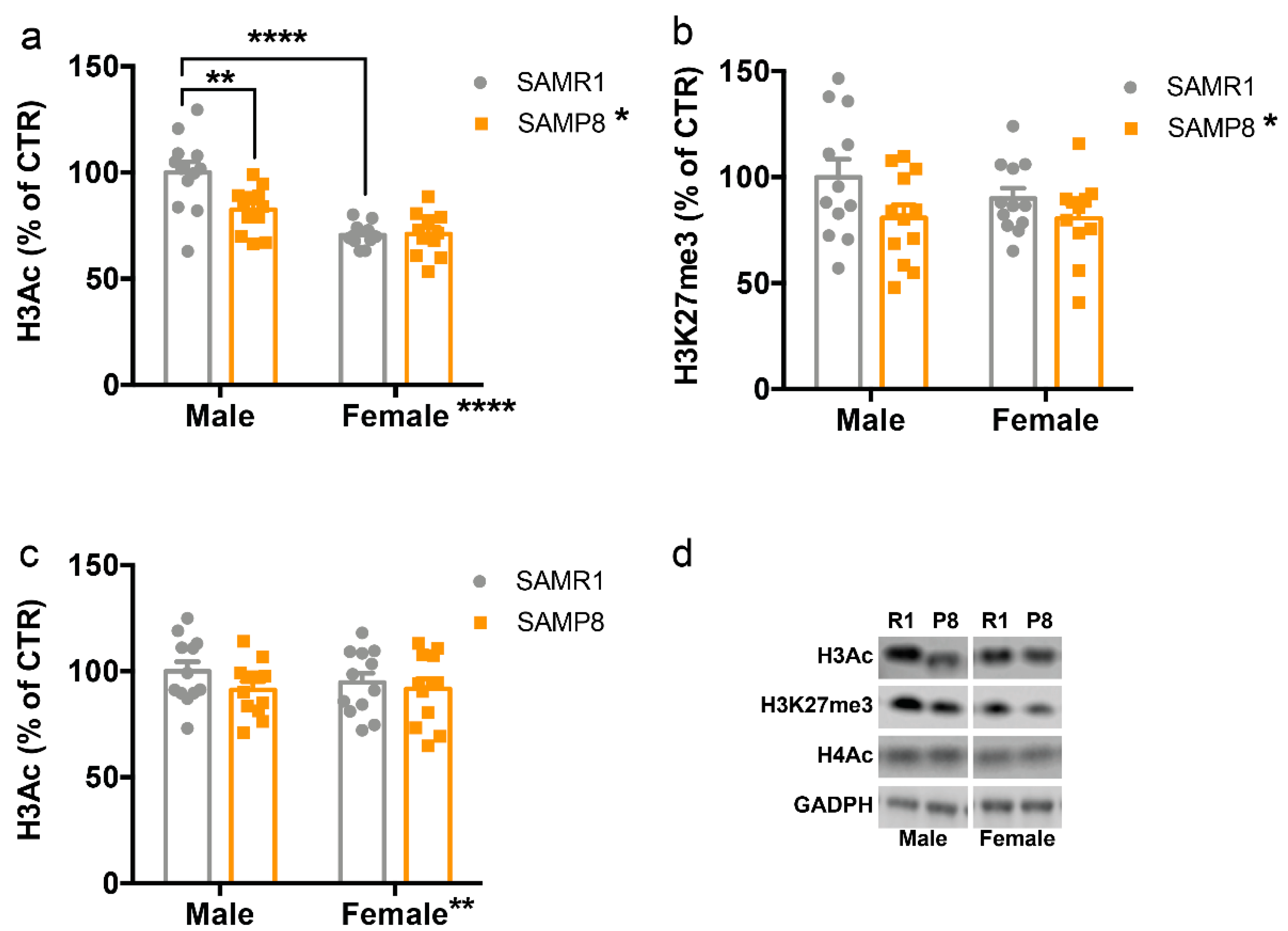
2.2. Histone Post-Translational Modifications Are Altered in the Dorsal Hippocampus of 9-Month-Old SAMP8 Mice
2.3. HDACs Protein Levels Are Altered in the Dorsal Hippocampus of 9-Month-Old SAMP8 Mice
2.4. Histone Demethylase Levels Are Increased in the Dorsal Hippocampus of 9-Month-Old SAMP8 Mice
2.5. Dnmt3a Levels Are Reduced in the Dorsal Hippocampus of 9-Month-Old SAMP8 Mice
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Western Blot Analysis
4.3. RNA Isolation and Reverse Transcription
4.4. Quantitative PCR
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mangialasche, F.; Solomon, A.; Winblad, B.; Mecocci, P.; Kivipelto, M. Alzheimer’s Disease: Clinical Trials and Drug Development. Lancet Neurol. 2010, 9, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a Risk Factor for Neurodegenerative Disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Petrov, D.; Ettcheto, M.; Abad, S.; Sánchez-López, E.; García, M.L.; Olloquequi, J.; Beas-Zarate, C.; Auladell, C.; Camins, A. Current Research Therapeutic Strategies for Alzheimer’s Disease Treatment. Neural Plast. 2016, 2016, 8501693. [Google Scholar] [CrossRef] [PubMed]
- Rabinovici, G.D. Late-Onset Alzheimer Disease. Continuum 2019, 25, 14–33. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic Modifications: Basic Mechanisms and Role in Cardiovascular Disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Lacal, I.; Ventura, R. Epigenetic Inheritance: Concepts, Mechanisms and Perspectives. Front. Mol. Neurosci. 2018, 11, 292. [Google Scholar] [CrossRef]
- Rudenko, A.; Tsai, L.H. Epigenetic Regulation in Memory and Cognitive Disorders. Neuroscience 2014, 264, 51–63. [Google Scholar] [CrossRef]
- Maity, S.; Farrell, K.; Navabpour, S.; Narayanan, S.N.; Jarome, T.J. Epigenetic Mechanisms in Memory and Cognitive Decline Associated with Aging and Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 12280. [Google Scholar] [CrossRef]
- Biswas, S.; Rao, C.M. Epigenetic Tools (The Writers, The Readers and The Erasers) and Their Implications in Cancer Therapy. Eur. J. Pharmacol. 2018, 837, 8–24. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, Erasing and Reading Histone Lysine Methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.; Workman, J.L. Histone Exchange, Chromatin Structure and the Regulation of Transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Santana, D.A.; Smith, M.D.A.C.; Chen, E.S. Histone Modifications in Alzheimer’s Disease. Genes 2023, 14, 347. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Xie, J.; Xie, L.; Wei, H.; Li, X.J.; Lin, L. Dynamic Regulation of DNA Methylation and Brain Functions. Biology 2023, 12, 152. [Google Scholar] [CrossRef]
- Akbarian, S.; Beeri, M.S.; Haroutunian, V. Epigenetic Determinants of Healthy and Diseased Brain Aging and Cognition. JAMA Neurol. 2013, 70, 711–718. [Google Scholar] [CrossRef]
- Berson, A.; Nativio, R.; Berger, S.L.; Bonini, N.M. Epigenetic Regulation in Neurodegenerative Diseases. Trends Neurosci. 2018, 41, 587–598. [Google Scholar] [CrossRef]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau Promotes Neurodegeneration through Global Chromatin Relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef]
- Klein, H.U.; McCabe, C.; Gjoneska, E.; Sullivan, S.E.; Kaskow, B.J.; Tang, A.; Smith, R.V.; Xu, J.; Pfenning, A.R.; Bernstein, B.E.; et al. Epigenome-Wide Study Uncovers Large-Scale Changes in Histone Acetylation Driven by Tau Pathology in Aging and Alzheimer’s Human Brains. Nat. Neurosci. 2019, 22, 37–46. [Google Scholar] [CrossRef]
- Mansuroglu, Z.; Benhelli-Mokrani, H.; Marcato, V.; Sultan, A.; Violet, M.; Chauderlier, A.; Delattre, L.; Loyens, A.; Talahari, S.; Bégard, S.; et al. Loss of Tau Protein Affects the Structure, Transcription and Repair of Neuronal Pericentromeric Heterochromatin. Sci. Rep. 2016, 6, 33047. [Google Scholar] [CrossRef]
- Sanchez-Varo, R.; Mejias-Ortega, M.; Fernandez-Valenzuela, J.J.; Nuñez-Diaz, C.; Caceres-Palomo, L.; Vegas-Gomez, L.; Sanchez-Mejias, E.; Trujillo-Estrada, L.; Garcia-Leon, J.A.; Moreno-Gonzalez, I.; et al. Transgenic Mouse Models of Alzheimer’s Disease: An Integrative Analysis. Int. J. Mol. Sci. 2022, 23, 5404. [Google Scholar] [CrossRef] [PubMed]
- Chishti, M.A.; Yang, D.S.; Janus, C.; Phinney, A.L.; Horne, P.; Pearson, J.; Strome, R.; Zuker, N.; Loukides, J.; French, J.; et al. Early-Onset Amyloid Deposition and Cognitive Deficits in Transgenic Mice Expressing a Double Mutant Form of Amyloid Precursor Protein 695. J. Biol. Chem. 2001, 276, 21562–21570. [Google Scholar] [CrossRef] [PubMed]
- Akiguchi, I.; Pallàs, M.; Budka, H.; Akiyama, H.; Ueno, M.; Han, J.; Yagi, H.; Nishikawa, T.; Chiba, Y.; Sugiyama, H.; et al. SAMP8 Mice as a Neuropathological Model of Accelerated Brain Aging and Dementia: Toshio Takeda’s Legacy and Future Directions. Neuropathology 2017, 37, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Karuppagounder, V.; Arumugam, S.; Babu, S.S.; Palaniyandi, S.S.; Watanabe, K.; Cooke, J.P.; Thandavarayan, R.A. The Senescence Accelerated Mouse Prone 8 (SAMP8): A Novel Murine Model for Cardiac Aging. Ageing Res. Rev. 2017, 35, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, J.; Shi, J.-S. SAMP8 Mice as a Model of Age-Related Cognition Decline with Underlying Mechanisms in Alzheimer’s Disease. J. Alzheimers Dis. 2020, 75, 385–395. [Google Scholar] [CrossRef]
- Morley, J.E.; Armbrecht, H.J.; Farr, S.A.; Kumar, V.B. The Senescence Accelerated Mouse (SAMP8) as a Model for Oxidative Stress and Alzheimer’s Disease. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 650–656. [Google Scholar] [CrossRef]
- Cosín-Tomás, M.; Alvarez-López, M.J.; Sanchez-Roige, S.; Lalanza, J.F.; Bayod, S.; Sanfeliu, C.; Pallàs, M.; Escorihuela, R.M.; Kaliman, P. Epigenetic Alterations in Hippocampus of SAMP8 Senescent Mice and Modulation by Voluntary Physical Exercise. Front. Aging Neurosci. 2014, 6, 51. [Google Scholar] [CrossRef]
- Griñan-Ferré, C.; Puigoriol-Illamola, D.; Palomera-ávalos, V.; Pérez-Cáceres, D.; Companys-Alemany, J.; Camins, A.; Ortuño-Sahagún, D.; Teresa Rodrigo, M.; Pallàs, M. Environmental Enrichment Modified Epigenetic Mechanisms in SAMP8 Mouse Hippocampus by Reducing Oxidative Stress and Inflammaging and Achieving Neuroprotection. Front. Aging Neurosci. 2016, 8, 241. [Google Scholar] [CrossRef]
- Cosín-Tomás, M.; Álvarez-López, M.J.; Companys-Alemany, J.; Kaliman, P.; González-Castillo, C.; Ortuño-Sahagún, D.; Pallàs, M.; Griñán-Ferré, C. Temporal Integrative Analysis of MRNA and MicroRNAs Expression Profiles and Epigenetic Alterations in Female SAMP8, a Model of Age-Related Cognitive Decline. Front. Genet. 2018, 9, 596. [Google Scholar] [CrossRef]
- Vasilopoulou, F.; Bellver-Sanchis, A.; Companys-Alemany, J.; Jarne-Ferrer, J.; Irisarri, A.; Palomera-Ávalos, V.; Gonzalez-Castillo, C.; Ortuño-Sahagún, D.; Sanfeliu, C.; Pallàs, M.; et al. Cognitive Decline and BPSD Are Concomitant with Autophagic and Synaptic Deficits Associated with G9a Alterations in Aged SAMP8 Mice. Cells 2022, 11, 2603. [Google Scholar] [CrossRef]
- Musazzi, L.; Carini, G.; Barbieri, S.S.; Maggi, S.; Veronese, N.; Popoli, M.; Barbon, A.; Ieraci, A. Phenotypic Frailty Assessment in SAMP8 Mice: Sex Differences and Potential Role of MiRNAs as Peripheral Biomarkers. J. Gerontol. A Biol. Sci. Med. Sci. 2023; Online ahead of print. [Google Scholar] [CrossRef]
- Szarowicz, C.A.; Steece-Collier, K.; Caulfield, M.E. New Frontiers in Neurodegeneration and Regeneration Associated with Brain-Derived Neurotrophic Factor and the Rs6265 Single Nucleotide Polymorphism. Int. J. Mol. Sci. 2022, 23, 8011. [Google Scholar] [CrossRef] [PubMed]
- Novoa, C.; Salazar, P.; Cisternas, P.; Gherardelli, C.; Vera-Salazar, R.; Zolezzi, J.M.; Inestrosa, N.C. Inflammation Context in Alzheimer’s Disease, a Relationship Intricate to Define. Biol. Res. 2022, 55, 39. [Google Scholar] [CrossRef] [PubMed]
- Sbai, O.; Djelloul, M.; Auletta, A.; Ieraci, A.; Vascotto, C.; Perrone, L. AGE-TXNIP Axis Drives Inflammation in Alzheimer’s by Targeting Aβ to Mitochondria in Microglia. Cell Death Dis. 2022, 13, 302. [Google Scholar] [CrossRef] [PubMed]
- Maurya, S.K.; Bhattacharya, N.; Mishra, S.; Bhattacharya, A.; Banerjee, P.; Senapati, S.; Mishra, R. Microglia Specific Drug Targeting Using Natural Products for the Regulation of Redox Imbalance in Neurodegeneration. Front. Pharm. 2021, 12, 654489. [Google Scholar] [CrossRef]
- Ulku, I.; Liebsch, F.; Akerman, S.C.; Schulz, J.F.; Kulic, L.; Hock, C.; Pietrzik, C.; Di Spiezio, A.; Thinakaran, G.; Saftig, P.; et al. Mechanisms of Amyloid-Β34 Generation Indicate a Pivotal Role for BACE1 in Amyloid Homeostasis. Sci. Rep. 2023, 13, 2216. [Google Scholar] [CrossRef]
- Li, J.; Hart, R.P.; Mallimo, E.M.; Swerdel, M.R.; Kusnecov, A.W.; Herrup, K. EZH2-Mediated H3K27 Trimethylation Mediates Neurodegeneration in Ataxia-Telangiectasia. Nat. Neurosci. 2013, 16, 1745–1753. [Google Scholar] [CrossRef] [PubMed]
- Ziemka-Nalecz, M.; Jaworska, J.; Sypecka, J.; Zalewska, T. Histone Deacetylase Inhibitors: A Therapeutic Key in Neurological Disorders? J. Neuropathol. Exp. Neurol. 2018, 77, 855–870. [Google Scholar] [CrossRef] [PubMed]
- Narayan, P.; Dragunow, M. Alzheimer’s Disease and Histone Code Alterations. Adv. Exp. Med. Biol. 2017, 978, 321–336. [Google Scholar] [CrossRef]
- Persico, G.; Casciaro, F.; Amatori, S.; Rusin, M.; Cantatore, F.; Perna, A.; Auber, L.A.; Fanelli, M.; Giorgio, M. Histone H3 Lysine 4 and 27 Trimethylation Landscape of Human Alzheimer’s Disease. Cells 2022, 11, 734. [Google Scholar] [CrossRef]
- Lin, K.A.; Doraiswamy, P.M. When Mars Versus Venus Is Not a Cliché: Gender Differences in the Neurobiology of Alzheimer’s Disease. Front. Neurol. 2015, 5, 288. [Google Scholar] [CrossRef]
- Nebel, R.A.; Aggarwal, N.T.; Barnes, L.L.; Gallagher, A.; Goldstein, J.M.; Kantarci, K.; Mallampalli, M.P.; Mormino, E.C.; Scott, L.; Yu, W.H.; et al. Understanding the Impact of Sex and Gender in Alzheimer’s Disease: A Call to Action. Alzheimers Dement. 2018, 14, 1171–1183. [Google Scholar] [CrossRef] [PubMed]
- Sohn, D.; Shpanskaya, K.; Lucas, J.E.; Petrella, J.R.; Saykin, A.J.; Tanzi, R.E.; Samatova, N.F.; Doraiswamy, P.M. Sex Differences in Cognitive Decline in Subjects with High Likelihood of Mild Cognitive Impairment Due to Alzheimer’s Disease. Sci. Rep. 2018, 8, 7490. [Google Scholar] [CrossRef] [PubMed]
- Morley, J.E.; Farr, S.A.; Kumar, V.B.; Armbrecht, H.J. The SAMP8 Mouse: A Model to Develop Therapeutic Interventions for Alzheimers Disease. Curr. Pharm. Des. 2012, 18, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.R.; Zhou, W.X.; Zhang, Y.X. The Behavioral, Pathological and Therapeutic Features of the Senescence-Accelerated Mouse Prone 8 Strain as an Alzheimer’s Disease Animal Model. Ageing Res. Rev. 2014, 13, 13–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Schrag, M.; Crofton, A.; Trivedi, R.; Vinters, H.; Kirsch, W. Targeted Proteomics for Quantification of Histone Acetylation in Alzheimer’s Disease. Proteomics 2012, 12, 1261–1268. [Google Scholar] [CrossRef]
- Nativio, R.; Donahue, G.; Berson, A.; Lan, Y.; Amlie-Wolf, A.; Tuzer, F.; Toledo, J.B.; Gosai, S.J.; Gregory, B.D.; Torres, C.; et al. Dysregulation of the Epigenetic Landscape of Normal Aging in Alzheimer’s Disease. Nat. Neurosci. 2018, 21, 497–505. [Google Scholar] [CrossRef]
- Marzi, S.J.; Leung, S.K.; Ribarska, T.; Hannon, E.; Smith, A.R.; Pishva, E.; Poschmann, J.; Moore, K.; Troakes, C.; Al-Sarraj, S.; et al. A Histone Acetylome-Wide Association Study of Alzheimer’s Disease Identifies Disease-Associated H3K27ac Differences in the Entorhinal Cortex. Nat. Neurosci. 2018, 21, 1618–1627. [Google Scholar] [CrossRef]
- Francis, Y.I.; Fà, M.; Ashraf, H.; Zhang, H.; Staniszewski, A.; Latchman, D.S.; Arancio, O. Dysregulation of Histone Acetylation in the APP/PS1 Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2009, 18, 131–139. [Google Scholar] [CrossRef]
- Govindarajan, N.; Agis-Balboa, R.C.; Walter, J.; Sananbenesi, F.; Fischer, A. Sodium Butyrate Improves Memory Function in an Alzheimer’s Disease Mouse Model When Administered at an Advanced Stage of Disease Progression. J. Alzheimer’s Dis. 2011, 26, 187–197. [Google Scholar] [CrossRef]
- Griñán-Ferré, C.; Izquierdo, V.; Otero, E.; Puigoriol-Illamola, D.; Corpas, R.; Sanfeliu, C.; Ortuño-Sahagún, D.; Pallàs, M. Environmental Enrichment Improves Cognitive Deficits, AD Hallmarks and Epigenetic Alterations Presented in 5xFAD Mouse Model. Front. Cell Neurosci. 2018, 12, 224. [Google Scholar] [CrossRef]
- Gräff, J.; Rei, D.; Guan, J.S.; Wang, W.Y.; Seo, J.; Hennig, K.M.; Nieland, T.J.F.; Fass, D.M.; Kao, P.F.; Kahn, M.; et al. An Epigenetic Blockade of Cognitive Functions in the Neurodegenerating Brain. Nature 2012, 483, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Alzheimer’s Disease Is a Synaptic Failure. Science 2002, 298, 789–791. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.S.; Haggarty, S.J.; Giacometti, E.; Dannenberg, J.H.; Joseph, N.; Gao, J.; Nieland, T.J.F.; Zhou, Y.; Wang, X.; Mazitschek, R.; et al. HDAC2 Negatively Regulates Memory Formation and Synaptic Plasticity. Nature 2009, 459, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Akhtar, M.W.; Adachi, M.; Mahgoub, M.; Bassel-Duby, R.; Kavalali, E.T.; Olson, E.N.; Monteggia, L.M. An Essential Role for Histone Deacetylase 4 in Synaptic Plasticity and Memory Formation. J. Neurosci. 2012, 32, 10879–10886. [Google Scholar] [CrossRef]
- Colussi, C.; Aceto, G.; Ripoli, C.; Bertozzi, A.; Li Puma, D.D.; Paccosi, E.; D’Ascenzo, M.; Grassi, C. Cytoplasmic HDAC4 Recovers Synaptic Function in the 3× Tg Mouse Model of Alzheimer’s Disease. Neuropathol. Appl. Neurobiol. 2023, 49, e12861. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb Complex PRC2 and Its Mark in Life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Agger, K.; Cloos, P.A.C.; Christensen, J.; Pasini, D.; Rose, S.; Rappsilber, J.; Issaeva, I.; Canaani, E.; Salcini, A.E.; Helin, K. UTX and JMJD3 Are Histone H3K27 Demethylases Involved in HOX Gene Regulation and Development. Nature 2007, 449, 731–734. [Google Scholar] [CrossRef]
- Hong, S.H.; Cho, Y.W.; Yu, L.R.; Yu, H.; Veenstra, T.D.; Ge, K. Identification of JmjC Domain-Containing UTX and JMJD3 as Histone H3 Lysine 27 Demethylases. Proc. Natl. Acad. Sci. USA 2007, 104, 18439–18444. [Google Scholar] [CrossRef]
- Södersten, E.; Feyder, M.; Lerdrup, M.; Gomes, A.L.; Kryh, H.; Spigolon, G.; Caboche, J.; Fisone, G.; Hansen, K. Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism. PLoS Genet. 2014, 10, e1004574. [Google Scholar] [CrossRef]
- Seong, I.S.; Woda, J.M.; Song, J.J.; Lloret, A.; Abeyrathne, P.D.; Woo, C.J.; Gregory, G.; Lee, J.M.; Wheeler, V.C.; Walz, T.; et al. Huntingtin Facilitates Polycomb Repressive Complex 2. Hum. Mol. Genet. 2010, 19, 573–583. [Google Scholar] [CrossRef]
- Pačesová, A.; Holubová, M.; Hrubá, L.; Strnadová, V.; Neprašová, B.; Pelantová, H.; Kuzma, M.; Železná, B.; Kuneš, J.; Maletínská, L. Age-Related Metabolic and Neurodegenerative Changes in SAMP8 Mice. Aging 2022, 14, 7300–7327. [Google Scholar] [CrossRef] [PubMed]
- Takeda, T. Senescence-Accelerated Mouse (SAM) with Special References to Neurodegeneration Models, SAMP8 and SAMP10 Mice. Neurochem. Res. 2009, 34, 639–659. [Google Scholar] [CrossRef] [PubMed]
- Ieraci, A.; Beggiato, S.; Ferraro, L.; Barbieri, S.S.; Popoli, M. Kynurenine Pathway Is Altered in BDNF Val66Met Knock-in Mice: Effect of Physical Exercise. Brain Behav. Immun. 2020, 89, 440–450. [Google Scholar] [CrossRef]
- Barbieri, S.S.; Sandrini, L.; Musazzi, L.; Popoli, M.; Ieraci, A. Apocynin Prevents Anxiety-Like Behavior and Histone Deacetylases Overexpression Induced by Sub-Chronic Stress in Mice. Biomolecules 2021, 11, 885. [Google Scholar] [CrossRef] [PubMed]
- Mallei, A.; Ieraci, A.; Corna, S.; Tardito, D.; Lee, F.S.; Popoli, M. Global Epigenetic Analysis of BDNF Val66Met Mice Hippocampus Reveals Changes in Dendrite and Spine Remodeling Genes. Hippocampus 2018, 28, 783–795. [Google Scholar] [CrossRef] [PubMed]




| Gene | Primer Forward | Primer Reverse | Size bp |
|---|---|---|---|
| Bdnf | TCGTTCCTTTCGAGTTAGCC | TTGGTAAACGGCACAAAAC | 97 |
| Gfap | GGCAGAAGCTCCAAGATGAAAC | TCCAGCGATTCAACCTTTCTCT | 127 |
| Iba1 | AGAGATCTGCCATCTTGAGAAT | TGACTCTGGCTCACGACTGTT | 195 |
| Utx | TGCCTTACCTGCAGCGAAA | ACCTTTGTGAAGCCCCTGAGT | 119 |
| Jmjd3 | TGGTTCACTTCGGCTCAACTTAG | TCTCGTCTGAGGGCTGCTGTA | 110 |
| Ezh2 | CAACCCTGTGACCATCCACG | CGACATCCAGGAAAGCGGTT | 122 |
| Eed | CGGGAGACGAAAATGACGAT | CTTTTCCTTCCTGGTGCATTTG | 100 |
| Suz12 | CCGGTGAAGAAGCCGAAAAA | TATTGGTGCGATAGATTTCGAGTT | 119 |
| Dnmt1 | GGACACAGGTGCCCGCGA | ATGAACCCCAGATGTTGACCA | 136 |
| Dnmt3a Gapdh | AGATCATGTACGTCGGGGAC CGTGCCGCCTGGAGAAACC | CAATCACCAGGTCGAATGGG CGTGCCGCCTGGAGAAACC | 78 144 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravanelli, F.; Musazzi, L.; Barbieri, S.S.; Rovati, G.; Popoli, M.; Barbon, A.; Ieraci, A. Differential Epigenetic Changes in the Dorsal Hippocampus of Male and Female SAMP8 Mice: A Preliminary Study. Int. J. Mol. Sci. 2023, 24, 13084. https://doi.org/10.3390/ijms241713084
Ravanelli F, Musazzi L, Barbieri SS, Rovati G, Popoli M, Barbon A, Ieraci A. Differential Epigenetic Changes in the Dorsal Hippocampus of Male and Female SAMP8 Mice: A Preliminary Study. International Journal of Molecular Sciences. 2023; 24(17):13084. https://doi.org/10.3390/ijms241713084
Chicago/Turabian StyleRavanelli, Federico, Laura Musazzi, Silvia Stella Barbieri, Gianenrico Rovati, Maurizio Popoli, Alessandro Barbon, and Alessandro Ieraci. 2023. "Differential Epigenetic Changes in the Dorsal Hippocampus of Male and Female SAMP8 Mice: A Preliminary Study" International Journal of Molecular Sciences 24, no. 17: 13084. https://doi.org/10.3390/ijms241713084
APA StyleRavanelli, F., Musazzi, L., Barbieri, S. S., Rovati, G., Popoli, M., Barbon, A., & Ieraci, A. (2023). Differential Epigenetic Changes in the Dorsal Hippocampus of Male and Female SAMP8 Mice: A Preliminary Study. International Journal of Molecular Sciences, 24(17), 13084. https://doi.org/10.3390/ijms241713084









