Abandoning the Isochore Theory Can Help Explain Genome Compositional Organization in Fish
Abstract
:1. Introduction
2. Results
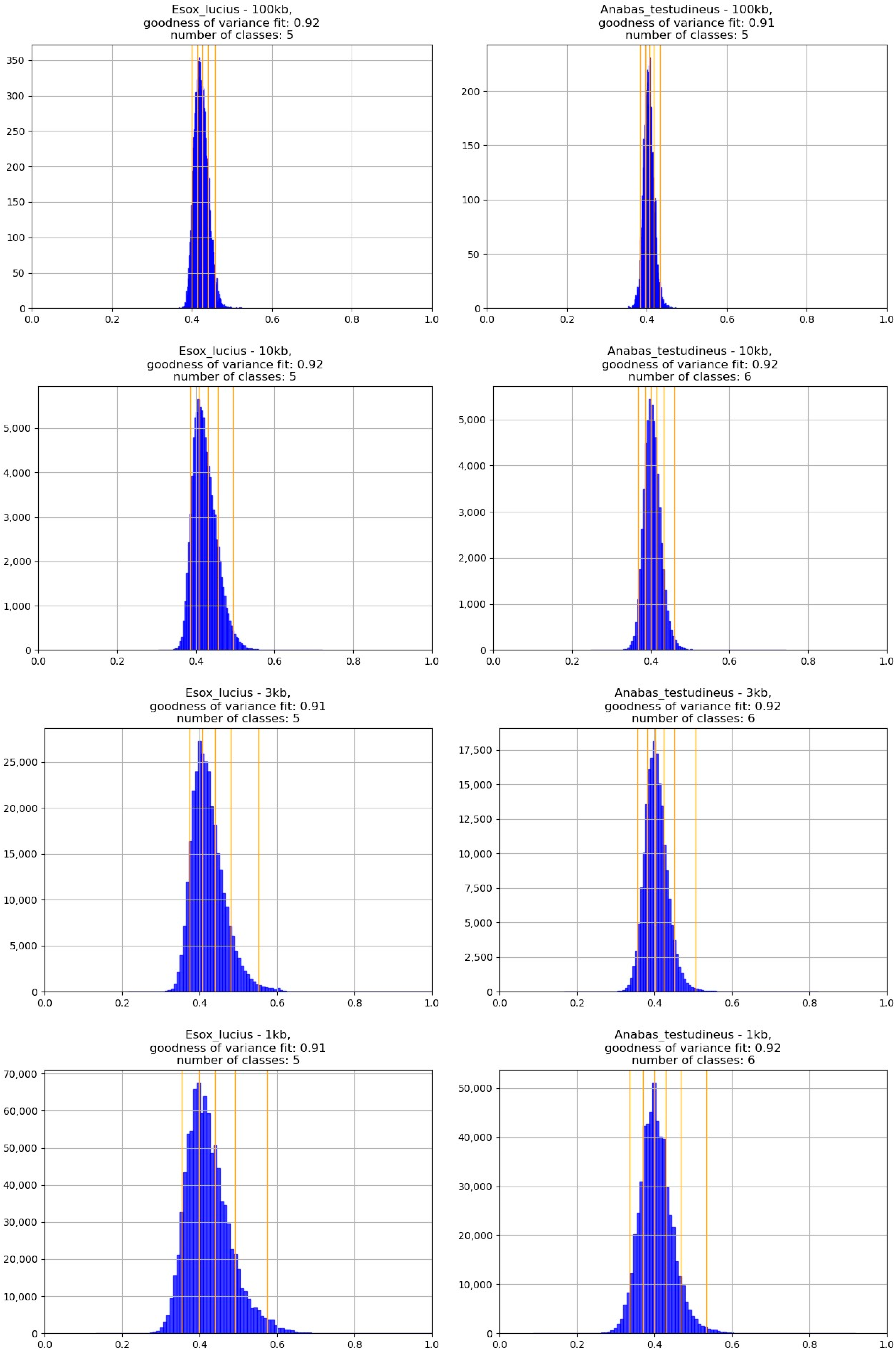
2.1. IsoSegmenter Results Are Inconsistent with Histograms of GC% under the Same Conditions
2.2. The Range of GC% Values Highly Varies with the Sliding Window Size
2.3. The Number of Natural GC% Clusters Varies with the Sliding Window Sequence Size
3. Discussion
3.1. Transposons as One of the Ways out of the Blind Alley of Isochores
3.2. When the Sequence Size Really Matters
4. Materials and Methods
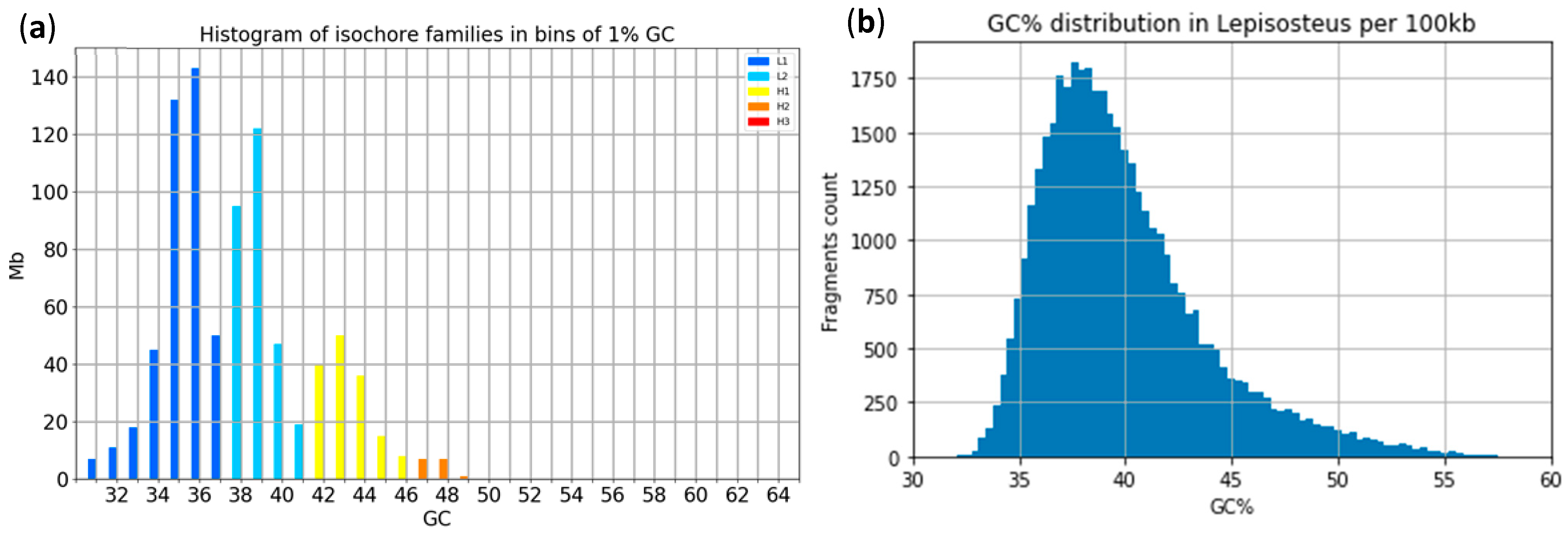
4.1. Isochore Families and GC% Histograms in the Spotted Gar (and in Other Species)
4.2. Fisher–Jenks’ Algorithm for Natural Breaks and the Goodness of Variance Fit (GVF)
4.3. Species Studied
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
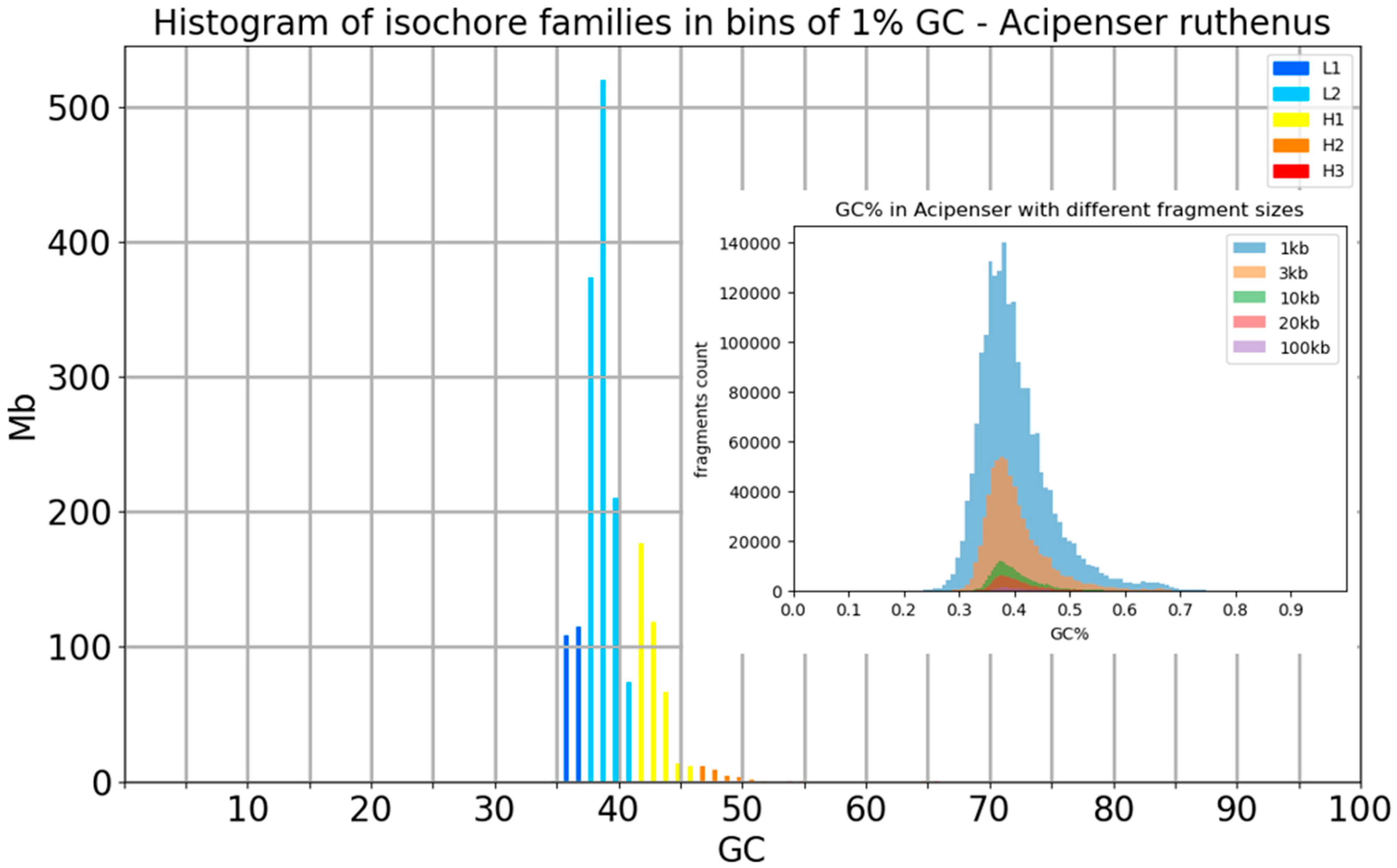
Appendix B
References
- Elhaik, E.; Graur, D. A Comparative Study and a Phylogenetic Exploration of the Compositional Architectures of Mammalian Nuclear Genomes. PLoS Comput. Biol. 2014, 10, e1003925. [Google Scholar] [CrossRef]
- Elhaik, E.; Graur, D.; Josić, K.; Landan, G. Identifying Compositionally Homogeneous and Nonhomogeneous Domains within the Human Genome Using a Novel Segmentation Algorithm. Nucleic Acids Res. 2010, 38, e158. [Google Scholar] [CrossRef]
- Bohlin, J.; Pettersson, J.H.-O. Evolution of Genomic Base Composition: From Single Cell Microbes to Multicellular Animals. Comput. Struct. Biotechnol. J. 2019, 17, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Pelleri, M.C.; Antonaros, F.; Strippoli, P.; Caracausi, M.; Vitale, L. On the Length, Weight and GC Content of the Human Genome. BMC Res. Notes 2019, 12, 106. [Google Scholar] [CrossRef]
- Symonová, R.; Majtánová, Z.; Arias-Rodriguez, L.; Mořkovský, L.; Kořínková, T.; Cavin, L.; Pokorná, M.J.; Doležálková, M.; Flajšhans, M.; Normandeau, E.; et al. Genome Compositional Organization in Gars Shows More Similarities to Mammals than to Other Ray-Finned Fish: Cytogenomics of Gars. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 607–619. [Google Scholar] [CrossRef]
- Bernardi, G. The Vertebrate Genome: Isochores and Evolution. Mol. Biol. Evol. 1993, 10, 186–204. [Google Scholar] [CrossRef]
- Bernardi, G.; Olofsson, B.; Filipski, J.; Zerial, M.; Salinas, J.; Cuny, G.; Meunier-Rotival, M.; Rodier, F. The Mosaic Genome of Warm-Blooded Vertebrates. Science 1985, 228, 953–958. [Google Scholar] [CrossRef]
- Macaya, G.; Thiery, J.-P.; Bernardi, G. An Approach to the Organization of Eukaryotic Genomes at a Macromolecular Level. J. Mol. Biol. 1976, 108, 237–254. [Google Scholar] [CrossRef]
- Cozzi, P.; Milanesi, L.; Bernardi, G. Segmenting the Human Genome into Isochores. Evol. Bioinform. Online 2015, 11, 253–261. [Google Scholar] [CrossRef]
- Bernardi, G. Structural and Evolutionary Genomics Natural Selection in Genome Evolution; Elsevier: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Costantini, M.; Auletta, F.; Bernardi, G. Isochore Patterns and Gene Distributions in Fish Genomes. Genomics 2007, 90, 364–371. [Google Scholar] [CrossRef]
- Cammarano, R.; Costantini, M.; Bernardi, G. The Isochore Patterns of Invertebrate Genomes. BMC Genom. 2009, 10, 538. [Google Scholar] [CrossRef]
- Costantini, M.; Filippo, M.D.; Auletta, F.; Bernardi, G. Isochore Pattern and Gene Distribution in the Chicken Genome. Gene 2007, 400, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.-P.; Macaya, G.; Bernardi, G. An Analysis of Eukaryotic Genomes by Density Gradient Centrifugation. J. Mol. Biol. 1976, 108, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Vizard, D.L.; Rinehart, F.P.; Rubin, C.M.; Schmid, C.W. Intramolecular Base Composition Heterogeneity of Human DNA. Nucleic Acids Res. 1977, 4, 3753–3768. [Google Scholar] [CrossRef] [PubMed]
- Corneo, G.; Nelli, L.C.; Meazza, D.; Ginelli, E. Repeated Nucleotide Sequences in Human Main Band DNA. Biochim. Et Biophys. Acta (BBA) Nucleic Acids Protein Synth. 1980, 607, 438–444. [Google Scholar] [CrossRef]
- Graur, D. Slaying (Yet Again) the Brain-Eating Zombie Called the “Isochore Theory”: A Segmentation Algorithm Used to “Confirm” the Existence of Isochores Creates “Isochores” Where None Exist. Int. J. Mol. Sci. 2022, 23, 6558. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.; Dagan, T.; Stone, L.; Graur, D. GC Composition of the Human Genome: In Search of Isochores. Mol. Biol. Evol. 2005, 22, 1260–1272. [Google Scholar] [CrossRef]
- Elhaik, E. Compositional domains in fishes. Personal communication via emails, 2022. [Google Scholar]
- Elhaik, E.; Landan, G.; Graur, D. Can GC Content at Third-Codon Positions Be Used as a Proxy for Isochore Composition? Mol. Biol. Evol. 2009, 26, 1829–1833. [Google Scholar] [CrossRef]
- Honeybee Genome Sequencing Consortium. The Honeybee Genome Sequencing Consortium Insights into Social Insects from the Genome of the Honeybee Apis Mellifera. Nature 2006, 443, 931–949. [Google Scholar] [CrossRef]
- The Bovine Genome Sequencing and Analysis Consortium; Elsik, C.G.; Tellam, R.L.; Worley, K.C.; Gibbs, R.A.; Muzny, D.M.; Weinstock, G.M.; Adelson, D.L.; Eichler, E.E.; Elnitski, L.; et al. The Genome Sequence of Taurine Cattle: A Window to Ruminant Biology and Evolution. Science 2009, 324, 522–528. [Google Scholar] [CrossRef]
- Kirkness, E.F.; Haas, B.J.; Sun, W.; Braig, H.R.; Perotti, M.A.; Clark, J.M.; Lee, S.H.; Robertson, H.M.; Kennedy, R.C.; Elhaik, E.; et al. Genome Sequences of the Human Body Louse and Its Primary Endosymbiont Provide Insights into the Permanent Parasitic Lifestyle. Proc. Natl. Acad. Sci. USA 2010, 107, 12168–12173. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, G.; Hughes, S.; Mouchiroud, D. The Major Compositional Transitions in the Vertebrate Genome. J. Mol. Evol. 1997, 44, S44–S51. [Google Scholar] [CrossRef] [PubMed]
- Cruveiller, S.; D’Onofrio, G.; Bernardi, G. The Compositional Transition between the Genomes of Cold- and Warm-Blooded Vertebrates: Codon Frequencies in Orthologous Genes. Gene 2000, 261, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, G. The Neoselectionist Theory of Genome Evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 8385–8390. [Google Scholar] [CrossRef] [PubMed]
- Matoulek, D.; Ježek, B.; Vohnoutová, M.; Symonová, R. Advances in Vertebrate (Cyto)Genomics Shed New Light on Fish Compositional Genome Evolution. Genes 2023, 14, 244. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, G. Misunderstandings about Isochores. Part 1. Gene 2001, 276, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Clay, O.; Bernardi, G. How Not to Search for Isochores: A Reply to Cohen et Al. Mol. Biol. Evol. 2005, 22, 2315–2317. [Google Scholar] [CrossRef]
- Costantini, M.; Greif, G.; Alvarez-Valin, F.; Bernardi, G. The Anolis Lizard Genome: An Amniote Genome without Isochores? Genome Biol. Evol. 2016, 8, 1048–1055. [Google Scholar] [CrossRef]
- Fujita, M.K.; Edwards, S.V.; Ponting, C.P. The Anolis Lizard Genome: An Amniote Genome without Isochores. Genome Biol. Evol. 2011, 3, 974–984. [Google Scholar] [CrossRef]
- Arhondakis, S.; Milanesi, M.; Castrignanò, T.; Gioiosa, S.; Valentini, A.; Chillemi, G. Evidence of Distinct Gene Functional Patterns in GC-poor and GC-rich Isochores in Bos Taurus. Anim. Genet. 2020, 51, 358–368. [Google Scholar] [CrossRef]
- Majtánová, Z.; Symonová, R.; Arias-Rodriguez, L.; Sallan, L.; Ráb, P. “Holostei versus Halecostomi” Problem: Insight from Cytogenetics of Ancient Nonteleost Actinopterygian Fish, Bowfin Amia Calva: Molecular Cytogenetics of Amia Calva. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The Spotted Gar Genome Illuminates Vertebrate Evolution and Facilitates Human-Teleost Comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.R. Animal Genome Size Database. 2022. Available online: https://www.genomesize.com/ (accessed on 16 August 2023).
- Kim, J.; Lee, C.; Ko, B.J.; Yoo, D.A.; Won, S.; Phillippy, A.M.; Fedrigo, O.; Zhang, G.; Howe, K.; Wood, J.; et al. False Gene and Chromosome Losses in Genome Assemblies Caused by GC Content Variation and Repeats. Genome Biol. 2022, 23, 204. [Google Scholar] [CrossRef] [PubMed]
- Borůvková, V.; Howell, W.M.; Matoulek, D.; Symonová, R. Quantitative Approach to Fish Cytogenetics in the Context of Vertebrate Genome Evolution. Genes 2021, 12, 312. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, J.; Cabau, C.; Nguyen, T.; Jouanno, E.; Severac, D.; Braasch, I.; Journot, L.; Pontarotti, P.; Klopp, C.; Postlethwait, J.H.; et al. Gene Evolution and Gene Expression after Whole Genome Duplication in Fish: The PhyloFish Database. BMC Genom. 2016, 17, 368. [Google Scholar] [CrossRef] [PubMed]
- NCBI. National Library of Medicine (US) NCBI/Genome; NCBI: Bethesda, MD, USA, 2004. [Google Scholar]
- Wilcox, J.J.S.; Arca-Ruibal, B.; Samour, J.; Mateuta, V.; Idaghdour, Y.; Boissinot, S. Linked-Read Sequencing of Eight Falcons Reveals a Unique Genomic Architecture in Flux. Genome Biol. Evol. 2022, 14, evac090. [Google Scholar] [CrossRef] [PubMed]
- Ayad, L.A.K.; Dourou, A.-M.; Arhondakis, S.; Pissis, S.P. IsoXpressor: A Tool to Assess Transcriptional Activity within Isochores. Genome Biol. Evol. 2020, 12, 1573–1578. [Google Scholar] [CrossRef]
- Thibaut, Y.; Tang, N.; Tran, H.N.; Vaurijoux, A.; Villagrasa, C.; Incerti, S.; Perrot, Y. Nanodosimetric Calculations of Radiation-Induced DNA Damage in a New Nucleus Geometrical Model Based on the Isochore Theory. Int. J. Mol. Sci. 2022, 23, 3770. [Google Scholar] [CrossRef]
- Mugal, C.F.; Weber, C.C.; Ellegren, H. GC-Biased Gene Conversion Links the Recombination Landscape and Demography to Genomic Base Composition: GC-Biased Gene Conversion Drives Genomic Base Composition across a Wide Range of Species. BioEssays 2015, 37, 1317–1326. [Google Scholar] [CrossRef]
- Ng, S.B.; Turner, E.H.; Robertson, P.D.; Flygare, S.D.; Bigham, A.W.; Lee, C.; Shaffer, T.; Wong, M.; Bhattacharjee, A.; Eichler, E.E.; et al. Targeted Capture and Massively Parallel Sequencing of 12 Human Exomes. Nature 2009, 461, 272–276. [Google Scholar] [CrossRef]
- Symonová, R.; Suh, A. Nucleotide Composition of Transposable Elements Likely Contributes to AT/GC Compositional Homogeneity of Teleost Fish Genomes. Mob. DNA 2019, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Matoulek, D.; Borůvková, V.; Ocalewicz, K.; Symonová, R. GC and Repeats Profiling along Chromosomes—The Future of Fish Compositional Cytogenomics. Genes 2020, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.W.; Hawkins, M.B.; Parey, E.; Wcisel, D.J.; Ota, T.; Kawasaki, K.; Funk, E.; Losilla, M.; Fitch, O.E.; Pan, Q.; et al. The Bowfin Genome Illuminates the Developmental Evolution of Ray-Finned Fishes. Nat. Genet. 2021, 53, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and Diversity of Transposable Elements in Vertebrate Genomes. Genome Biol. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef]
- Vohnoutová, M.; Žifčáková, L.; Symonová, R. Hidden Compositional Heterogeneity of Fish Chromosomes in the Era of Polished Genome Assemblies. Fishes 2023, 8, 185. [Google Scholar] [CrossRef]
- Knytl, M.; Kalous, L.; Symonová, R.; Rylková, K.; Ráb, P. Chromosome Studies of European Cyprinid Fishes: Cross-Species Painting Reveals Natural Allotetraploid Origin of a Carassius Female with 206 Chromosomes. Cytogenet. Genome Res 2013, 139, 276–283. [Google Scholar] [CrossRef]
- Knytl, M.; Fornaini, N. Measurement of Chromosomal Arms and FISH Reveal Complex Genome Architecture and Standardized Karyotype of Model Fish, Genus Carassius. Cells 2021, 10, 2343. [Google Scholar] [CrossRef]
- Knytl, M.; Kalous, L.; Rab, P. Karyotype and Chromosome Banding of Endangered Crucian Carp, Carassius carassius (Linnaeus, 1758) (Teleostei, Cyprinidae). Comp. Cytogenet. 2013, 7, 205–213. [Google Scholar] [CrossRef]
- Bertollo, L.A.C.; Fontes, M.S.; Fenocchio, A.S.; Cano, J. The X1X2Y Sex Chromosome System in the Fish Hoplias malabaricus. I. G-, C- and Chromosome Replication Banding. Chromosome Res. 1997, 5, 493–499. [Google Scholar] [CrossRef]
- Gold, J.R.; Li, Y.C. Trypsin G-Banding of North American Cyprinid Chromosomes: Phylogenetic Considerations, Implications for Fish Chromosome Structure, and Chromosomal Polymorphism. Cytologia 1991, 56, 199–208. [Google Scholar] [CrossRef]
- Medrano, L.; Bernardi, G.; Couturier, J.; Dutrillaux, B.; Bernardi, G. Chromosome Banding and Genome Compartmentalization in Fishes. Chromosoma 1988, 96, 178–183. [Google Scholar] [CrossRef]
- Wiberg, U.H. Sex Determination in the European Eel (Anguilla anguilla, L.). Cytogenet Genome Res. 1983, 36, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Gaffaroglu, M.; Majtánová, Z.; Symonová, R.; Pelikánová, Š.; Unal, S.; Lajbner, Z.; Ráb, P. Present and Future Salmonid Cytogenetics. Genes 2020, 11, 1462. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Wang, K.; Yang, L.; Pan, H.; Jiang, H.; Wei, Q.; Fang, M.; Yu, H.; Zhu, C.; Cai, Y.; et al. Tracing the Genetic Footprints of Vertebrate Landing in Non-Teleost Ray-Finned Fishes. Cell 2021, 184, 1377–1391.e14. [Google Scholar] [CrossRef]
- Fisher, W.D. On Grouping for Maximum Homogeneity. J. Am. Stat. Assoc. 1958, 53, 789–798. [Google Scholar] [CrossRef]
- Coulson, M.R.C. In The Matter of Class Intervals for Choropleth Maps: With Particular Reference to the Work of George F Jenks. Cartogr. Int. J. Geogr. Inf. Geovisualiz. 1987, 24, 16–39. [Google Scholar] [CrossRef]

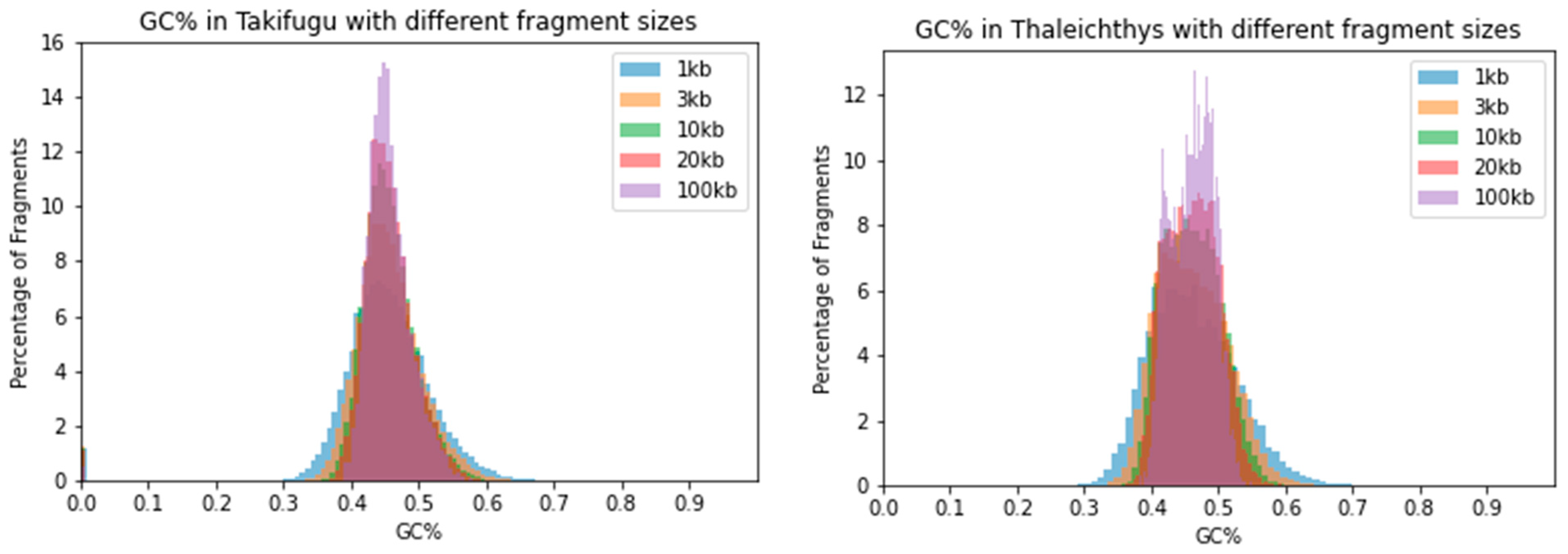
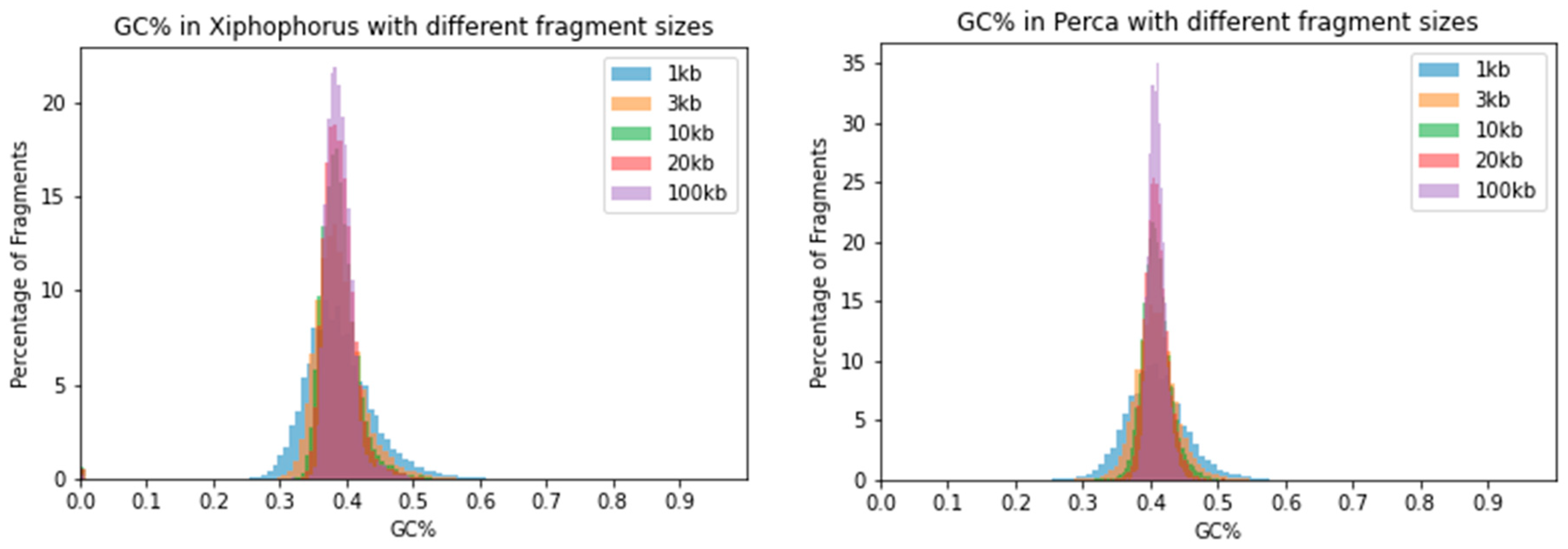
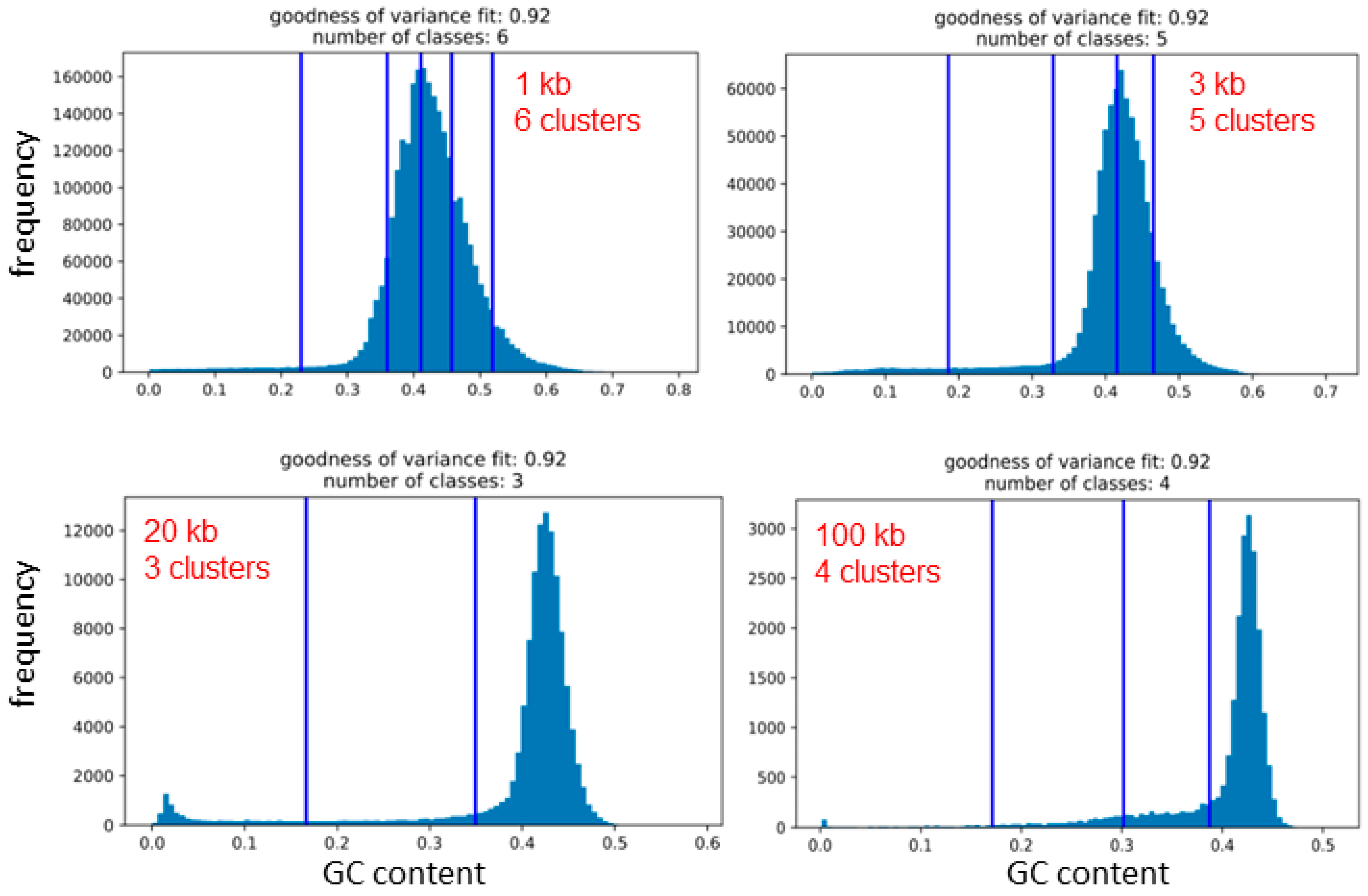
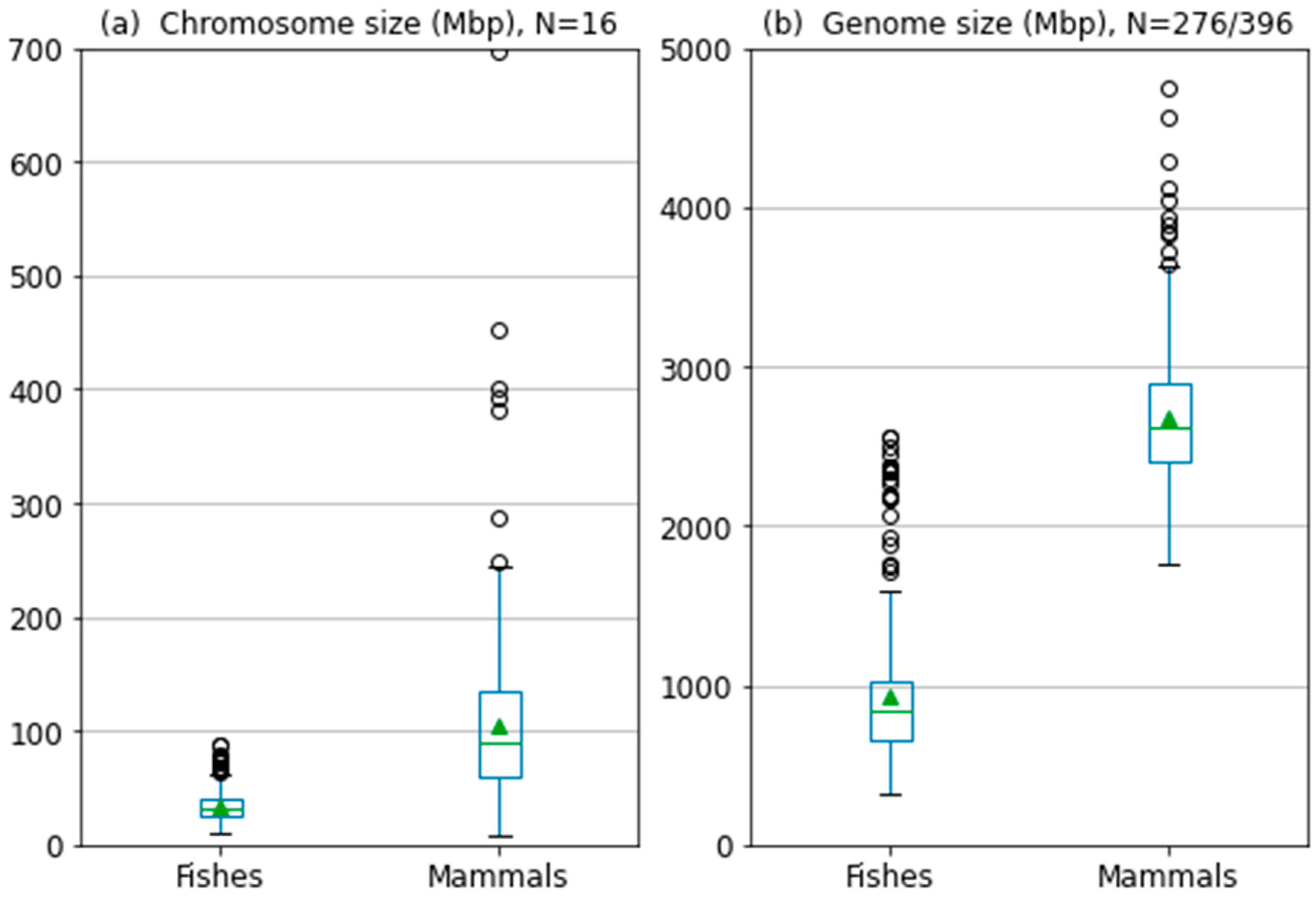
| Species | 1 kb | 3 kb | 10 kb | 20 kb | 100 kb |
|---|---|---|---|---|---|
| Acipenser ruthenus | 5 | 5 | 5 | 6 | 5 |
| Amblyraja radiata | 6 | 6 | 6 | 6 | 4 |
| Amia calva | 5 | 5 | 5 | 5 | 5 |
| Anguilla anguilla | 5 | 5 | 5 | 4 | 4 |
| Atractosteus spatula | 5 | 6 | 6 | 6 | 5 |
| Branchiostoma floridae | 6 | 6 | 5 | 5 | 5 |
| Erpetoichthys calabaricus | 6 | 6 | 6 | 6 | 5 |
| Esox lucius | 5 | 5 | 5 | 6 | 6 |
| Lethenteron reissneri | 5 | 6 | 5 | 5 | 5 |
| Perca fluviatilis | 6 | 6 | 7 | 7 | 6 |
| Salmo salar | 6 | 5 | 4 | 3 | 4 |
| Xiphophorus maculatus | 5 | 6 | 6 | 6 | 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vohnoutová, M.; Sedláková, A.; Symonová, R. Abandoning the Isochore Theory Can Help Explain Genome Compositional Organization in Fish. Int. J. Mol. Sci. 2023, 24, 13167. https://doi.org/10.3390/ijms241713167
Vohnoutová M, Sedláková A, Symonová R. Abandoning the Isochore Theory Can Help Explain Genome Compositional Organization in Fish. International Journal of Molecular Sciences. 2023; 24(17):13167. https://doi.org/10.3390/ijms241713167
Chicago/Turabian StyleVohnoutová, Marta, Anastázie Sedláková, and Radka Symonová. 2023. "Abandoning the Isochore Theory Can Help Explain Genome Compositional Organization in Fish" International Journal of Molecular Sciences 24, no. 17: 13167. https://doi.org/10.3390/ijms241713167
APA StyleVohnoutová, M., Sedláková, A., & Symonová, R. (2023). Abandoning the Isochore Theory Can Help Explain Genome Compositional Organization in Fish. International Journal of Molecular Sciences, 24(17), 13167. https://doi.org/10.3390/ijms241713167







