Circulating microRNAs as Potential Biomarkers in Pancreatic Cancer—Advances and Challenges
Abstract
:1. Introduction
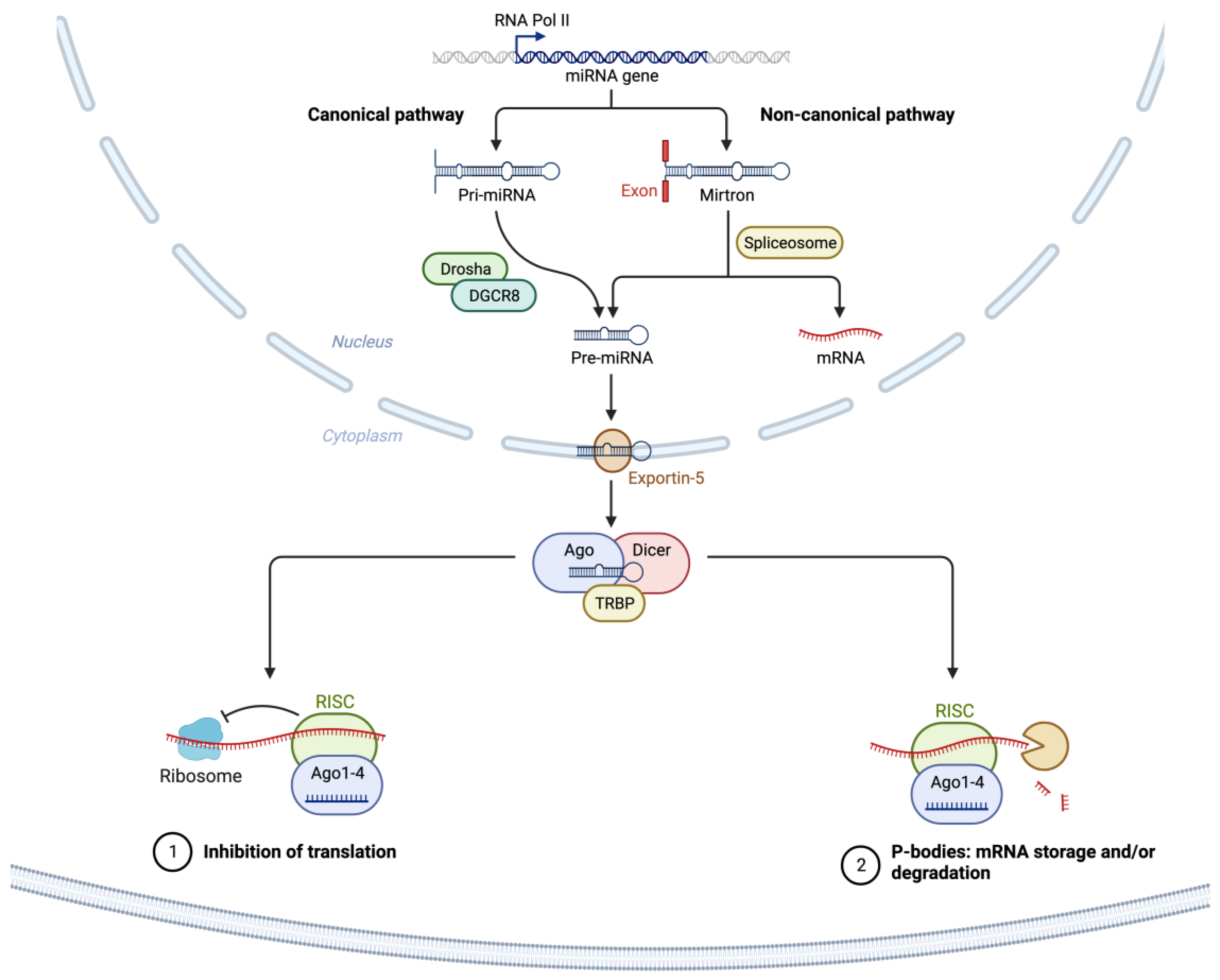
2. microRNAs
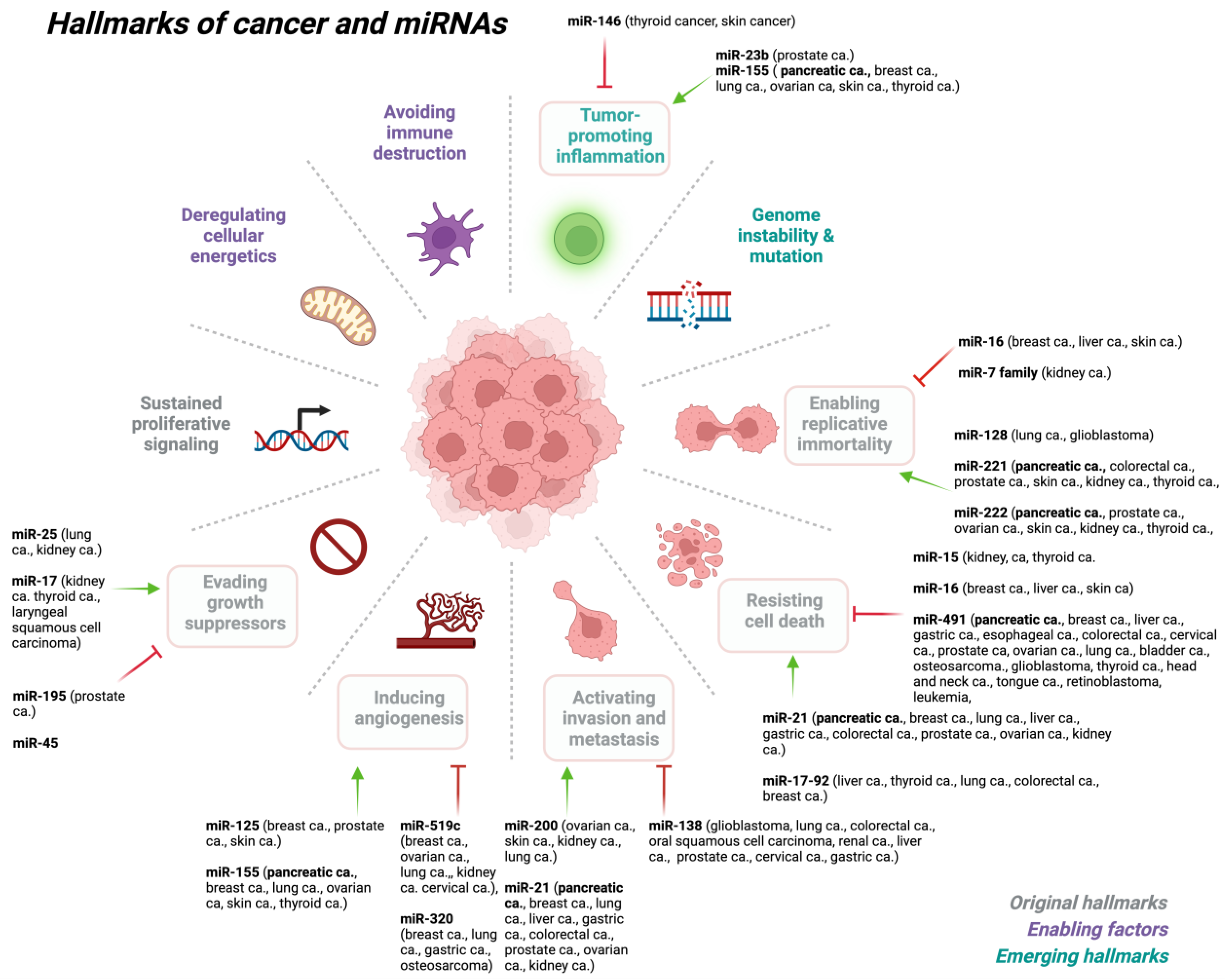
3. miRNA Dysregulation in Cancer
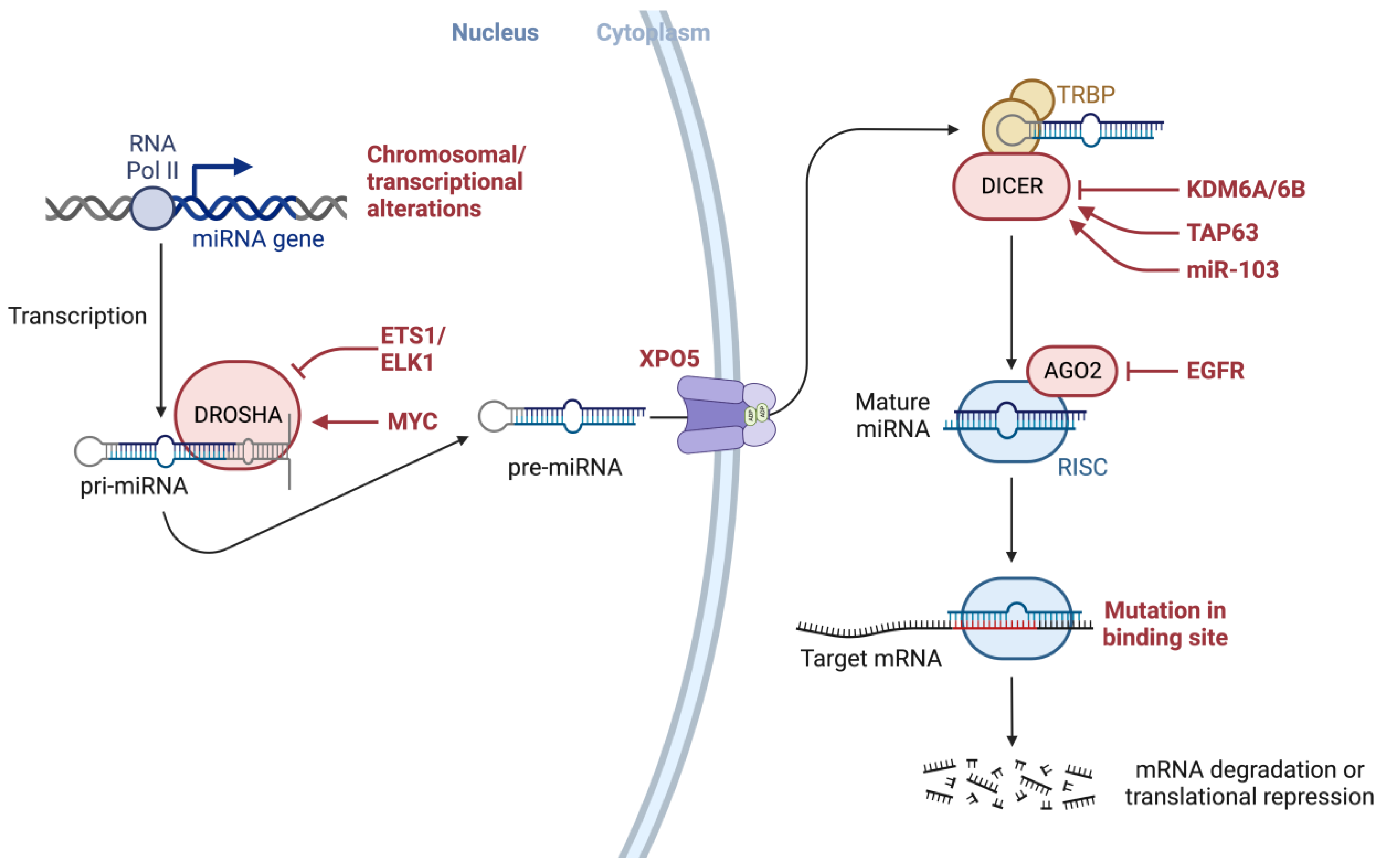
4. Oncogenic Mutations in Key Factors Involved in the miRNA Biogenesis
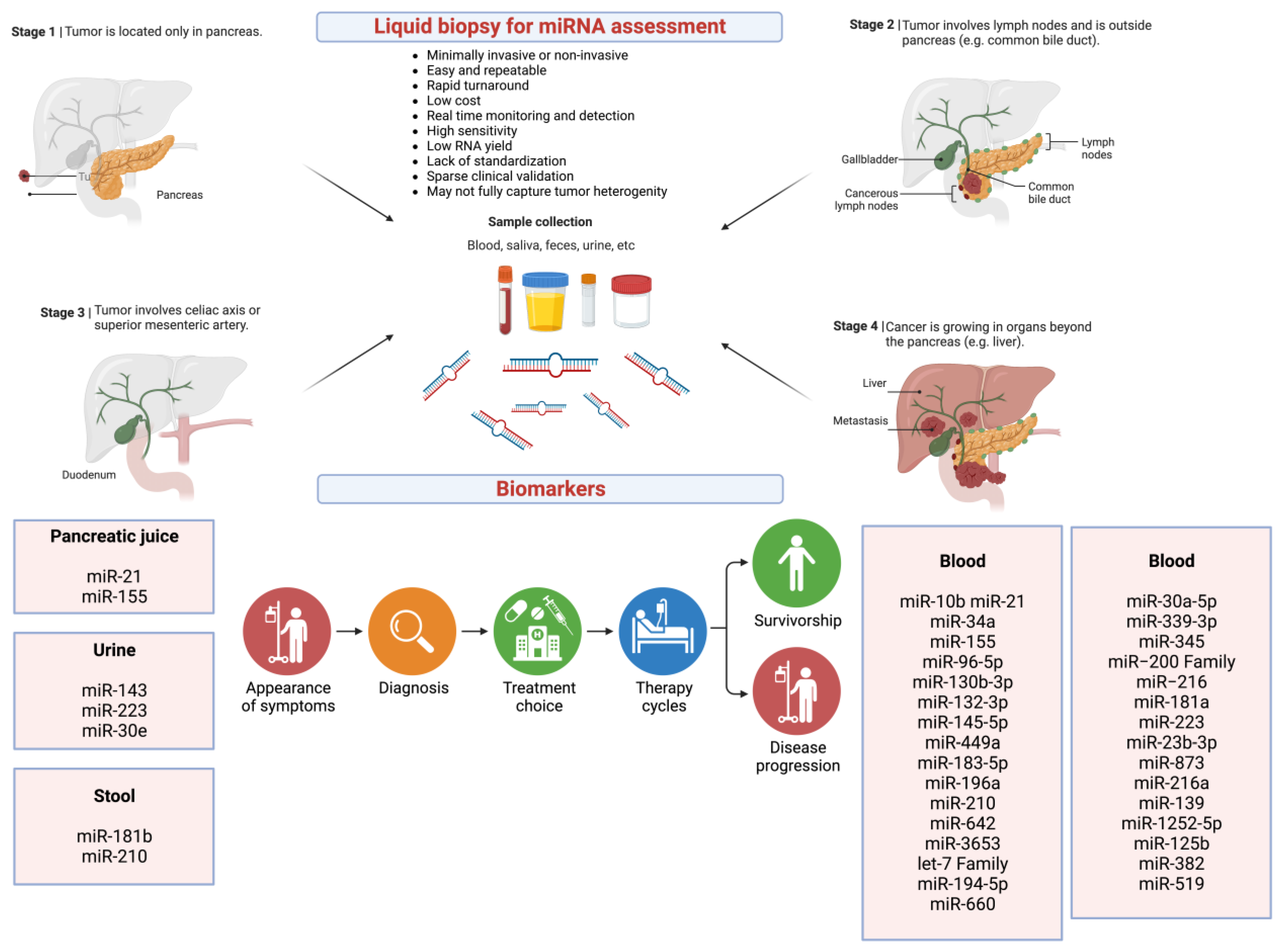
5. Liquid Biopsy in Pancreatic Cancer
6. miRNAs as Potential Biomarkers
7. Circulating miRNAs Detected in Different Body Fluids
8. Circulating miRNAs as Potential Biomarkers in Pancreatic Cancer
9. Challenges Involving miRNAs as Biomarkers in Clinical Applications
9.1. Heterogeneity of Methodologies, miRNA Source Specimens, and Lack of Controls
9.2. Sample Source and Physiological Variations and Low RNA Input
9.3. Methodological and Technical Variations in Detecting miRNAs
9.4. Variations in Data Analysis and Bioinformatics Tools in the Analysis of miRNAs
9.5. Poor Experimental Design in Clinical Studies
10. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singhi, A.D.; Koay, E.J.; Chari, S.T.; Maitra, A. Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology 2019, 156, 2024–2040. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Advancing on pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 447. [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Raufi, A.G.; May, M.S.; Hadfield, M.J.; Seyhan, A.A.; El-Deiry, W.S. Advances in Liquid Biopsy Technology and Implications for Pancreatic Cancer. Int. J. Mol. Sci. 2023, 24, 4238. [Google Scholar] [CrossRef]
- SEER Cancer Statistics. Available online: https://seer.cancer.gov (accessed on 22 June 2022).
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.L.; Chone, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Moletta, L.; Serafini, S.; Valmasoni, M.; Pierobon, E.S.; Ponzoni, A.; Sperti, C. Surgery for Recurrent Pancreatic Cancer: Is It Effective? Cancers 2019, 11, 991. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Goldstein, D.; El-Maraghi, R.H.; Hammel, P.; Heinemann, V.; Kunzmann, V.; Sastre, J.; Scheithauer, W.; Siena, S.; Tabernero, J.; Teixeira, L.; et al. nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: Long-term survival from a phase III trial. J. Natl. Cancer Inst. 2015, 107, dju413. [Google Scholar] [CrossRef]
- Chhoda, A.; Lu, L.; Clerkin, B.M.; Risch, H.; Farrell, J.J. Current Approaches to Pancreatic Cancer Screening. Am. J. Pathol. 2019, 189, 22–35. [Google Scholar] [CrossRef]
- Luo, G.; Jin, K.; Deng, S.; Cheng, H.; Fan, Z.; Gong, Y.; Qian, Y.; Huang, Q.; Ni, Q.; Liu, C.; et al. Roles of CA19-9 in pancreatic cancer: Biomarker, predictor and promoter. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188409. [Google Scholar] [PubMed]
- Ni, X.G.; Bai, X.F.; Mao, Y.L.; Shao, Y.F.; Wu, J.X.; Shan, Y.; Wang, C.F.; Wang, J.; Tian, Y.T.; Liu, Q.; et al. The clinical value of serum CEA, CA19-9, and CA242 in the diagnosis and prognosis of pancreatic cancer. Eur. J. Surg. Oncol. 2005, 31, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Hogendorf, P. The Role of microRNA in Pancreatic Cancer. Biomedicines 2021, 9, 1322. [Google Scholar] [CrossRef]
- Macconaill, L.E.; Garraway, L.A. Clinical implications of the cancer genome. J. Clin. Oncol. 2010, 28, 5219–5228. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Rands, C.M.; Meader, S.; Ponting, C.P.; Lunter, G. 8.2% of the Human genome is constrained: Variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 2014, 10, e1004525. [Google Scholar] [CrossRef]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Halldorsson, B.V.; Eggertsson, H.P.; Moore, K.H.S.; Hauswedell, H.; Eiriksson, O.; Ulfarsson, M.O.; Palsson, G.; Hardarson, M.T.; Oddsson, A.; Jensson, B.O.; et al. The sequences of 150,119 genomes in the UK Biobank. Nature 2022, 607, 732–740. [Google Scholar]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar]
- Li, Y.; Kowdley, K.V. MicroRNAs in Common Human Diseases. Genom. Proteom. Bioinform. 2012, 10, 246–253. [Google Scholar] [CrossRef]
- De Guire, V.; Robitaille, R.; Tetreault, N.; Guerin, R.; Menard, C.; Bambace, N.; Sapieha, P. Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: Promises and challenges. Clin. Biochem. 2013, 46, 846–860. [Google Scholar]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar]
- Gareev, I.; de Jesus Encarnacion Ramirez, M.; Goncharov, E.; Ivliev, D.; Shumadalova, A.; Ilyasova, T.; Wang, C. MiRNAs and lncRNAs in the regulation of innate immune signaling. Noncoding RNA Res. 2023, 8, 534–541. [Google Scholar] [PubMed]
- Pauley, K.M.; Cha, S.; Chan, E.K. MicroRNA in autoimmunity and autoimmune diseases. J. Autoimmun. 2009, 32, 189–194. [Google Scholar] [PubMed]
- Sullivan, C.S.; Ganem, D. MicroRNAs and viral infection. Mol. Cell 2005, 20, 3–7. [Google Scholar]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar]
- Barbu, M.G.; Condrat, C.E.; Thompson, D.C.; Bugnar, O.L.; Cretoiu, D.; Toader, O.D.; Suciu, N.; Voinea, S.C. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front. Cell Dev. Biol. 2020, 8, 143. [Google Scholar] [PubMed]
- Abu-Izneid, T.; AlHajri, N.; Ibrahim, A.M.; Javed, M.N.; Salem, K.M.; Pottoo, F.H.; Kamal, M.A. Micro-RNAs in the regulation of immune response against SARS CoV-2 and other viral infections. J. Adv. Res. 2021, 30, 133–145. [Google Scholar]
- Elfimova, N.; Schlattjan, M.; Sowa, J.P.; Dienes, H.P.; Canbay, A.; Odenthal, M. Circulating microRNAs: Promising candidates serving as novel biomarkers of acute hepatitis. Front. Physiol. 2012, 3, 476. [Google Scholar] [PubMed]
- Li, Y.J.; Xu, M.; Gao, Z.H.; Wang, Y.Q.; Yue, Z.; Zhang, Y.X.; Li, X.X.; Zhang, C.; Xie, S.Y.; Wang, P.Y. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS ONE 2013, 8, e63648. [Google Scholar]
- Scott, K.A.; Hoban, A.E.; Clarke, G.; Moloney, G.M.; Dinan, T.G.; Cryan, J.F. Thinking small: Towards microRNA-based therapeutics for anxiety disorders. Expert. Opin. Investig. Drugs 2015, 24, 529–542. [Google Scholar]
- Liu, W.; Liu, C.; Zhu, J.; Shu, P.; Yin, B.; Gong, Y.; Qiang, B.; Yuan, J.; Peng, X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 2012, 33, 522–534. [Google Scholar]
- Weir, D.W.; Sturrock, A.; Leavitt, B.R. Development of biomarkers for Huntington’s disease. Lancet Neurol. 2011, 10, 573–590. [Google Scholar]
- Wang, R.; Li, N.; Zhang, Y.; Ran, Y.; Pu, J. Circulating microRNAs are promising novel biomarkers of acute myocardial infarction. Intern. Med. 2011, 50, 1789–1795. [Google Scholar]
- Recchioni, R.; Marcheselli, F.; Olivieri, F.; Ricci, S.; Procopio, A.D.; Antonicelli, R. Conventional and novel diagnostic biomarkers of acute myocardial infarction: A promising role for circulating microRNAs. Biomarkers 2013, 18, 547–558. [Google Scholar]
- Nunez Lopez, Y.O.; Coen, P.M.; Goodpaster, B.H.; Seyhan, A.A. Gastric bypass surgery with exercise alters plasma microRNAs that predict improvements in cardiometabolic risk. Int. J. Obes. 2017, 41, 1121–1130. [Google Scholar]
- Nunez Lopez, Y.O.; Garufi, G.; Pasarica, M.; Seyhan, A.A. Elevated and Correlated Expressions of miR-24, miR-30d, miR-146a, and SFRP-4 in Human Abdominal Adipose Tissue Play a Role in Adiposity and Insulin Resistance. Int. J. Endocrinol. 2018, 2018, 7351902. [Google Scholar] [PubMed]
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008, 22, 4126–4135. [Google Scholar] [PubMed]
- Gambardella, S.; Rinaldi, F.; Lepore, S.M.; Viola, A.; Loro, E.; Angelini, C.; Vergani, L.; Novelli, G.; Botta, A. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J. Transl. Med. 2010, 8, 48. [Google Scholar] [PubMed]
- Seyhan, A.A. microRNAs with different functions and roles in disease development and as potential biomarkers of diabetes: Progress and challenges. Mol. Biosyst. 2015, 11, 1217–1234. [Google Scholar] [PubMed]
- Seyhan, A.A.; Nunez Lopez, Y.O.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci. Rep. 2016, 6, 31479. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Garufi, G.; Seyhan, A.A. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst. 2016, 13, 106–121. [Google Scholar] [CrossRef] [PubMed]
- Nunez Lopez, Y.O.; Pittas, A.G.; Pratley, R.E.; Seyhan, A.A. Circulating levels of miR-7, miR-152 and miR-192 respond to vitamin D supplementation in adults with prediabetes and correlate with improvements in glycemic control. J. Nutr. Biochem. 2017, 49, 117–122. [Google Scholar] [CrossRef]
- Kluiver, J.; Poppema, S.; de Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Kroesen, B.J.; van den Berg, A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005, 207, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Ventura, A.; Jacks, T. MicroRNAs and cancer: Short RNAs go a long way. Cell 2009, 136, 586–591. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Lawler, S.; Chiocca, E.A. Emerging functions of microRNAs in glioblastoma. J. Neurooncol. 2009, 92, 297–306. [Google Scholar] [CrossRef]
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. Epigenetic alteration and microRNA dysregulation in cancer. Front. Genet. 2013, 4, 258. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Graveel, C.R.; Calderone, H.M.; Westerhuis, J.J.; Winn, M.E.; Sempere, L.F. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer 2015, 7, 59–79. [Google Scholar] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Lulla, A.R.; Slifker, M.J.; Zhou, Y.; Lev, A.; Einarson, M.B.; Dicker, D.T.; El-Deiry, W.S. miR-6883 Family miRNAs Target CDK4/6 to Induce G(1) Phase Cell-Cycle Arrest in Colon Cancer Cells. Cancer Res. 2017, 77, 6902–6913. [Google Scholar] [CrossRef]
- Biswas, S. MicroRNAs as Therapeutic Agents: The Future of the Battle against Cancer. Curr. Top. Med. Chem. 2018, 18, 2544–2554. [Google Scholar] [CrossRef]
- Hu, W.; Tan, C.; He, Y.; Zhang, G.; Xu, Y.; Tang, J. Functional miRNAs in breast cancer drug resistance. Oncotargets Ther. 2018, 11, 1529–1541. [Google Scholar] [CrossRef]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive Analysis of Human microRNA-mRNA Interactome. Front. Genet. 2019, 10, 933. [Google Scholar] [CrossRef]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Annese, T.; Tamma, R.; De Giorgis, M.; Ribatti, D. microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 2020, 10, 581007. [Google Scholar] [CrossRef]
- Fathi, M.; Ghafouri-Fard, S.; Abak, A.; Taheri, M. Emerging roles of miRNAs in the development of pancreatic cancer. Biomed. Pharmacother. 2021, 141, 111914. [Google Scholar] [CrossRef] [PubMed]
- Galka-Marciniak, P.; Urbanek-Trzeciak, M.O.; Nawrocka, P.M.; Kozlowski, P. A pan-cancer atlas of somatic mutations in miRNA biogenesis genes. Nucleic Acids Res. 2021, 49, 601–620. [Google Scholar] [CrossRef] [PubMed]
- Pajares, M.J.; Alemany-Cosme, E.; Goni, S.; Bandres, E.; Palanca-Ballester, C.; Sandoval, J. Epigenetic Regulation of microRNAs in Cancer: Shortening the Distance from Bench to Bedside. Int. J. Mol. Sci. 2021, 22, 7350. [Google Scholar] [CrossRef]
- Inoue, J.; Inazawa, J. Cancer-associated miRNAs and their therapeutic potential. J. Hum. Genet. 2021, 66, 937–945. [Google Scholar] [CrossRef]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Szyłło, K.; Hogendorf, P. miRNAs in Cancer (Review of Literature). Int. J. Mol. Sci. 2022, 23, 2805. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, Q.; Qian, J.; Li, M.; Gu, C.; Yang, Y. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol. Ther. 2022, 234, 108123. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Ding, J.; Cao, Y.; Qi, C.; Zong, Z. Dysregulated microRNAs participate in the crosstalk between colorectal cancer and atrial fibrillation. Hum. Cell 2023, 36, 1336–1342. [Google Scholar] [CrossRef]
- Ricarte-Filho, J.C.; Casado-Medrano, V.; Reichenberger, E.; Spangler, Z.; Scheerer, M.; Isaza, A.; Baran, J.; Patel, T.; MacFarland, S.P.; Brodeur, G.M.; et al. DICER1 RNase IIIb domain mutations trigger widespread miRNA dysregulation and MAPK activation in pediatric thyroid cancer. Front. Endocrinol. 2023, 14, 1083382. [Google Scholar] [CrossRef]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- Hata, A.; Lieberman, J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal. 2015, 8, re3. [Google Scholar] [CrossRef]
- Hata, A.; Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 121–134. [Google Scholar] [CrossRef]
- Bortoletto, A.S.; Parchem, R.J. KRAS Hijacks the miRNA Regulatory Pathway in Cancer. Cancer Res. 2023, 83, 1563–1572. [Google Scholar] [CrossRef]
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H. MicroRNA expression in zebrafish embryonic development. Science 2005, 309, 310–311. [Google Scholar] [CrossRef]
- Aboobaker, A.A.; Tomancak, P.; Patel, N.; Rubin, G.M.; Lai, E.C. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. USA 2005, 102, 18017–18022. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.C.; Harland, R.M. Expression of microRNAs during embryonic development of Xenopus tropicalis. Gene Expr. Patterns 2008, 8, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Li, L.J.; Leng, R.X.; Fan, Y.G.; Pan, H.F.; Ye, D.Q. Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and circRNAs. Exp. Cell Res. 2017, 361, 1–8. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Li, S.C.; Chan, W.C.; Hu, L.Y.; Lai, C.H.; Hsu, C.N.; Lin, W.C. Identification of homologous microRNAs in 56 animal genomes. Genomics 2010, 96, 1–9. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Lizano, E.; Houben, A.J.S.; Bezdan, D.; Báñez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1000 novel human microRNAs. Genome Biol. 2014, 15, R57. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Arif, M.A.; Seumel, G.I.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Transcriptional control of gene expression by microRNAs. Cell 2010, 140, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Cui, Q.; Wang, J.; Zhou, Y. TransmiR v2.0: An updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2018, 47, D253–D258. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Meister, G. microRNA-guided posttranscriptional gene regulation. Biol. Chem. 2005, 386, 1205–1218. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef]
- van Rooij, E. The art of microRNA research. Circ. Res. 2011, 108, 219–234. [Google Scholar] [CrossRef]
- Guo, Z.; Maki, M.; Ding, R.; Yang, Y.; Zhang, B.; Xiong, L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci. Rep. 2014, 4, 5150. [Google Scholar] [CrossRef] [PubMed]
- de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Beck, J.D.; Reidenbach, D.; Salomon, N.; Sahin, U.; Türeci, Ö.; Vormehr, M.; Kranz, L.M. mRNA therapeutics in cancer immunotherapy. Mol. Cancer 2021, 20, 69. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, D.; Schaefer, A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology 2013, 38, 39–54. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell Pharmacol. 2011, 3, 83–92. [Google Scholar]
- Akgul, B.; Erdogan, I. Intracytoplasmic Re-localization of miRISC Complexes. Front. Genet. 2018, 9, 403. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Uhlmann, S.; Mannsperger, H.; Zhang, J.D.; Horvat, E.; Schmidt, C.; Küblbeck, M.; Henjes, F.; Ward, A.; Tschulena, U.; Zweig, K.; et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 2012, 8, 570. [Google Scholar] [CrossRef]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef]
- Saetrom, P.; Heale, B.S.; Snøve, O., Jr.; Aagaard, L.; Alluin, J.; Rossi, J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007, 35, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Desterro, J.; Bak-Gordon, P.; Carmo-Fonseca, M. Targeting mRNA processing as an anticancer strategy. Nat. Rev. Drug Discov. 2020, 19, 112–129. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012, 122, 4059–4076. [Google Scholar] [CrossRef] [PubMed]
- Stahlhut, C.; Slack, F.J. MicroRNAs and the cancer phenotype: Profiling, signatures and clinical implications. Genome Med. 2013, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Dvinge, H.; Git, A.; Gräf, S.; Salmon-Divon, M.; Curtis, C.; Sottoriva, A.; Zhao, Y.; Hirst, M.; Armisen, J.; Miska, E.A.; et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013, 497, 378–382. [Google Scholar] [CrossRef]
- Kim, T.M.; Huang, W.; Park, R.; Park, P.J.; Johnson, M.D. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011, 71, 3387–3399. [Google Scholar] [CrossRef] [PubMed]
- Manterola, L.; Guruceaga, E.; Gállego Pérez-Larraya, J.; González-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Samprón, N.; Barrena, C.; et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014, 16, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Kasinski, A.L.; Slack, F.J. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 2011, 11, 849–864. [Google Scholar] [CrossRef]
- Shankar, S.; Tien, J.C.-Y.; Siebenaler, R.F.; Chugh, S.; Dommeti, V.L.; Zelenka-Wang, S.; Wang, X.-M.; Apel, I.J.; Waninger, J.; Eyunni, S.; et al. An essential role for Argonaute 2 in EGFR-KRAS signaling in pancreatic cancer development. Nat. Commun. 2020, 11, 2817. [Google Scholar] [CrossRef]
- Eder, M.; Scherr, M. MicroRNA and lung cancer. N. Engl. J. Med. 2005, 352, 2446–2448. [Google Scholar] [CrossRef]
- Gonzales-Aloy, E.; Connerty, P.; Salik, B.; Liu, B.; Woo, A.J.; Haber, M.; Norris, M.D.; Wang, J.; Wang, J.Y. miR-101 suppresses the development of MLL-rearranged acute myeloid leukemia. Haematologica 2019, 104, e296–e299. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Cheng, R.; Yang, F.; Yu, M.; Wang, C.; Cui, S.; Hong, Y.; Liang, H.; Liu, M.; et al. The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer. Mol. Cancer 2018, 17, 11. [Google Scholar] [CrossRef]
- Zhu, S.; Cheng, X.; Wang, R.; Tan, Y.; Ge, M.; Li, D.; Xu, Q.; Sun, Y.; Zhao, C.; Chen, S.; et al. Restoration of microRNA function impairs MYC-dependent maintenance of MLL leukemia. Leukemia 2020, 34, 2484–2488. [Google Scholar] [CrossRef]
- Fu, Z.; Wang, L.; Li, S.; Chen, F.; Au-Yeung, K.K.; Shi, C. MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 2021, 12, 736323. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Gambari, R.; Brognara, E.; Spandidos, D.A.; Fabbri, E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Νew trends in the development of miRNA therapeutic strategies in oncology (Review). Int. J. Oncol. 2016, 49, 5–32. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C.S. OncomiRs: The discovery and progress of microRNAs in cancers. Mol. Cancer 2007, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [CrossRef]
- Zhou, K.; Liu, M.; Cao, Y. New Insight into microRNA Functions in Cancer: Oncogene-microRNA-Tumor Suppressor Gene Network. Front. Mol. Biosci. 2017, 4, 46. [Google Scholar] [CrossRef]
- Shenouda, S.K.; Alahari, S.K. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis. Rev. 2009, 28, 369–378. [Google Scholar] [CrossRef]
- Daoud, A.Z.; Mulholland, E.J.; Cole, G.; McCarthy, H.O. MicroRNAs in Pancreatic Cancer: Biomarkers, prognostic, and therapeutic modulators. BMC Cancer 2019, 19, 1130. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Vardapour, R.; Kehl, T.; Kneitz, S.; Ludwig, N.; Meese, E.; Lenhof, H.P.; Gessler, M. The DGCR8 E518K mutation found in Wilms tumors leads to a partial miRNA processing defect that alters gene expression patterns and biological processes. Carcinogenesis 2022, 43, 82–93. [Google Scholar] [CrossRef]
- Rivera, B.; Nadaf, J.; Fahiminiya, S.; Apellaniz-Ruiz, M.; Saskin, A.; Chong, A.S.; Sharma, S.; Wagener, R.; Revil, T.; Condello, V.; et al. DGCR8 microprocessor defect characterizes familial multinodular goiter with schwannomatosis. J. Clin. Investig. 2020, 130, 1479–1490. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Canberk, S.; Macedo, S.; Soares, P.; Vinagre, J. DGCR8 Microprocessor Subunit Mutation and Expression Deregulation in Thyroid Lesions. Int. J. Mol. Sci. 2022, 23, 14812. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Lv, Z.; Ding, H.; Fang, X.; Sun, M. Association of miRNA biosynthesis genes DROSHA and DGCR8 polymorphisms with cancer susceptibility: A systematic review and meta-analysis. Biosci. Rep. 2018, 38, BSR20180072. [Google Scholar] [CrossRef] [PubMed]
- Li, B.K.; Vasiljevic, A.; Dufour, C.; Yao, F.; Ho, B.L.B.; Lu, M.; Hwang, E.I.; Gururangan, S.; Hansford, J.R.; Fouladi, M.; et al. Pineoblastoma segregates into molecular sub-groups with distinct clinico-pathologic features: A Rare Brain Tumor Consortium registry study. Acta Neuropathol. 2020, 139, 223–241. [Google Scholar] [CrossRef]
- Rotunno, M.; Zhao, Y.; Bergen, A.W.; Koshiol, J.; Burdette, L.; Rubagotti, M.; Linnoila, R.I.; Marincola, F.M.; Bertazzi, P.A.; Pesatori, A.C.; et al. Inherited polymorphisms in the RNA-mediated interference machinery affect microRNA expression and lung cancer survival. Br. J. Cancer 2010, 103, 1870–1874. [Google Scholar] [CrossRef]
- Permuth-Wey, J.; Kim, D.; Tsai, Y.Y.; Lin, H.Y.; Chen, Y.A.; Barnholtz-Sloan, J.; Birrer, M.J.; Bloom, G.; Chanock, S.J.; Chen, Z.; et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011, 71, 3896–3903. [Google Scholar] [CrossRef]
- Felicio, P.S.; Grasel, R.S.; Campacci, N.; de Paula, A.E.; Galvão, H.C.R.; Torrezan, G.T.; Sabato, C.S.; Fernandes, G.C.; Souza, C.P.; Michelli, R.D.; et al. Whole-exome sequencing of non-BRCA1/BRCA2 mutation carrier cases at high-risk for hereditary breast/ovarian cancer. Hum. Mutat. 2021, 42, 290–299. [Google Scholar] [CrossRef]
- Liang, D.; Meyer, L.; Chang, D.W.; Lin, J.; Pu, X.; Ye, Y.; Gu, J.; Wu, X.; Lu, K. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010, 70, 9765–9776. [Google Scholar] [CrossRef]
- Mohammadpour-Gharehbagh, A.; Heidari, Z.; Eskandari, M.; Aryan, A.; Salimi, S. Association between Genetic Polymorphisms in microRNA Machinery Genes and Risk of Papillary Thyroid Carcinoma. Pathol. Oncol. Res. 2020, 26, 1235–1241. [Google Scholar] [CrossRef]
- Guénard, F.; Labrie, Y.; Ouellette, G.; Beauparlant, C.J.; Durocher, F. Genetic sequence variations of BRCA1-interacting genes AURKA, BAP1, BARD1 and DHX9 in French Canadian families with high risk of breast cancer. J. Hum. Genet. 2009, 54, 152–161. [Google Scholar] [CrossRef]
- Melo, S.A.; Moutinho, C.; Ropero, S.; Calin, G.A.; Rossi, S.; Spizzo, R.; Fernandez, A.F.; Davalos, V.; Villanueva, A.; Montoya, G.; et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 2010, 18, 303–315. [Google Scholar] [CrossRef]
- Liao, Y.; Liao, Y.; Li, J.; Liu, L.; Li, J.; Wan, Y.; Peng, L. Genetic variants in miRNA machinery genes associated with clinicopathological characteristics and outcomes of gastric cancer patients. Int. J. Biol. Markers 2018, 33, 301–307. [Google Scholar] [CrossRef]
- Palculict, T.B.; Ruteshouser, E.C.; Fan, Y.; Wang, W.; Strong, L.; Huff, V. Identification of germline DICER1 mutations and loss of heterozygosity in familial Wilms tumour. J. Med. Genet. 2016, 53, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Schultz, K.A.; Pacheco, M.C.; Yang, J.; Williams, G.M.; Messinger, Y.; Hill, D.A.; Dehner, L.P.; Priest, J.R. Ovarian sex cord-stromal tumors, pleuropulmonary blastoma and DICER1 mutations: A report from the International Pleuropulmonary Blastoma Registry. Gynecol. Oncol. 2011, 122, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.K.; Sabbaghian, N.; Xu, B.; Addidou-Kalucki, S.; Bernard, C.; Zou, D.; Reeve, A.E.; Eccles, M.R.; Cole, C.; Choong, C.S.; et al. Biallelic DICER1 mutations occur in Wilms tumours. J. Pathol. 2013, 230, 154–164. [Google Scholar] [CrossRef]
- Caruso, S.; Calderaro, J.; Letouzé, E.; Nault, J.C.; Couchy, G.; Boulai, A.; Luciani, A.; Zafrani, E.S.; Bioulac-Sage, P.; Seror, O.; et al. Germline and somatic DICER1 mutations in familial and sporadic liver tumors. J. Hepatol. 2017, 66, 734–742. [Google Scholar] [CrossRef]
- Heravi-Moussavi, A.; Anglesio, M.S.; Cheng, S.W.; Senz, J.; Yang, W.; Prentice, L.; Fejes, A.P.; Chow, C.; Tone, A.; Kalloger, S.E.; et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N. Engl. J. Med. 2012, 366, 234–242. [Google Scholar] [CrossRef]
- Witkowski, L.; Mattina, J.; Schönberger, S.; Murray, M.J.; Choong, C.S.; Huntsman, D.G.; Reis-Filho, J.S.; McCluggage, W.G.; Nicholson, J.C.; Coleman, N.; et al. DICER1 hotspot mutations in non-epithelial gonadal tumours. Br. J. Cancer 2013, 109, 2744–2750. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, D.; Li, Y.; Bian, L.; Ma, T.; Xie, M. DICER1 mutations in a patient with an ovarian Sertoli-Leydig tumor, well-differentiated fetal adenocarcinoma of the lung, and familial multinodular goiter. Eur. J. Med. Genet. 2014, 57, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Brenneman, M.; Field, A.; Yang, J.; Williams, G.; Doros, L.; Rossi, C.; Schultz, K.A.; Rosenberg, A.; Ivanovich, J.; Turner, J.; et al. Temporal order of RNase IIIb and loss-of-function mutations during development determines phenotype in pleuropulmonary blastoma/DICER1 syndrome: A unique variant of the two-hit tumor suppression model. F1000Research 2015, 4, 214. [Google Scholar] [CrossRef]
- Garre, P.; Pérez-Segura, P.; Díaz-Rubio, E.; Caldés, T.; de la Hoya, M. Reassessing the TARBP2 mutation rate in hereditary nonpolyposis colorectal cancer. Nat. Genet. 2010, 42, 817–818. [Google Scholar] [CrossRef]
- Torrezan, G.T.; Ferreira, E.N.; Nakahata, A.M.; Barros, B.D.; Castro, M.T.; Correa, B.R.; Krepischi, A.C.; Olivieri, E.H.; Cunha, I.W.; Tabori, U.; et al. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat. Commun. 2014, 5, 4039. [Google Scholar] [CrossRef]
- Caramuta, S.; Lee, L.; Ozata, D.M.; Akçakaya, P.; Xie, H.; Höög, A.; Zedenius, J.; Bäckdahl, M.; Larsson, C.; Lui, W.O. Clinical and functional impact of TARBP2 over-expression in adrenocortical carcinoma. Endocr. Relat. Cancer 2013, 20, 551–564. [Google Scholar] [CrossRef]
- Chang, S.S.; Smith, I.; Glazer, C.; Hennessey, P.; Califano, J.A. EIF2C is overexpressed and amplified in head and neck squamous cell carcinoma. ORL J. Otorhinolaryngol. Relat. Spec. 2010, 72, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, L.; Barlogie, B.; Stephens, O.; Wu, X.; Williams, D.R.; Cartron, M.A.; van Rhee, F.; Nair, B.; Waheed, S.; et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc. Natl. Acad. Sci. USA 2010, 107, 7904–7909. [Google Scholar] [CrossRef] [PubMed]
- Casey, M.C.; Prakash, A.; Holian, E.; McGuire, A.; Kalinina, O.; Shalaby, A.; Curran, C.; Webber, M.; Callagy, G.; Bourke, E.; et al. Quantifying Argonaute 2 (Ago2) expression to stratify breast cancer. BMC Cancer 2019, 19, 712. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Huang, J.; Zhou, Y.; Zhou, Y.; Yu, L.; Li, H.; Hou, L.; Zhu, L.; Ge, D.; Zeng, Y.; et al. Germline and somatic variations influence the somatic mutational signatures of esophageal squamous cell carcinomas in a Chinese population. BMC Genom. 2018, 19, 538. [Google Scholar] [CrossRef]
- Kim, M.S.; Oh, J.E.; Kim, Y.R.; Park, S.W.; Kang, M.R.; Kim, S.S.; Ahn, C.H.; Yoo, N.J.; Lee, S.H. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J. Pathol. 2010, 221, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Szafranska, A.E.; Davison, T.S.; John, J.; Cannon, T.; Sipos, B.; Maghnouj, A.; Labourier, E.; Hahn, S.A. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 2007, 26, 4442–4452. [Google Scholar] [CrossRef]
- Jiao, L.R.; Frampton, A.E.; Jacob, J.; Pellegrino, L.; Krell, J.; Giamas, G.; Tsim, N.; Vlavianos, P.; Cohen, P.; Ahmad, R.; et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS ONE 2012, 7, e32068. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Okayama, H.; Harris, C.C. The role of microRNAs in colorectal cancer. Cancer J. 2012, 18, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.L.; Tsai, Y.M.; Lien, C.T.; Kuo, P.L.; Hung, A.J. The Roles of MicroRNA in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 1611. [Google Scholar] [CrossRef]
- Le Rolle, A.F.; Chiu, T.K.; Zeng, Z.; Shia, J.; Weiser, M.R.; Paty, P.B.; Chiu, V.K. Oncogenic KRAS activates an embryonic stem cell-like program in human colon cancer initiation. Oncotarget 2016, 7, 2159–2174. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Middleton, J.; Jeon, Y.J.; Magee, P.; Veneziano, D.; Laganà, A.; Leong, H.S.; Sahoo, S.; Fassan, M.; Booton, R.; et al. KRAS induces lung tumorigenesis through microRNAs modulation. Cell Death Dis. 2018, 9, 219. [Google Scholar] [CrossRef]
- Buscail, L.; Bournet, B.; Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 153–168. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Haigis, K.M. KRAS Alleles: The Devil Is in the Detail. Trends Cancer 2017, 3, 686–697. [Google Scholar] [CrossRef]
- Wang, M.T.; Fer, N.; Galeas, J.; Collisson, E.A.; Kim, S.E.; Sharib, J.; McCormick, F. Blockade of leukemia inhibitory factor as a therapeutic approach to KRAS driven pancreatic cancer. Nat. Commun. 2019, 10, 3055. [Google Scholar] [CrossRef]
- Drosten, M.; Barbacid, M. Targeting the MAPK Pathway in KRAS-Driven Tumors. Cancer Cell 2020, 37, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Shui, B.; La Rocca, G.; Ventura, A.; Haigis, K.M. Interplay between K-RAS and miRNAs. Trends Cancer 2022, 8, 384–396. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, A.J.; Hoshino, D.; Hong, N.H.; Cha, D.J.; Franklin, J.L.; Coffey, R.J.; Patton, J.G.; Weaver, A.M. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep. 2016, 15, 978–987. [Google Scholar] [CrossRef]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Schmich, F.; Srivatsa, S.; Weidner, J.; Beerenwinkel, N.; Spang, A. Context-dependent deposition and regulation of mRNAs in P-bodies. Elife 2018, 7, e29815. [Google Scholar] [CrossRef]
- Zhang, B.; Herman, P.K. It is all about the process(ing): P-body granules and the regulation of signal transduction. Curr. Genet. 2020, 66, 73–77. [Google Scholar] [CrossRef]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef]
- Nakajima, E.C.; Drezner, N.; Li, X.; Mishra-Kalyani, P.S.; Liu, Y.; Zhao, H.; Bi, Y.; Liu, J.; Rahman, A.; Wearne, E.; et al. FDA Approval Summary: Sotorasib for KRAS G12C-Mutated Metastatic NSCLC. Clin. Cancer Res. 2022, 28, 1482–1486. [Google Scholar] [CrossRef]
- Dhillon, S. Adagrasib: First Approval. Drugs 2023, 83, 275–285. [Google Scholar] [CrossRef]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal. Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef]
- Goonetilleke, K.S.; Siriwardena, A.K. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncol. 2007, 33, 266–270. [Google Scholar] [CrossRef]
- Kabiraj, L.; Kundu, A. Potential role of microRNAs in pancreatic cancer manifestation: A review. J. Egypt. Natl. Cancer Inst. 2022, 34, 26. [Google Scholar] [CrossRef]
- Qi, Z.H.; Xu, H.X.; Zhang, S.R.; Xu, J.Z.; Li, S.; Gao, H.L.; Jin, W.; Wang, W.Q.; Wu, C.T.; Ni, Q.X.; et al. The Significance of Liquid Biopsy in Pancreatic Cancer. J. Cancer 2018, 9, 3417–3426. [Google Scholar] [CrossRef] [PubMed]
- Ganepola, G.A.; Rutledge, J.R.; Suman, P.; Yiengpruksawan, A.; Chang, D.H. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J. Gastrointest. Oncol. 2014, 6, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.A.; Dehlendorff, C.; Jensen, B.V.; Bjerregaard, J.K.; Nielsen, K.R.; Bojesen, S.E.; Calatayud, D.; Nielsen, S.E.; Yilmaz, M.; Holländer, N.H.; et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA 2014, 311, 392–404. [Google Scholar] [CrossRef]
- Sadri, F.; Hosseini, S.F.; Aghayei, A.; Fereidouni, M.; Rezaei, Z. The Tumor Suppressor Roles and Mechanisms of MiR-491 in Human Cancers. DNA Cell Biol. 2022, 41, 810–823. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, J.G.; Shi, Y.; Qin, Q.; Liu, Y.; Wang, B.; Tian, K.; Deng, S.C.; Li, X.; Zhu, S.; et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS ONE 2013, 8, e73803. [Google Scholar]
- Mardin, W.A.; Mees, S.T. MicroRNAs: Novel diagnostic and therapeutic tools for pancreatic ductal adenocarcinoma? Ann. Surg. Oncol. 2009, 16, 3183–3189. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Flatmark, K.; Hoye, E.; Fromm, B. microRNAs as cancer biomarkers. Scand. J. Clin. Lab. Investig. Suppl. 2016, 245, S80–S83. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, P.H. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim. Biophys. Acta 2016, 1862, 1617–1627. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.M.H. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020, 248, 117473. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, H.; Carney, G.E. Evidence and potential in vivo functions for biofluid miRNAs: From expression profiling to functional testing: Potential roles of extracellular miRNAs as indicators of physiological change and as agents of intercellular information exchange. Bioessays 2016, 38, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef]
- Kanlikilicer, P.; Rashed, M.H.; Bayraktar, R.; Mitra, R.; Ivan, C.; Aslan, B.; Zhang, X.; Filant, J.; Silva, A.M.; Rodriguez-Aguayo, C.; et al. Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells. Cancer Res. 2016, 76, 7194–7207. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell communication: MicroRNAs as hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef]
- Rashed, M.H.; Kanlikilicer, P.; Rodriguez-Aguayo, C.; Pichler, M.; Bayraktar, R.; Bayraktar, E.; Ivan, C.; Filant, J.; Silva, A.; Aslan, B.; et al. Exosomal miR-940 maintains SRC-mediated oncogenic activity in cancer cells: A possible role for exosomal disposal of tumor suppressor miRNAs. Oncotarget 2017, 8, 20145–20164. [Google Scholar] [CrossRef]
- Paškevičiūtė, M.; Petrikaitė, V. Overcoming transporter-mediated multidrug resistance in cancer: Failures and achievements of the last decades. Drug Deliv. Transl. Res. 2019, 9, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Märte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles 2018, 7, 1481321. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Du, D.; Wang, X.; Liu, D.; Xu, W.; Luo, Y.; Lin, Y. Recent Advances in Biosensors for Detecting Cancer-Derived Exosomes. Trends Biotechnol. 2019, 37, 1236–1254. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.J.; Wei, R.; Li, F.; Liao, S.Y.; Tse, H.F. Mesenchymal stromal cell-derived exosomes in cardiac regeneration and repair. Stem Cell Rep. 2021, 16, 1662–1673. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Ofrecio, J.M.; Luo, Z.; Ji, Y.; Jin, Z.; Ly, C.; Olefsky, J.M. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021, 33, 781–790.e5. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Budakoti, M.; Panwar, A.S.; Molpa, D.; Singh, R.K.; Büsselberg, D.; Mishra, A.P.; Coutinho, H.D.M.; Nigam, M. Micro-RNA: The darkhorse of cancer. Cell. Signal. 2021, 83, 109995. [Google Scholar] [CrossRef]
- Kong, Y.W.; Ferland-McCollough, D.; Jackson, T.J.; Bushell, M. microRNAs in cancer management. Lancet Oncol. 2012, 13, e249–e258. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Ping, J.; Wen, D. MicroRNA-186 regulates the invasion and metastasis of bladder cancer via vascular endothelial growth factor C. Exp. Ther. Med. 2017, 14, 3253–3258. [Google Scholar] [CrossRef]
- Geva, G.A.; Gielchinsky, I.; Aviv, N.; Max, K.E.A.; Gofrit, O.N.; Gur-Wahnon, D.; Ben-Dov, I.Z. Urine cell-free microRNA as biomarkers for transitional cell carcinoma. BMC Res. Notes 2017, 10, 641. [Google Scholar] [CrossRef] [PubMed]
- Fedorko, M.; Juracek, J.; Stanik, M.; Svoboda, M.; Poprach, A.; Buchler, T.; Pacik, D.; Dolezel, J.; Slaby, O. Detection of let-7 miRNAs in urine supernatant as potential diagnostic approach in non-metastatic clear-cell renal cell carcinoma. Biochem. Med. 2017, 27, 411–417. [Google Scholar] [CrossRef]
- Butz, H.; Nofech-Mozes, R.; Ding, Q.; Khella, H.W.Z.; Szabó, P.M.; Jewett, M.; Finelli, A.; Lee, J.; Ordon, M.; Stewart, R.; et al. Exosomal MicroRNAs Are Diagnostic Biomarkers and Can Mediate Cell-Cell Communication in Renal Cell Carcinoma. Eur. Urol. Focus. 2016, 2, 210–218. [Google Scholar] [CrossRef]
- Erbes, T.; Hirschfeld, M.; Rücker, G.; Jaeger, M.; Boas, J.; Iborra, S.; Mayer, S.; Gitsch, G.; Stickeler, E. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer 2015, 15, 193. [Google Scholar] [CrossRef]
- Debernardi, S.; Massat, N.J.; Radon, T.P.; Sangaralingam, A.; Banissi, A.; Ennis, D.P.; Dowe, T.; Chelala, C.; Pereira, S.P.; Kocher, H.M.; et al. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. Am. J. Cancer Res. 2015, 5, 3455–3466. [Google Scholar]
- Shams, R.; Saberi, S.; Zali, M.; Sadeghi, A.; Ghafouri-Fard, S.; Aghdaei, H.A. Identification of potential microRNA panels for pancreatic cancer diagnosis using microarray datasets and bioinformatics methods. Sci. Rep. 2020, 10, 7559. [Google Scholar] [CrossRef]
- Al-Shaheri, F.N.; Alhamdani, M.S.S.; Bauer, A.S.; Giese, N.; Buchler, M.W.; Hackert, T.; Hoheisel, J.D. Blood biomarkers for differential diagnosis and early detection of pancreatic cancer. Cancer Treat. Rev. 2021, 96, 102193. [Google Scholar] [CrossRef]
- Marin, A.M.; Mattar, S.B.; Amatuzzi, R.F.; Chammas, R.; Uno, M.; Zanette, D.L.; Aoki, M.N. Plasma Exosome-Derived microRNAs as Potential Diagnostic and Prognostic Biomarkers in Brazilian Pancreatic Cancer Patients. Biomolecules 2022, 12, 769. [Google Scholar] [CrossRef] [PubMed]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschênes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Chang, P.; LeBlanc, A.; Li, D.; Abbruzzesse, J.L.; Frazier, M.L.; Killary, A.M.; Sen, S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. 2009, 2, 807–813. [Google Scholar] [CrossRef]
- Havasi, A.; Sur, D.; Cainap, S.S.; Lungulescu, C.V.; Gavrilas, L.I.; Cainap, C.; Vlad, C.; Balacescu, O. Current and New Challenges in the Management of Pancreatic Neuroendocrine Tumors: The Role of miRNA-Based Approaches as New Reliable Biomarkers. Int. J. Mol. Sci. 2022, 23, 1109. [Google Scholar] [CrossRef]
- Yang, J.; Xu, R.; Wang, C.; Qiu, J.; Ren, B.; You, L. Early screening and diagnosis strategies of pancreatic cancer: A comprehensive review. Cancer Commun. 2021, 41, 1257–1274. [Google Scholar] [CrossRef]
- Ouyang, H.; Gore, J.; Deitz, S.; Korc, M. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-β actions. Oncogene 2014, 33, 4664–4674. [Google Scholar] [CrossRef]
- Preis, M.; Gardner, T.B.; Gordon, S.R.; Pipas, J.M.; Mackenzie, T.A.; Klein, E.E.; Longnecker, D.S.; Gutmann, E.J.; Sempere, L.F.; Korc, M. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2011, 17, 5812–5821. [Google Scholar] [CrossRef]
- Giovannetti, E.; Funel, N.; Peters, G.J.; Del Chiaro, M.; Erozenci, L.A.; Vasile, E.; Leon, L.G.; Pollina, L.E.; Groen, A.; Falcone, A.; et al. MicroRNA-21 in pancreatic cancer: Correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010, 70, 4528–4538. [Google Scholar] [CrossRef]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef]
- Guo, S.; Fesler, A.; Wang, H.; Ju, J. microRNA based prognostic biomarkers in pancreatic Cancer. Biomark. Res. 2018, 6, 18. [Google Scholar] [CrossRef]
- Drakaki, A.; Iliopoulos, D. MicroRNA-gene signaling pathways in pancreatic cancer. Biomed. J. 2013, 36, 200–208. [Google Scholar] [PubMed]
- Ohuchida, K.; Mizumoto, K.; Kayashima, T.; Fujita, H.; Moriyama, T.; Ohtsuka, T.; Ueda, J.; Nagai, E.; Hashizume, M.; Tanaka, M. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann. Surg. Oncol. 2011, 18, 2381–2387. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; VandenBoom, T.G., 2nd; Kong, D.; Wang, Z.; Ali, S.; Philip, P.A.; Sarkar, F.H. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009, 69, 6704–6712. [Google Scholar] [CrossRef] [PubMed]
- Paik, W.H.; Song, B.J.; Kim, H.W.; Kim, H.R.; Hwang, J.H. MicroRNA-200c as a Prognostic Biomarker for Pancreatic Cancer. Korean J. Gastroenterol. 2015, 66, 215–220. [Google Scholar] [CrossRef]
- Yang, X.L.; Ma, Y.S.; Liu, Y.S.; Jiang, X.H.; Ding, H.; Shi, Y.; Jia, C.Y.; Lu, G.X.; Zhang, D.D.; Wang, H.M.; et al. microRNA-873 inhibits self-renewal and proliferation of pancreatic cancer stem cells through pleckstrin-2-dependent PI3K/AKT pathway. Cell Signal. 2021, 84, 110025. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.A.; Andersen, K.K.; Roslind, A.; Willenbrock, H.; Wøjdemann, M.; Johansen, J.S. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer--five microRNAs in a prognostic index. World J. Surg. 2012, 36, 2699–2707. [Google Scholar] [CrossRef]
- Pai, P.; Rachagani, S.; Are, C.; Batra, S.K. Prospects of miRNA-based therapy for pancreatic cancer. Curr. Drug Targets 2013, 14, 1101–1109. [Google Scholar] [CrossRef]
- Liu, Y.A.; Zhang, Y.; Zheng, Z.; Li, K.; Wu, X.H.; Du, Q.G.; Ye, X.; Wang, L.; Zhu, L. MicroRNA-216b reduces growth, migration and invasion of pancreatic ductal adenocarcinoma cells by directly targeting ρ-associated coiled-coil containing protein kinase 1. Oncol. Lett. 2018, 15, 6745–6751. [Google Scholar] [CrossRef]
- Yi, J.; Li, S.; Wang, C.; Cao, N.; Qu, H.; Cheng, C.; Wang, Z.; Wang, L.; Zhou, L. Potential applications of polyphenols on main ncRNAs regulations as novel therapeutic strategy for cancer. Biomed. Pharmacother. 2019, 113, 108703. [Google Scholar] [CrossRef]
- Kang, H.; Ma, D.; Zhang, J.; Zhao, J.; Yang, M. MicroRNA-18a induces epithelial-mesenchymal transition like cancer stem cell phenotype via regulating RKIP pathway in pancreatic cancer. Ann. Transl. Med. 2020, 8, 433. [Google Scholar] [CrossRef]
- Zhu, J.; Lv, J.; Chen, J.; Zhang, X.; Ji, Y. Down-regulated microRNA-223 or elevated ZIC1 inhibits the development of pancreatic cancer via inhibiting PI3K/Akt/mTOR signaling pathway activation. Cell Cycle 2020, 19, 2851–2865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, D.; Zhang, G.; Wu, X.; Zhou, L.; Lin, Y.; Ding, J.; An, F.; Zhan, Q. MicroRNA-23b-3p promotes pancreatic cancer cell tumorigenesis and metastasis via the JAK/PI3K and Akt/NF-kappaB signaling pathways. Oncol. Lett. 2020, 20, 160. [Google Scholar]
- Wang, W.; Wang, J.; Yang, C.; Wang, J. MicroRNA-216a targets WT1 expression and regulates KRT7 transcription to mediate the progression of pancreatic cancer-A transcriptome analysis. IUBMB Life 2021, 73, 866–882. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, Y.; Chen, Q.; Dai, Y.; Li, T. MicroRNA-139 inhibits pancreatic-cancer carcinogenesis by suppressing RalB via the Ral/RAC/PI3K pathway. Arch. Biochem. Biophys. 2021, 704, 108719. [Google Scholar] [CrossRef]
- Xue, Y.; Wu, T.; Sheng, Y.; Zhong, Y.; Hu, B.; Bao, C. MicroRNA-1252-5p, regulated by Myb, inhibits invasion and epithelial-mesenchymal transition of pancreatic cancer cells by targeting NEDD9. Aging 2021, 13, 18924–18945. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhong, Y.; Wu, T.; Sheng, Y.; Dai, Y.; Xu, L.; Bao, C. Anti-Proliferative and Apoptosis-Promoting Effect of microRNA-125b on Pancreatic Cancer by Targeting NEDD9 via PI3K/AKT Signaling. Cancer Manag. Res. 2020, 12, 7363–7373. [Google Scholar] [CrossRef]
- Wan, X.; Guo, D.; Zhu, Q.; Qu, R. microRNA-382 suppresses the progression of pancreatic cancer through the PI3K/Akt signaling pathway by inhibition of Anxa3. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G309–G322. [Google Scholar] [CrossRef]
- Nong, K.; Zhang, D.; Chen, C.; Yang, Y.; Yang, Y.; Liu, S.; Cai, H. MicroRNA-519 inhibits hypoxia-induced tumorigenesis of pancreatic cancer by regulating immune checkpoint PD-L1. Oncol. Lett. 2020, 19, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Zubair, H.; Srivastava, S.K.; Singh, S.; Singh, A.P. Insights into the Role of microRNAs in Pancreatic Cancer Pathogenesis: Potential for Diagnosis, Prognosis, and Therapy. Adv. Exp. Med. Biol. 2015, 889, 71–87. [Google Scholar]
- Mikamori, M.; Yamada, D.; Eguchi, H.; Hasegawa, S.; Kishimoto, T.; Tomimaru, Y.; Asaoka, T.; Noda, T.; Wada, H.; Kawamoto, K.; et al. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2017, 7, 42339. [Google Scholar] [CrossRef]
- Greither, T.; Grochola, L.F.; Udelnow, A.; Lautenschlager, C.; Wurl, P.; Taubert, H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int. J. Cancer 2010, 126, 73–80. [Google Scholar] [CrossRef]
- Papaconstantinou, I.G.; Manta, A.; Gazouli, M.; Lyberopoulou, A.; Lykoudis, P.M.; Polymeneas, G.; Voros, D. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas 2013, 42, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Chen, H.; Wang, L.; Wang, F.; Wang, P.; Ning, Z.; Li, Y.; Liu, L.; Chen, Z.; Meng, Z. Low serum miR-373 predicts poor prognosis in patients with pancreatic cancer. Cancer Biomark. 2017, 20, 95–100. [Google Scholar] [CrossRef]
- Yuan, W.; Tang, W.; Xie, Y.; Wang, S.; Chen, Y.; Qi, J.; Qiao, Y.; Ma, J. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget 2016, 7, 80033–80045. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Pérez-Boza, J.; Curado, J.; Devaux, Y. Challenges of microRNA-based biomarkers in clinical application for cardiovascular diseases. Clin. Transl. Med. 2022, 12, e585. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ma, K.; Hao, W.; Jiao, Y.; Li, P.; Chen, J.; Xu, C.; Xu, F.J.; Lau, W.B.; Du, J.; et al. mir15a/mir16-1 cluster and its novel targeting molecules negatively regulate cardiac hypertrophy. Clin. Transl. Med. 2020, 10, e242. [Google Scholar] [CrossRef]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef]
- Urbanek, M.O.; Nawrocka, A.U.; Krzyzosiak, W.J. Small RNA Detection by in Situ Hybridization Methods. Int. J. Mol. Sci. 2015, 16, 13259–13286. [Google Scholar] [CrossRef]
- Tiberio, P.; Callari, M.; Angeloni, V.; Daidone, M.G.; Appierto, V. Challenges in Using Circulating miRNAs as Cancer Biomarkers. BioMed. Res. Int. 2015, 2015, 731479. [Google Scholar] [CrossRef]
- Godoy, P.M.; Bhakta, N.R.; Barczak, A.J.; Cakmak, H.; Fisher, S.; MacKenzie, T.C.; Patel, T.; Price, R.W.; Smith, J.F.; Woodruff, P.G.; et al. Large Differences in Small RNA Composition Between Human Biofluids. Cell Rep. 2018, 25, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Politz, J.C.; Zhang, F.; Pederson, T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. USA 2006, 103, 18957–18962. [Google Scholar] [CrossRef]
- Sweetman, D.; Rathjen, T.; Jefferson, M.; Wheeler, G.; Smith, T.G.; Wheeler, G.N.; Münsterberg, A.; Dalmay, T. FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev. Dyn. 2006, 235, 2185–2191. [Google Scholar] [CrossRef]
- Tuddenham, L.; Wheeler, G.; Ntounia-Fousara, S.; Waters, J.; Hajihosseini, M.K.; Clark, I.; Dalmay, T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006, 580, 4214–4217. [Google Scholar] [CrossRef] [PubMed]
- Beyret, E.; Lin, H. Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev. Biol. 2011, 355, 215–226. [Google Scholar] [CrossRef]
- Song, R.; Ro, S.; Michaels, J.D.; Park, C.; McCarrey, J.R.; Yan, W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat. Genet. 2009, 41, 488–493. [Google Scholar] [CrossRef]
- Huan, T.; Chen, G.; Liu, C.; Bhattacharya, A.; Rong, J.; Chen, B.H.; Seshadri, S.; Tanriverdi, K.; Freedman, J.E.; Larson, M.G.; et al. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell 2018, 17, e12687. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.; Woldhuis, R.R.; Boudewijn, I.M.; van den Berg, A.; Kluiver, J.; Kok, K.; Terpstra, M.M.; Guryev, V.; de Vries, M.; Vermeulen, C.J.; et al. Age-related gene and miRNA expression changes in airways of healthy individuals. Sci. Rep. 2019, 9, 3765. [Google Scholar] [CrossRef]
- Chandrasekar, V.; Dreyer, J.L. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell Neurosci. 2009, 42, 350–362. [Google Scholar] [CrossRef]
- Hollander, J.A.; Im, H.I.; Amelio, A.L.; Kocerha, J.; Bali, P.; Lu, Q.; Willoughby, D.; Wahlestedt, C.; Conkright, M.D.; Kenny, P.J. Striatal microRNA controls cocaine intake through CREB signalling. Nature 2010, 466, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.D.; Yelamanchili, S.V.; Marcondes, M.C.; Fox, H.S. Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1. FASEB J. 2013, 27, 3720–3729. [Google Scholar] [CrossRef]
- Kong, W.; Yang, H.; He, L.; Zhao, J.J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell Biol. 2008, 28, 6773–6784. [Google Scholar] [CrossRef]
- Kocerha, J.; Kouri, N.; Baker, M.; Finch, N.; DeJesus-Hernandez, M.; Gonzalez, J.; Chidamparam, K.; Josephs, K.A.; Boeve, B.F.; Graff-Radford, N.R.; et al. Altered microRNA expression in frontotemporal lobar degeneration with TDP-43 pathology caused by progranulin mutations. BMC Genom. 2011, 12, 527. [Google Scholar] [CrossRef]
- Huang, C.J.; Huang, W.Y.; Chen, C.Y.; Chao, Y.J.; Chiang, N.J.; Shan, Y.S. Cancer-cell-derived cell-free DNA can predict distant metastasis earlier in pancreatic cancer: A prospective cohort study. Ther. Adv. Med. Oncol. 2022, 14, 17588359221106558. [Google Scholar] [CrossRef] [PubMed]
- Pietri, E.; Balsano, R.; Coriano, M.; Gelsomino, F.; Leonardi, F.; Bui, S.; Gnetti, L.; Valle, R.D.; Garajová, I. The implication of liquid biopsies to predict chemoresistance in pancreatic cancer. Cancer Drug Resist. 2021, 4, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Kay Li, Q.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef]
- Sugimori, M.; Sugimori, K.; Tsuchiya, H.; Suzuki, Y.; Tsuyuki, S.; Kaneta, Y.; Hirotani, A.; Sanga, K.; Tozuka, Y.; Komiyama, S.; et al. Quantitative monitoring of circulating tumor DNA in patients with advanced pancreatic cancer undergoing chemotherapy. Cancer Sci. 2020, 111, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Toledano-Fonseca, M.; Cano, M.T.; Inga, E.; Rodríguez-Alonso, R.; Gómez-España, M.A.; Guil-Luna, S.; Mena-Osuna, R.; de la Haba-Rodríguez, J.R.; Rodríguez-Ariza, A.; Aranda, E. Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer. Cancers 2020, 12, 1754. [Google Scholar] [CrossRef]
- Uesato, Y.; Sasahira, N.; Ozaka, M.; Sasaki, T.; Takatsuki, M.; Zembutsu, H. Evaluation of circulating tumor DNA as a biomarker in pancreatic cancer with liver metastasis. PLoS ONE 2020, 15, e0235623. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, Q.; Li, X.; Su, W.; Li, G.; Ma, T.; Gao, S.; Lou, J.; Que, R.; Zheng, L.; et al. Monitoring Tumor Burden in Response to FOLFIRINOX Chemotherapy Via Profiling Circulating Cell-Free DNA in Pancreatic Cancer. Mol. Cancer Ther. 2019, 18, 196–203. [Google Scholar] [CrossRef]
- Koshiol, J.; Wang, E.; Zhao, Y.; Marincola, F.; Landi, M.T. Strengths and limitations of laboratory procedures for microRNA detection. Cancer Epidemiol. Biomark. Prev. 2010, 19, 907–911. [Google Scholar] [CrossRef]
- Wong, R.K.Y.; MacMahon, M.; Woodside, J.V.; Simpson, D.A. A comparison of RNA extraction and sequencing protocols for detection of small RNAs in plasma. BMC Genom. 2019, 20, 446. [Google Scholar] [CrossRef]
- Giraldez, M.D.; Spengler, R.M.; Etheridge, A.; Godoy, P.M.; Barczak, A.J.; Srinivasan, S.; De Hoff, P.L.; Tanriverdi, K.; Courtright, A.; Lu, S.; et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018, 36, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Leshkowitz, D.; Horn-Saban, S.; Parmet, Y.; Feldmesser, E. Differences in microRNA detection levels are technology and sequence dependent. RNA 2013, 19, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Washietl, S.; Will, S.; Hendrix, D.A.; Goff, L.A.; Rinn, J.L.; Berger, B.; Kellis, M. Computational analysis of noncoding RNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 759–778. [Google Scholar] [CrossRef] [PubMed]
- Godoy, P.M.; Barczak, A.J.; DeHoff, P.; Srinivasan, S.; Etheridge, A.; Galas, D.; Das, S.; Erle, D.J.; Laurent, L.C. Comparison of Reproducibility, Accuracy, Sensitivity, and Specificity of miRNA Quantification Platforms. Cell Rep. 2019, 29, 4212–4222.e5. [Google Scholar] [CrossRef]
- Yaylak, B.; Akgül, B. Experimental MicroRNA Detection Methods. In miRNomics: MicroRNA Biology and Computational Analysis; Allmer, J., Yousef, M., Eds.; Springer: New York, NY, USA, 2022; pp. 33–55. [Google Scholar]
- Jet, T.; Gines, G.; Rondelez, Y.; Taly, V. Advances in multiplexed techniques for the detection and quantification of microRNAs. Chem. Soc. Rev. 2021, 50, 4141–4161. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Martínez-Camblor, P.; Bär, C.; Duarte, K.; Girerd, N.; Fellström, B.; Schmieder, R.E.; Jardine, A.G.; Massy, Z.A.; Holdaas, H.; et al. Improved cardiovascular risk prediction in patients with end-stage renal disease on hemodialysis using machine learning modeling and circulating microribonucleic acids. Theranostics 2020, 10, 8665–8676. [Google Scholar] [CrossRef]
- Brown, C.; Mantzaris, M.; Nicolaou, E.; Karanasiou, G.; Papageorgiou, E.; Curigliano, G.; Cardinale, D.; Filippatos, G.; Memos, N.; Naka, K.K.; et al. A systematic review of miRNAs as biomarkers for chemotherapy-induced cardiotoxicity in breast cancer patients reveals potentially clinically informative panels as well as key challenges in miRNA research. Cardio-Oncology 2022, 8, 16. [Google Scholar] [CrossRef]
- Roldo, C.; Missiaglia, E.; Hagan, J.P.; Falconi, M.; Capelli, P.; Bersani, S.; Calin, G.A.; Volinia, S.; Liu, C.G.; Scarpa, A.; et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006, 24, 4677–4684. [Google Scholar] [CrossRef]
- Zimmermann, N.; Knief, J.; Kacprowski, T.; Lazar-Karsten, P.; Keck, T.; Billmann, F.; Schmid, S.; Luley, K.; Lehnert, H.; Brabant, G.; et al. MicroRNA analysis of gastroenteropancreatic neuroendocrine tumors and metastases. Oncotarget 2018, 9, 28379–28390. [Google Scholar] [CrossRef]
- Cavalcanti, E.; Galleggiante, V.; Coletta, S.; Stasi, E.; Chieppa, M.; Armentano, R.; Serino, G. Altered miRNAs Expression Correlates With Gastroenteropancreatic Neuroendocrine Tumors Grades. Front. Oncol. 2020, 10, 1187. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kim, H.; Kim, H.W.; Lee, J.C.; Paik, K.H.; Kang, J.; Kim, J.; Yoon, Y.S.; Han, H.S.; Sohn, I.; et al. High Expression of MicroRNA-196a Indicates Poor Prognosis in Resected Pancreatic Neuroendocrine Tumor. Medicine 2015, 94, e2224. [Google Scholar] [CrossRef] [PubMed]
- Gill, P.; Kim, E.; Chua, T.C.; Clifton-Bligh, R.J.; Nahm, C.B.; Mittal, A.; Gill, A.J.; Samra, J.S. MiRNA-3653 Is a Potential Tissue Biomarker for Increased Metastatic Risk in Pancreatic Neuroendocrine Tumours. Endocr. Pathol. 2019, 30, 128–133. [Google Scholar] [CrossRef]
- Cao, D.; Di, M.; Liang, J.; Shi, S.; Tan, Q.; Wang, Z. MicroRNA-183 in Cancer Progression. J. Cancer 2020, 11, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Vicentini, C.; Calore, F.; Nigita, G.; Fadda, P.; Simbolo, M.; Sperandio, N.; Luchini, C.; Lawlor, R.T.; Croce, C.M.; Corbo, V.; et al. Exosomal miRNA signatures of pancreatic lesions. BMC Gastroenterol. 2020, 20, 137. [Google Scholar] [CrossRef]
- Mao, L.; Liu, S.; Hu, L.; Jia, L.; Wang, H.; Guo, M.; Chen, C.; Liu, Y.; Xu, L. miR-30 Family: A Promising Regulator in Development and Disease. Biomed. Res. Int. 2018, 2018, 9623412. [Google Scholar] [CrossRef]
| miRNA | Expression Status | Type of Cancer | Function | Clinical Implications | References |
|---|---|---|---|---|---|
| miR-10b | Upregulated | Pancreatic cancer | TIP30—mRNA inhibition | Poor Survival | [227,228] |
| miR-21 | Upregulated | Pancreatic cancer Pancreatic neuroendocrine tumors | PTEN, PDCD4, IL-6R, CDK6 —mRNA inhibition Metastatic disease Proliferative activity | Worse Survival Tumor grade | [229,230,231,232,233] |
| miR-34a | Upregulated | Pancreatic cancer | NOTCH, BCL2, CDK6—mRNA inhibition Somatostatin, gastrin, and serotonin expression | Better Survival | [209,234,235,236] |
| miR-155 | Upregulated | Pancreatic cancer | TP53INP—mRNA inhibition | Poor Survival | [237,238] |
| miR-96-5p | Upregulated | Pancreatic neuroendocrine tumors | Oncogenic, FOXO1a inhibition | High tumor grade | [239] |
| miR-130b-3p | Upregulated | Pancreatic neuroendocrine tumors | NA | High tumor grade | [239] |
| miR-132-3p | Upregulated | Pancreatic neuroendocrine tumors | Tumor-suppressing and tumor-promoting function | Low tumor grade Vascular invasion Somatostatin expression | [236] |
| miR-145-5p | Upregulated | Pancreatic neuroendocrine tumors | High tumor grade Lymphatic invasion Serotonin expression | [236] | |
| miR-449a | Upregulated | Pancreatic neuroendocrine tumors | Oncogenic via histone deacetylases 3/4 | High tumor grade, mitotic and proliferative activity, lymph-node invasion | [236] |
| miR-183-5p | Upregulated | Pancreatic neuroendocrine tumors | Tumor suppressor | High tumor grade Tumor size Somatostatin-receptor expression | [236] |
| miR-196a | Upregulated | Pancreatic neuroendocrine tumors | NA | Advanced tumor Lymph-node invasion High mitotic and proliferative activity Recurrence | [240] |
| miR-210 | Upregulated | Pancreatic neuroendocrine tumors | Oncogenic | Metastatic disease | [232,241] |
| miR-642 | Upregulated | Pancreatic neuroendocrine tumors | Oncogenic | Proliferative activity | [241] |
| miR-3653 | Upregulated | Pancreatic neuroendocrine tumors | Oncogenic associated with ATRX mutations | Metastatic disease | [242] |
| let-7 Family | Downregulated | Pancreatic cancer | KRAS, HRAS, TRIM71 | Poor Survival | [243,244] |
| miR-194-5p | Downregulated | Pancreatic neuroendocrine tumors | NA | High tumor grade | [239] |
| miR-660 | Downregulated | Pancreatic neuroendocrine tumors | NA | Metastatic disease | [232] |
| miR-30a-5p | Downregulated | Pancreatic neuroendocrine tumors | NA | Metastatic disease | [245] |
| miR-339-3p | Downregulated | Pancreatic neuroendocrine tumors | NA | Metastatic disease | [232] |
| miR-345 | Downregulated | Pancreatic neuroendocrine tumors | NA | Metastatic disease | [232] |
| miR−200 Family | Downregulated | Pancreatic cancer | E-cadherin, ZEB1 | Better Survival | [234,246,247] |
| miR−216 | Downregulated | Pancreatic cancer | ROCK1 | Poor Survival | [248,249] |
| miR-181a | Upregulated | Pancreatic cancer | RKIP—mRNA inhibition and induces epithelial–mesenchymal transition like cancer stem cell phenotype | Poor Survival | [250] |
| miR-223 | Upregulated | Pancreatic cancer | ZIC1—mRNA inhibition via PI3K/Akt/mTOR signaling pathway | Poor Survival | [251] |
| miR-23b-3p | Upregulated | Pancreatic cancer | PTEN—mRNA inhibition via the JAK/PI3K and Akt/NF-kappa B signaling pathways | Poor Survival | [252] |
| miR-873 | Downregulated | Pancreatic cancer | PLEK2—mRNA inhibition via pleckstrin-2-dependent PI3K/AKT pathway | Better Survival | [253] |
| miR-216a | Downregulated | Pancreatic cancer | WT1—mRNA inhibition and regulates KRT7 transcription | Poor Survival | [254] |
| miR-139 | Downregulated | Pancreatic cancer | RalB—mRNA inhibition via the Ral/RAC/PI3K pathway | Poor Survival | [255] |
| miR-1252-5p | Downregulated | Pancreatic cancer | NEDD9 mRNA inhibition. MicroRNA-1252-5p, regulated by Myb regulates NEDD9 mRNA inhibition | Poor Survival | [256] |
| miR-125b | Downregulated | Pancreatic cancer | NEDD9—mRNA inhibition via PI3K/AKT signaling pathway | Poor Survival | [257] |
| miR-382 | Downregulated | Pancreatic cancer | Anxa3—mRNA inhibition via PI3K/Akt signaling pathway | Poor Survival | [258] |
| miR-519 | Downregulated | Pancreatic cancer | PD-L1—mRNA inhibition | Poor Survival | [259] |
| Study Title | Conditions | Interventions | Study Type | Phase | Status | NCT Number |
|---|---|---|---|---|---|---|
| Detection of MicroRNA-25 in the Diagnosis of Pancreatic Cancer | Carcinoma, Pancreatic Ductal | Diagnostic Test: Serum MicroRNA-25 detection | Observational | NA | Unknown | NCT03432624 |
| The Role of MicroRNA in the Diagnosis, Prognosis and Response to Treatment in Pancreatic Cancer | Pancreatic Cancer Stage III Pancreatic Cancer Stage IV Pancreatic Ductal Adenocarcinoma Pancreatic Neoplasms | Procedure: Blood draw | Observational | NA | Recruiting | NCT04406831 |
| Identify microRNAs in Cachexia in Pancreatic Carcinoma | Resectable Pancreatic Adenocarcinoma | Other: Pancreatic cancer microRNA and messenger RNA expression | Observational | NA | Recruiting | NCT05275075 |
| Lipidomics, Proteomics, Micro RNAs, and Volatile Organic Compounds (VOC) | Pancreatic Neoplasms | Other: blood and bile | Observational | NA | Active, not recruiting | NCT02531607 |
| U01-Biomarkers for Noninvasive and Early Detection of Pancreatic Cancer | Pancreatic Cancer | Observational | NA | Recruiting | NCT03886571 | |
| iDentification and vAlidation Model of Liquid biopsY Based cfDNA Methylation and pRotEin biomArKers for Pancreatic Cancer (DAYBREAK Study) | Cancer | Observational | NA | Recruiting | NCT05495685 | |
| Radiofrequency Ablation Combined With S-1 for Pancreatic Cancer with Liver Metastasis | Carcinoma, Pancreatic | Procedure: radiofrequency ablation Drug: S-1 | Interventional | Phase 2 | Unknown | NCT02634502 |
| BIOmarkers in Patients with Pancreatic Cancer (“BIOPAC”) | Pancreatic Cancer | Observational | NA | Recruiting | NCT03311776 | |
| AssesSment of Early-deteCtion basEd oN liquiD Biopsy in PANCEATIC Cancer (ASCEND-PANCREATIC) | Cancer | Observational | NA | Recruiting | NCT05556603 | |
| Validation of Useful Markers Generated by Next Generation Bio-data Based Genome Research and Cohort Study | BCL2 Gene mRNA Overexpression | Observational | NA | Completed | NCT02807896 | |
| Project CADENCE (CAncer Detected Early caN be CurEd) | Pancreatic Cancer Thoracic Cancer Ovarian Cancer Liver Cancer Prostate Cancer Gastric Cancer Colorectal Cancer Breast Cancer Esophageal Cancer | Observational | NA | Recruiting | NCT05633342 | |
| Metabolomics and Genetic Diagnosing Pancreatic Neuroendocrine Tumors in MEN1 Patients | Multiple Endocrine Neoplasia | Observational | NA | Recruiting | NCT03048266 | |
| Gemcitabine, Nab-Paclitaxel, and Bosentan for the Treatment of Unresectable Pancreatic Cancer | Stage III Pancreatic Cancer AJCC v8 | Drug: Bosentan | Interventional | Phase 1 | Recruiting | NCT04158635 |
| Atu027 Plus Gemcitabine in Advanced or Metastatic Pancreatic Cancer (Atu027-I-02) | Carcinoma, Pancreatic Ductal | Drug: Atu027 & gemcitabine in lead in safety period Drug: Atu027 & gemcitabine in treatment arm 1 | Interventional | Phase 1 | Completed | NCT01808638 |
| A Prospective Translational Tissue Collection Study in Early and Advanced Pancreatic Ductal Adenocarcinoma and Pancreatic Neuroendocrine Tumours to Enable Further Disease Characterisation and the Development of Potential Predictive and Prognostic Biomarkers | Pancreatic Adenocarcinoma | Observational | Unknown | NCT03840460 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seyhan, A.A. Circulating microRNAs as Potential Biomarkers in Pancreatic Cancer—Advances and Challenges. Int. J. Mol. Sci. 2023, 24, 13340. https://doi.org/10.3390/ijms241713340
Seyhan AA. Circulating microRNAs as Potential Biomarkers in Pancreatic Cancer—Advances and Challenges. International Journal of Molecular Sciences. 2023; 24(17):13340. https://doi.org/10.3390/ijms241713340
Chicago/Turabian StyleSeyhan, Attila A. 2023. "Circulating microRNAs as Potential Biomarkers in Pancreatic Cancer—Advances and Challenges" International Journal of Molecular Sciences 24, no. 17: 13340. https://doi.org/10.3390/ijms241713340






