Untousling the Role of Tousled-like Kinase 1 in DNA Damage Repair
Abstract
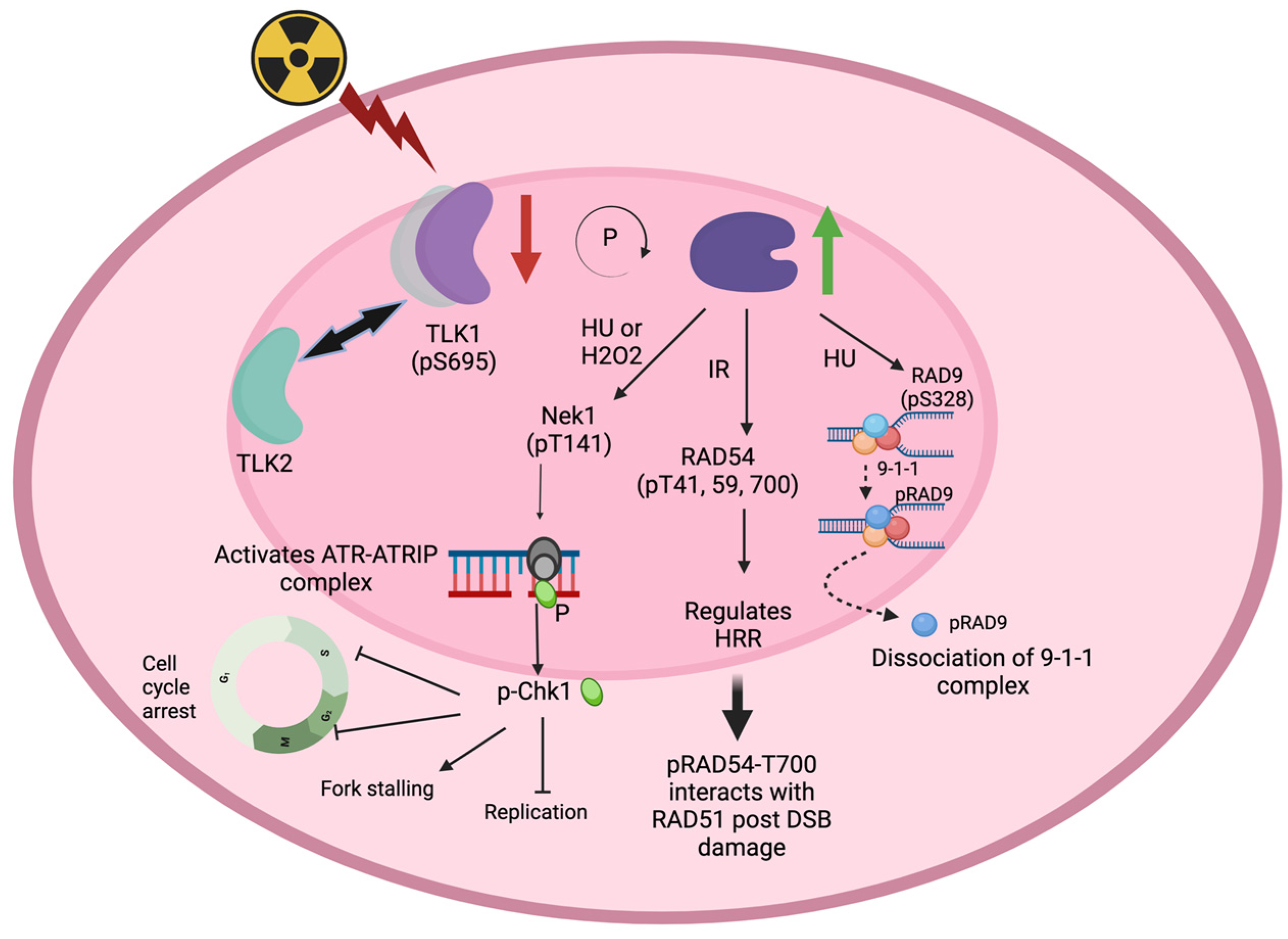
:1. Introduction
2. Role of TLK1 in DSB Repair
3. Role of TLK1 in Regulating HRR Factors
4. Role of TLK1 in Eukaryotic Recombination Repair
5. TLKs and DNA Damage and Checkpoint Functions
6. Role of TLK1 in Cancer
7. Conclusions: Targeting TLK1 for Cancer Treatment
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Silljé, H.H.W.; Takahashi, K.; Tanaka, K.; Van Houwe, G.; Nigg, E.A. Mammalian homologues of the plant Tousled gene code for cell-cycle-regulated kinases with maximal activities linked to ongoing DNA replication. EMBO J. 1999, 18, 5691–5702. [Google Scholar] [CrossRef]
- Li, Y.; DeFatta, R.; Anthony, C.; Sunavala, G.; De Benedetti, A. A translationally regulated Tousled kinase phosphorylates histone H3 and confers radioresistance when overexpressed. Oncogene 2001, 20, 726–738. [Google Scholar] [CrossRef]
- Groth, A.; Lukas, J.; Nigg, E.A.; Silljé, H.H.W.; Wernstedt, C.; Bartek, J.; Hansen, K. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J. 2003, 22, 1676–1687. [Google Scholar] [CrossRef]
- Ghosh, I.; Kwon, Y.; Shabestari, A.B.; Chikhale, R.; Chen, J.; Wiese, C.; Sung, P.; De Benedetti, A. TLK1-Mediated RAD54 Phosphorylation Spatio-Temporally Regulates Homologous Recombination Repair. Nucleic Acids Res. 2023. ahead of print. [Google Scholar] [CrossRef]
- Segura-Bayona, S.; Stracker, T.H. The Tousled-like kinases regulate genome and epigenome stability: Implications in development and disease. Cell. Mol. Life Sci. 2019, 76, 3827–3841. [Google Scholar] [CrossRef]
- Li, Z.; Gourguechon, S.; Wang, C.C. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J. Cell Sci. 2007, 120, 3883–3894. [Google Scholar] [CrossRef]
- Awate, S.; De Benedetti, A. TLK1B mediated phosphorylation of Rad9 regulates its nuclear/cytoplasmic localization and cell cycle checkpoint. BMC Mol. Biol. 2016, 17, 3. [Google Scholar] [CrossRef]
- Sunavala-Dossabhoy, G.; De Benedetti, A. Tousled homolog, TLK1, binds and phosphorylates Rad9; TLK1 acts as a molecular chaperone in DNA repair. DNA Repair 2009, 8, 87–102. [Google Scholar] [CrossRef]
- Klimovskaia, I.M.; Young, C.; Strømme, C.B.; Menard, P.; Jasencakova, Z.; Mejlvang, J.; Ask, K.; Ploug, M.; Nielsen, M.L.; Jensen, O.N.; et al. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nat. Commun. 2014, 5, 3394. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.P.; De Benedetti, A. TLK1B promotes repair of UV-damaged DNA through chromatin remodeling by Asf1. BMC Mol. Biol. 2006, 7, 37. [Google Scholar] [CrossRef]
- Singh, V.; Connelly, Z.M.; Shen, X.; De Benedetti, A. Identification of the proteome complement of humanTLK1 reveals it binds and phosphorylates NEK1 regulating its activity. Cell Cycle 2017, 16, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Mengwasser, K.E.; Adeyemi, R.O.; Leng, Y.; Choi, M.Y.; Clairmont, C.; D’Andrea, A.D.; Elledge, S.J. Genetic Screens Reveal FEN1 and APEX2 as BRCA2 Synthetic Lethal Targets. Mol. Cell 2019, 73, 885–899.e886. [Google Scholar] [CrossRef]
- Kiianitsa, K.; Solinger Jachen, A.; Heyer, W.-D. Terminal association of Rad54 protein with the Rad51–dsDNA filament. Proc. Natl. Acad. Sci. USA 2006, 103, 9767–9772. [Google Scholar] [CrossRef] [PubMed]
- Elisabeth, K.; Reinhard, K.; Hak, C.; Debabrata, S. Sustained Metaphase Arrest in Response to Ionizing Radiation in a Non-small Cell Lung Cancer Cell Line. Radiat. Res. 2008, 169, 46–58. [Google Scholar]
- Kelly, R.; Davey, S.K. Tousled-Like Kinase-Dependent Phosphorylation of Rad9 Plays a Role in Cell Cycle Progression and G2/M Checkpoint Exit. PLoS ONE 2013, 8, e85859. [Google Scholar] [CrossRef] [PubMed]
- Kodym, R.; Mayerhofer, T.; Ortmann, E. Purification and identification of a protein kinase activity modulated by ionizing radiation. Biochem. Biophys. Res. Commun. 2004, 313, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Bhoir, S.; Chikhale, R.V.; Hussain, J.; Dwyer, D.; Bryce, R.A.; Kirubakaran, S.; De Benedetti, A. Generation of Phenothiazine with Potent Anti-TLK1 Activity for Prostate Cancer Therapy. iScience 2020, 23, 101474. [Google Scholar] [CrossRef]
- Adkins, N.L.; Niu, H.; Sung, P.; Peterson, C.L. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 2013, 20, 836–842. [Google Scholar] [CrossRef]
- Uhrig, M.E.; Sharma, N.; Maxwell, P.; Selemenakis, P.; Wiese, C. RAD54L regulates replication fork progression and nascent strand degradation in BRCA1/2-deficient cells. bioRxiv 2023. bioRxiv:2023.07.26.550704. [Google Scholar]
- Ghosh, I.; Khalil, M.I.; Mirza, R.; King, J.; Olatunde, D.; De Benedetti, A. NEK1-Mediated Phosphorylation of YAP1 Is Key to Prostate Cancer Progression. Biomedicines 2023, 11, 734. [Google Scholar] [CrossRef] [PubMed]
- Lisby, M.; Barlow, J.H.; Burgess, R.C.; Rothstein, R. Choreography of the DNA Damage Response: Spatiotemporal Relationships among Checkpoint and Repair Proteins. Cell 2004, 118, 699–713. [Google Scholar]
- Delacroix, S.; Wagner, J.M.; Kobayashi, M.; Yamamoto, K.-i.; Karnitz, L.M. The Rad9–Hus1–Rad1 (9–1–1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007, 21, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Zhang, C.X.; Lieberman, H.B. Rad9 Has a Functional Role in Human Prostate Carcinogenesis. Cancer Res. 2008, 68, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Post, S.M.; Tomkinson, A.E.; Lee, E.Y.H.P. The human checkpoint Rad protein Rad17 is chromatin-associated throughout the cell cycle, localizes to DNA replication sites, and interacts with DNA polymerase ϵ. Nucleic Acids Res. 2003, 31, 5568–5575. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Chang, H.-H.; Wu, K.-J.; Tseng, S.-F.; Lin, C.-C.; Lin, C.-P.; Teng, S.-C. Extrachromosomal Telomeric Circles Contribute to Rad52-, Rad50-, and Polymerase δ-Mediated Telomere-Telomere Recombination in Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 327–336. [Google Scholar] [CrossRef]
- Canfield, C.; Rains, J.; De Benedetti, A. TLK1B promotes repair of DSBs via its interaction with Rad9 and Asf1. BMC Mol. Biol. 2009, 10, 110. [Google Scholar] [CrossRef]
- Brandsma, I.; Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef]
- Melo, J.; Toczyski, D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 2002, 14, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ho, C.K.; Ouyang, J.; Zou, L. Nek1 kinase associates with ATR-ATRIP and primes ATR for efficient DNA damage signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 2175–2180. [Google Scholar] [CrossRef] [PubMed]
- Day, M.; Rappas, M.; Ptasinska, K.; Boos, D.; Oliver, A.W.; Pearl, L.H. BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands. eLife 2018, 7, e39979. [Google Scholar] [CrossRef]
- Hammet, A.; Magill, C.; Heierhorst, J.; Jackson, S.P. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 2007, 8, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Goyal, N.; Rossi, M.J.; Mazina, O.M.; Chi, Y.; Moritz, R.L.; Clurman, B.E.; Mazin, A.V. RAD54 N-terminal domain is a DNA sensor that couples ATP hydrolysis with branch migration of Holliday junctions. Nat. Commun. 2018, 9, 34. [Google Scholar] [CrossRef]
- Maranon, D.G.; Sharma, N.; Huang, Y.; Selemenakis, P.; Wang, M.; Altina, N.; Zhao, W.; Wiese, C. NUCKS1 promotes RAD54 activity in homologous recombination DNA repair. J. Cell Biol. 2020, 219, e201911049. [Google Scholar] [CrossRef]
- Selemenakis, P.; Sharma, N.; Uhrig, M.E.; Katz, J.; Kwon, Y.; Sung, P.; Wiese, C. RAD51AP1 and RAD54L Can Underpin Two Distinct RAD51-Dependent Routes of DNA Damage Repair via Homologous Recombination. Front. Cell Dev. Biol. 2022, 10, 866601. [Google Scholar] [CrossRef]
- Sunavala-Dossabhoy, G.; Li, Y.; Williams, B.; De Benedetti, A. A dominant negative mutant of TLK1 causes chromosome missegregation and aneuploidy in normal breast epithelial cells. BMC Cell Biol. 2003, 4, 16. [Google Scholar] [CrossRef]
- Lee, S.-B.; Segura-Bayona, S.; Villamor-Payà, M.; Saredi, G.; Todd, M.A.M.; Attolini, C.S.-O.; Chang, T.-Y.; Stracker, T.H.; Groth, A. Tousled-like kinases stabilize replication forks and show synthetic lethality with checkpoint and PARP inhibitors. Sci. Adv. 2018, 4, eaat4985. [Google Scholar] [CrossRef]
- Timiri Shanmugam, P.S.; Nair, R.P.; De Benedetti, A.; Caldito, G.; Abreo, F.; Sunavala-Dossabhoy, G. Tousled kinase activator, gallic acid, promotes homologous recombinational repair and suppresses radiation cytotoxicity in salivary gland cells. Free. Radic. Biol. Med. 2016, 93, 217–226. [Google Scholar] [CrossRef]
- Ronald, S.; Awate, S.; Rath, A.; Carroll, J.; Galiano, F.; Dwyer, D.; Kleiner-Hancock, H.; Mathis, J.M.; Vigod, S.; De Benedetti, A. Phenothiazine Inhibitors of TLKs Affect Double-Strand Break Repair and DNA Damage Response Recovery and Potentiate Tumor Killing with Radiomimetic Therapy. Genes Cancer 2013, 4, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, L.; Cao, C.; Song, N.; Wang, Y.; Ma, S.; Zhang, Q.; Yu, N.; Ding, X.; Yang, F.; et al. USP52 acts as a deubiquitinase and promotes histone chaperone ASF1A stabilization. Nat. Commun. 2018, 9, 1285. [Google Scholar] [CrossRef] [PubMed]
- Sukackaite, R.; Cornacchia, D.; Jensen Malene, R.; Mas, P.J.; Blackledge, M.; Enervald, E.; Duan, G.; Auchynnikava, T.; Köhn, M.; Hart, D.J.; et al. Mouse Rif1 is a regulatory subunit of protein phosphatase 1 (PP1). Sci. Rep. 2017, 7, 2119. [Google Scholar] [CrossRef]
- Escribano-Díaz, C.; Orthwein, A.; Fradet-Turcotte, A.; Xing, M.; Young Jordan, T.F.; Tkáč, J.; Cook Michael, A.; Rosebrock Adam, P.; Munro, M.; Canny Marella, D.; et al. A Cell Cycle-Dependent Regulatory Circuit Composed of 53BP1-RIF1 and BRCA1-CtIP Controls DNA Repair Pathway Choice. Mol. Cell 2013, 49, 872–883. [Google Scholar] [CrossRef]
- Batenburg, N.L.; Walker, J.R.; Noordermeer, S.M.; Moatti, N.; Durocher, D.; Zhu, X.-D. ATM and CDK2 control chromatin remodeler CSB to inhibit RIF1 in DSB repair pathway choice. Nat. Commun. 2017, 8, 1921. [Google Scholar] [CrossRef]
- Tang, M.; Chen, Z.; Wang, C.; Feng, X.; Lee, N.; Huang, M.; Zhang, H.; Li, S.; Xiong, Y.; Chen, J. Histone chaperone ASF1 acts with RIF1 to promote DNA end-joining in BRCA1-deficient cells. J. Biol. Chem. 2022, 298, 101979. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Blackburn, E.H. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J. Cell Biol. 2004, 167, 819–830. [Google Scholar] [CrossRef]
- Segura-Bayona, S.; Villamor-Payà, M.; Attolini, C.S.-O.; Koenig, L.M.; Sanchiz-Calvo, M.; Boulton, S.J.; Stracker, T.H. Tousled-Like Kinases Suppress Innate Immune Signaling Triggered by Alternative Lengthening of Telomeres. Cell Rep. 2020, 32, 107983. [Google Scholar] [CrossRef]
- Carrera, P.; Moshkin, Y.M.; Grönke, S.; Silljé, H.H.W.; Nigg, E.A.; Jäckle, H.; Karch, F. Tousled-like kinase functions with the chromatin assembly pathway regulating nuclear divisions. Genes Dev. 2003, 17, 2578–2590. [Google Scholar] [CrossRef]
- Han, Z.; Riefler, G.M.; Saam, J.R.; Mango, S.E.; Schumacher, J.M. The C. elegans Tousled-like Kinase Contributes to Chromosome Segregation as a Substrate and Regulator of the Aurora B Kinase. Curr. Biol. 2005, 15, 894–904. [Google Scholar] [CrossRef]
- Korsholm, L.M.; Gál, Z.; Nieto, B.; Quevedo, O.; Boukoura, S.; Lund, C.C.; Larsen, D.H. Recent advances in the nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2020, 48, 9449–9461. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Kokuryo, T.; Yokoyama, Y.; Ito, S.; Nagino, M.; Hamaguchi, M.; Senga, T. Silencing of Tousled-like kinase 1 sensitizes cholangiocarcinoma cells to cisplatin-induced apoptosis. Cancer Lett. 2010, 296, 27–34. [Google Scholar] [CrossRef]
- Lairmore, T.C.; Abdulsattar, J.; De Benedetti, A.; Shi, R.; Huang, S.; Khalil, M.I.; Witt, S.N. Loss of tumor suppressor menin expression in high grade cholangiocarcinomas. BMC Res. Notes 2023, 16, 15. [Google Scholar] [CrossRef]
- Singh, V.; Jaiswal, P.K.; Ghosh, I.; Koul, H.K.; Yu, X.; De Benedetti, A. The TLK1-Nek1 axis promotes prostate cancer progression. Cancer Lett. 2019, 453, 131–141. [Google Scholar] [CrossRef]
- Singh, V.; Jaiswal, P.K.; Ghosh, I.; Koul, H.K.; Yu, X.; De Benedetti, A. Targeting the TLK1/NEK1 DDR axis with Thioridazine suppresses outgrowth of androgen independent prostate tumors. Int. J. Cancer 2019, 145, 1055–1067. [Google Scholar] [CrossRef]
- Ibrahim, K.; Abdul Murad, N.A.; Harun, R.; Jamal, R.; Ibrahim, K.; Abdul Murad, N.A.; Harun, R.; Jamal, R.; Ibrahim, K.; Abdul Murad, N.A.; et al. Knockdown of Tousled-like kinase 1 inhibits survival of glioblastoma multiforme cells. Int. J. Mol. Med. 2020, 46, 685–699. [Google Scholar] [CrossRef]
- Jiang, J.; Jia, P.; Zhao, Z.; Shen, B. Key regulators in prostate cancer identified by co-expression module analysis. BMC Genom. 2014, 15, 1015. [Google Scholar] [CrossRef]
- Kim, J.-A.; Anurag, M.; Veeraraghavan, J.; Schiff, R.; Li, K.; Wang, X.-S. Amplification of TLK2 Induces Genomic Instability via Impairing the G2–M Checkpoint. Mol. Cancer Res. 2016, 14, 920–927. [Google Scholar] [CrossRef]
- Lin, M.; Yao, Z.; Zhao, N.; Zhang, C. TLK2 enhances aggressive phenotypes of glioblastoma cells through the activation of SRC signaling pathway. Cancer Biol. Ther. 2019, 20, 101–108. [Google Scholar] [CrossRef]
- Van Roy, N.; Vandesompele, J.; Berx, G.; Staes, K.; Van Gele, M.; De Smet, E.; De Paepe, A.; Laureys, G.; van der Drift, P.; Versteeg, R.; et al. Localization of the 17q breakpoint of a constitutional 1;17 translocation in a patient with neuroblastoma within a 25-kb segment located between the ACCN1 and TLK2 genes and near the distal breakpoints of two microdeletions in neurofibromatosis type 1 patients. Genes Chromosom. Cancer 2002, 35, 113–120. [Google Scholar]
- Shaaban, M.; Othman, H.; Ibrahim, T.; Ali, M.; Abdelmoaty, M.; Abdel-Kawi, A.R.; Mostafa, A.; El Nakeeb, A.; Emam, H.; Refaat, A. Immune Checkpoint Regulators: A New Era Toward Promising Cancer Therapy. Curr. Cancer Drug Targets 2020, 20, 429–460. [Google Scholar] [CrossRef]
- Lentz, R.W.; Colton, M.D.; Mitra, S.S.; Messersmith, W.A. Innate Immune Checkpoint Inhibitors: The Next Breakthrough in Medical Oncology? Mol. Cancer Ther. 2021, 20, 961–974. [Google Scholar] [CrossRef]
- Mortuza, G.B.; Hermida, D.; Pedersen, A.-K.; Segura-Bayona, S.; López-Méndez, B.; Redondo, P.; Rüther, P.; Pozdnyakova, I.; Garrote, A.M.; Muñoz, I.G.; et al. Molecular basis of Tousled-Like Kinase 2 activation. Nat. Commun. 2018, 9, 2535. [Google Scholar] [CrossRef]
- Bhoir, S.; De Benedetti, A. Targeting Prostate Cancer, the ‘Tousled Way’. Int. J. Mol. Sci. 2023, 24, 11100. [Google Scholar] [CrossRef]
- Khalil, M.I.; De Benedetti, A. Tousled-like kinase 1: A novel factor with multifaceted role in mCRPC progression and development of therapy resistance. Cancer Drug Resist. 2022, 5, 93–101. [Google Scholar] [CrossRef]
- Singh, V.; Khalil, M.I.; De Benedetti, A. The TLK1/Nek1 axis contributes to mitochondrial integrity and apoptosis prevention via phosphorylation of VDAC1. Cell Cycle 2020, 19, 363–375. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghosh, I.; De Benedetti, A. Untousling the Role of Tousled-like Kinase 1 in DNA Damage Repair. Int. J. Mol. Sci. 2023, 24, 13369. https://doi.org/10.3390/ijms241713369
Ghosh I, De Benedetti A. Untousling the Role of Tousled-like Kinase 1 in DNA Damage Repair. International Journal of Molecular Sciences. 2023; 24(17):13369. https://doi.org/10.3390/ijms241713369
Chicago/Turabian StyleGhosh, Ishita, and Arrigo De Benedetti. 2023. "Untousling the Role of Tousled-like Kinase 1 in DNA Damage Repair" International Journal of Molecular Sciences 24, no. 17: 13369. https://doi.org/10.3390/ijms241713369
APA StyleGhosh, I., & De Benedetti, A. (2023). Untousling the Role of Tousled-like Kinase 1 in DNA Damage Repair. International Journal of Molecular Sciences, 24(17), 13369. https://doi.org/10.3390/ijms241713369







