Synthesis, Conformational Analysis and Antitumor Activity of the Naturally Occurring Antimicrobial Medium-Length Peptaibol Pentadecaibin and Spin-Labeled Analogs Thereof
Abstract
:1. Introduction
2. Results
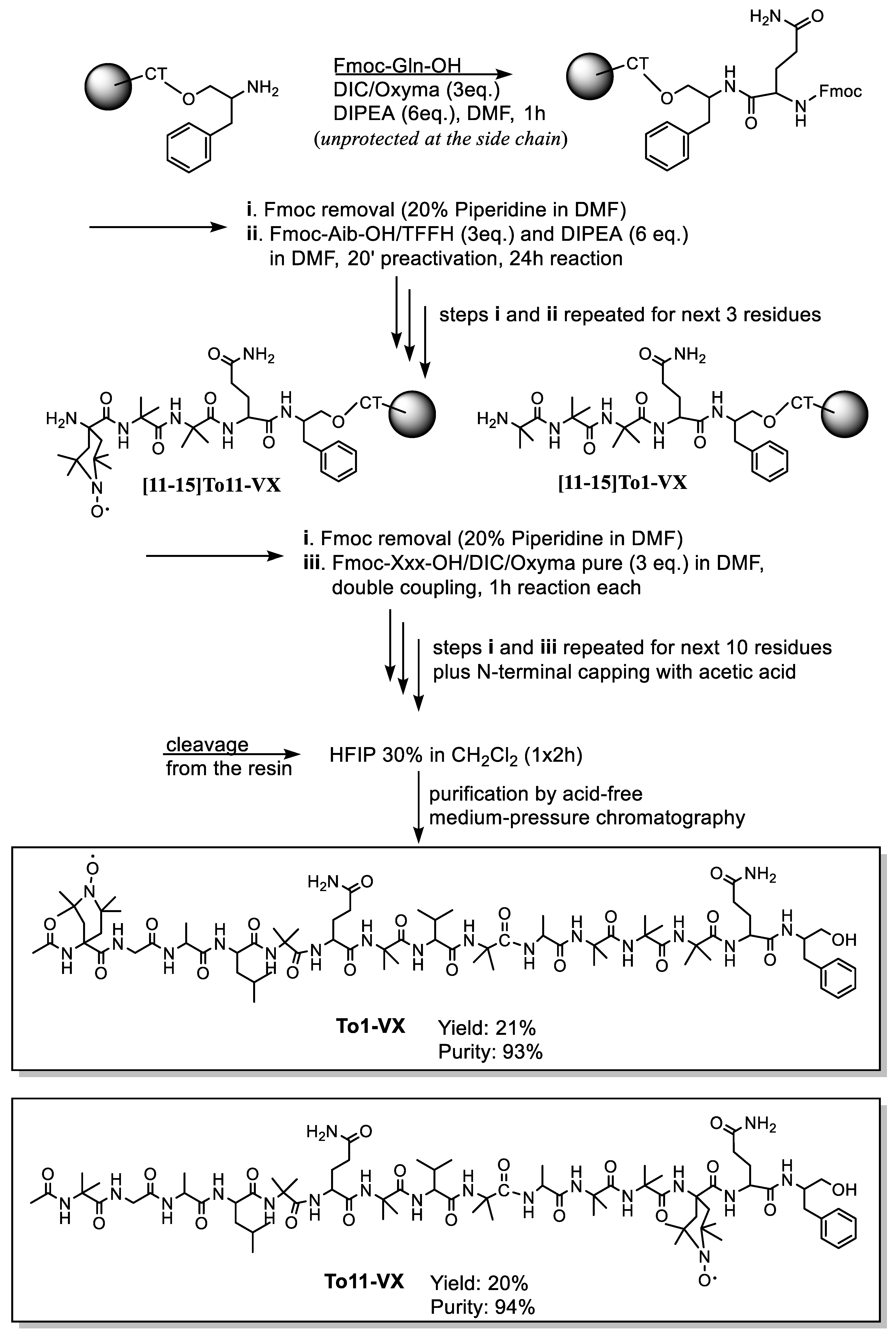
2.1. Peptide Synthesis
2.2. Electron Paramagnetic Resonance Characterization
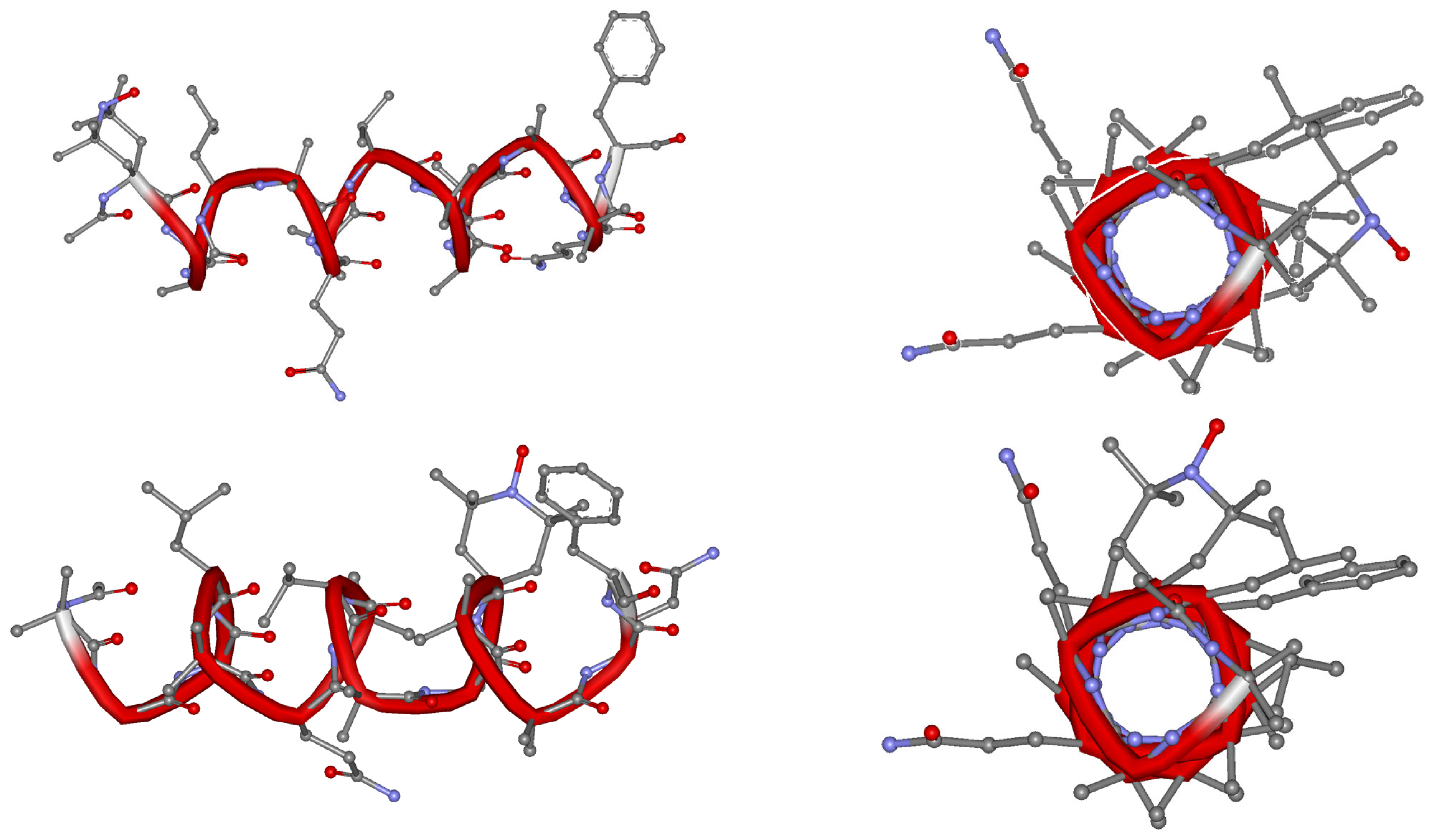
2.3. Circular Dichroism (CD)
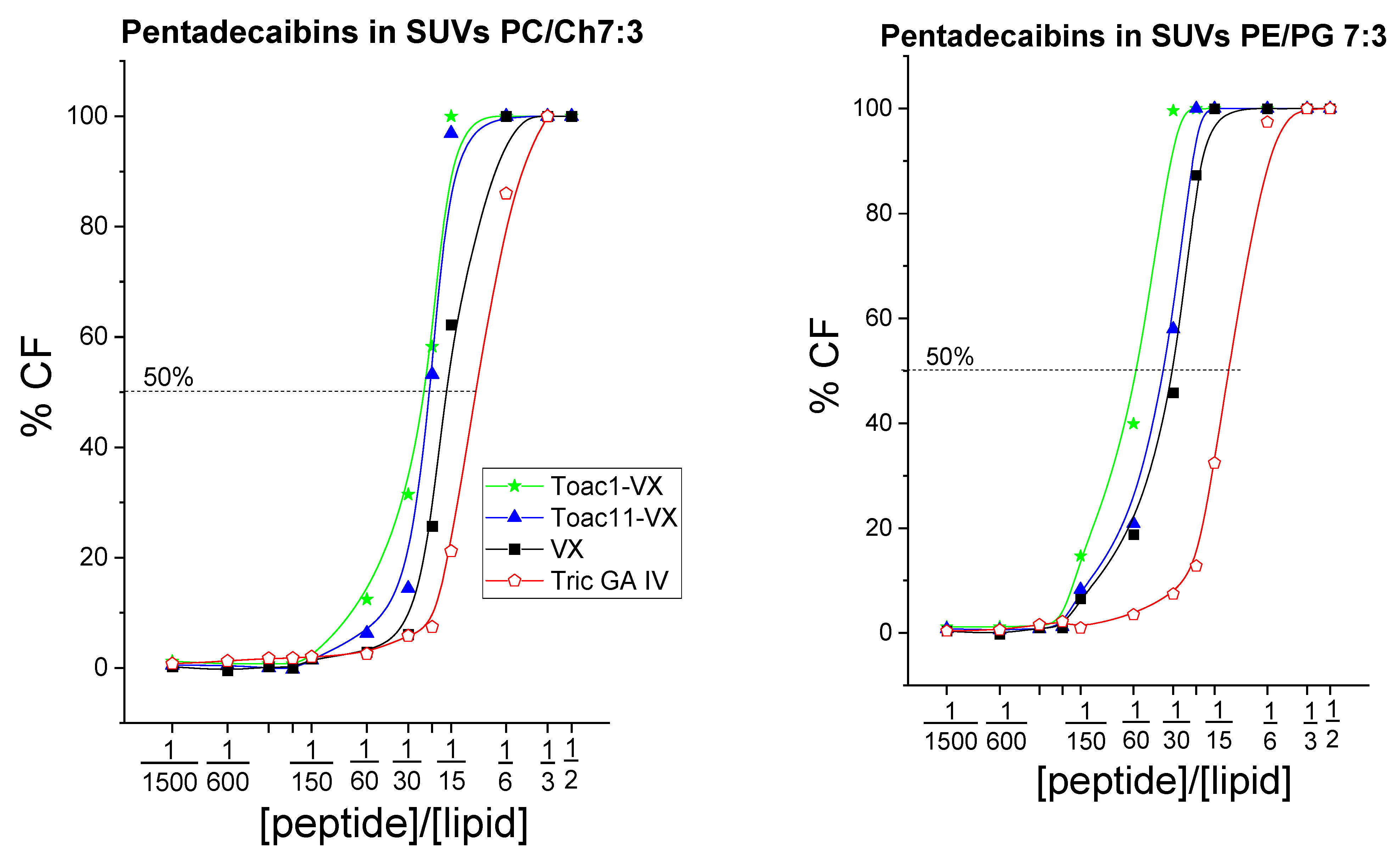
2.4. Peptide-Induced Leakage from Model Liposomes
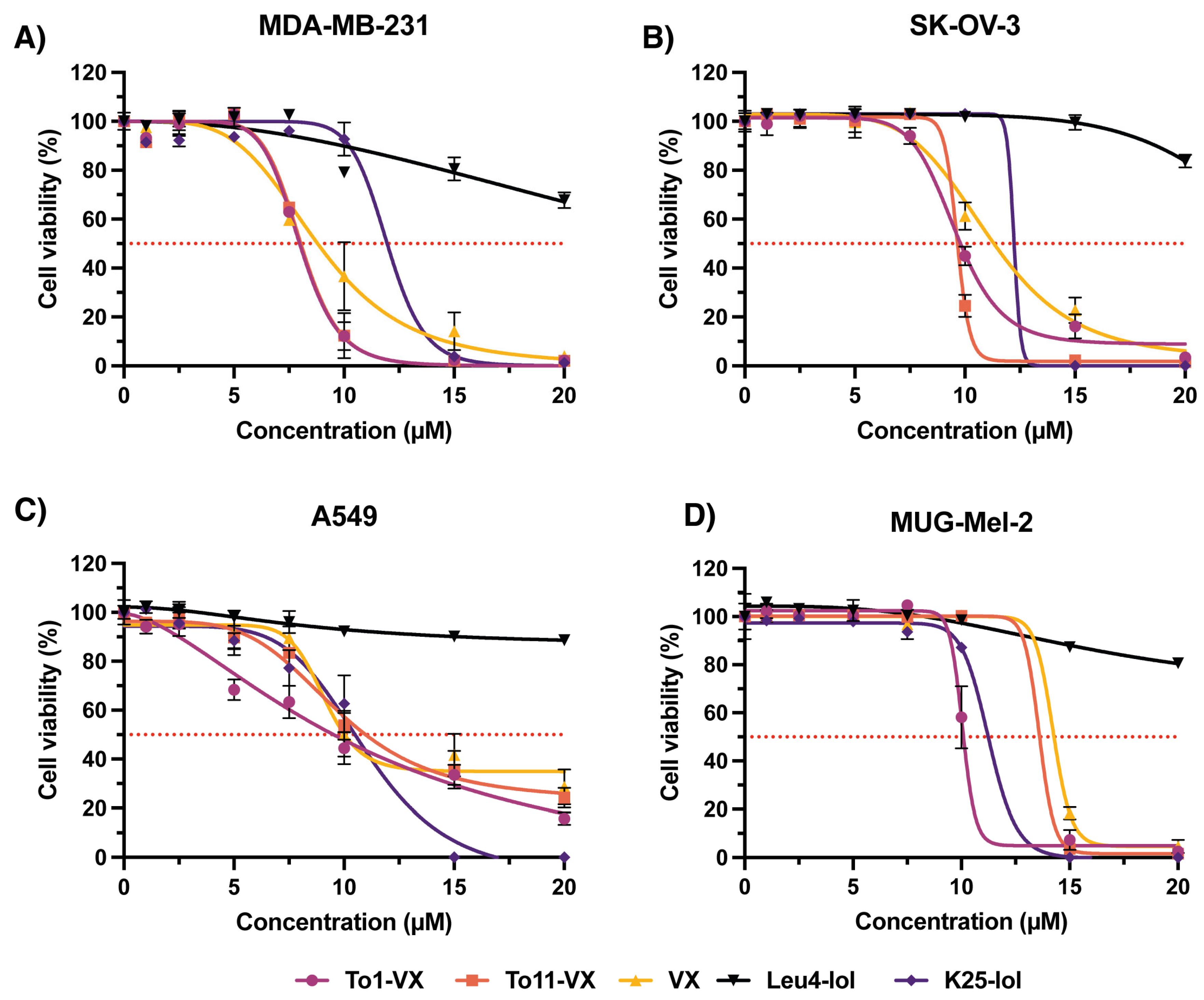
2.5. In Vitro Activity of Pentadecaibins against Human Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. EPR
4.3. CD Measurements
4.4. Leakage Assay
4.5. Antitumor Activity Studies
4.5.1. Cell Lines
4.5.2. In Vitro Cytotoxicity of Peptaibols
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 29 June 2023).
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020. Available online: https://gco.iarc.fr/today (accessed on 29 June 2023).
- McIntosh, S.A.; Alam, F.; Adams, L.; Boon, I.S.; Callaghan, J.; Conti, I.; Copson, E.; Carson, V.; Davidson, M.; Fitzgerald, H.; et al. Global funding for cancer research between 2016 and 2020: A content analysis of public and philanthropic investments. Lancet Oncol. 2023, 24, 636–645. [Google Scholar] [CrossRef]
- Nayab, S.; Aslam, M.A.; Rahman, S.U.; Sindhu, Z.U.D.; Sajid, S.; Zafar, N.; Razaq, M.; Kanwar, R.; Amanullah. A Review of Antimicrobial Peptides: Its Function, Mode of Action and Therapeutic Potential. Int. J. Pept. Res. Ther. 2022, 28, 46. [Google Scholar] [CrossRef]
- Bakare, O.O.; Gokul, A.; Wu, R.; Niekerk, L.A.; Klein, A.; Keyster, M. Biomedical Relevance of Novel Anticancer Peptides in the Sensitive Treatment of Cancer. Biomolecules 2021, 11, 1120. [Google Scholar] [CrossRef]
- Galdiero, E.; Lombardi, L.; Falanga, A.; Libralato, G.; Guida, M.; Carotenuto, R. Biofilms: Novel strategies based on antimicrobial peptides. Pharmaceutics 2019, 11, 322. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Vishwanatha, J.K. Role of Anti-Cancer Peptides as Immunomodulatory Agents: Potential and Design Strategy. Pharmaceutics 2022, 14, 2686. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Deslouches, B.; Di, Y.P. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 2017, 8, 46635–46651. [Google Scholar] [CrossRef] [PubMed]
- Del Genio, V.; Bellavita, R.; Falanga, A.; Hervé-Aubert, K.; Chourpa, I.; Galdiero, S. Peptides to Overcome the Limitations of Current Anticancer and Antimicrobial Nanotherapies. Pharmaceutics 2022, 14, 1235. [Google Scholar] [CrossRef]
- Vadevoo, S.M.P.; Gurung, S.; Lee, H.-S.; Gunassekaran, G.R.; Lee, S.-M.; Yoon, J.-W.; Lee, Y.-K.; Lee, B. Peptides as multifunctional players in cancer therapy. Exp. Mol. Med. 2023, 55, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Stoppacher, N.; Neumann, N.K.N.; Burgstaller, L.; Zeilinger, S.; Degenkolb, T.; Brückner, H.; Schuhmacher, R. The comprehensive peptaibiotics database. Chem. Biodivers. 2013, 10, 734–743. [Google Scholar] [CrossRef]
- Szekeres, A.; Leitgeb, B.; Kredics, L.; Antal, Z.; Hatvani, L.; Manczinger, L.; Vágvölgyi, C. Peptaibols and related peptaibiotics of Trichoderma. A review. Acta Microbiol. Immunol. Hung. 2005, 52, 137–168. [Google Scholar] [CrossRef]
- De Zotti, M.; Biondi, B.; Formaggio, F.; Toniolo, C.; Stella, L.; Park, Y.; Hahm, K.-S. Trichogin GA IV: An antibacterial and protease-resistant peptide. J. Pept. Sci. 2009, 15, 615–619. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Kodama, H.; Osada, S.; Kato, F.; Jelokhani-Niaraki, M.; Kondo, M. Effect of α,α-Dialkyl Amino Acids on the Protease Resistance of Peptides. Biosci. Biotechnol. Biochem. 2003, 67, 2269–2272. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Zhang, M.; Wu, P.; Li, T.; Li, W.; Zhang, L.; Yue, Q.; Chen, X.; Wei, X.; Xu, Y.; et al. Halovirs I–K, antibacterial and cytotoxic lipopeptaibols from the plant pathogenic fungus Paramyrothecium roridum NRRL 2183. J. Antibiot. 2022, 75, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Rebuffat, S.; Goulard, C.; Bodo, B.; Roquebert, M.-F. The peptaibol antibiotics from Trichoderma soil fungi: Structural diversity and membrane properties. Recent Res. Dev. Org. Bioorg. Chem. 1999, 3, 65–91. [Google Scholar]
- Bong-Sik, Y.; Ick-Dong, Y.; Young Ho, K.; Young-Sook, K.; Sang-Jun, L.; Kab-Sig, K.; Woon-Hyung, Y. Peptaivirins A and B, two new antiviral peptaibols against TMV infection. Tetrahedron Lett. 2000, 41, 1429–1431. [Google Scholar] [CrossRef]
- He, H.; Janso, J.E.; Yang, H.Y.; Bernan, V.S.; Lin, S.L.; Yu, K. Culicinin D, an antitumor peptaibol produced by the fungus Culicinomyces clavisporus, strain LL-12I252. J. Nat. Prod. 2006, 69, 736–741. [Google Scholar] [CrossRef]
- Gavryushina, I.A.; Georgieva, M.L.; Kuvarina, A.E.; Sadykova, V.S. Peptaibols as Potential Antifungal and Anticancer Antibiotics: Current and Foreseeable Development (Review). Appl. Biochem. Microbiol. 2021, 57, 556–563. [Google Scholar] [CrossRef]
- Casagrande, N.; Borghese, C.; Gabbatore, L.; Morbiato, L.; De Zotti, M.; Aldinucci, D. Analogs of a Natural Peptaibol Exert Anticancer Activity in Both Cisplatin- and Doxorubicin-Resistant Cells and in Multicellular Tumor Spheroids. Int. J. Mol. Sci. 2021, 22, 8362. [Google Scholar] [CrossRef]
- Moret, F.; Menilli, L.; Milani, C.; Di Cintio, G.; Dalla Torre, C.; Amendola, V.; De Zotti, M. Anticancer and Targeting Activity of Phytopharmaceutical Structural Analogs of a Natural Peptide from Trichoderma longibrachiatum and Related Peptide-Decorated Gold Nanoparticles. Int. J. Mol. Sci. 2023, 24, 5537. [Google Scholar] [CrossRef]
- Dalla Torre, C.; Sannio, F.; Battistella, M.; Docquier, J.-D.; De Zotti, M. Peptaibol Analogs Show Potent Antibacterial Activity against Multidrug Resistant Opportunistic Pathogens. Int. J. Mol. Sci. 2023, 24, 7997. [Google Scholar] [CrossRef]
- Lizio, M.G.; Campana, M.; De Poli, M.; Jefferies, D.F.; Cullen, W.; Andrushchenko, V.; Chmel, N.P.; Bouř, P.; Khalid, S.; Clayden, J.; et al. Insight into the Mechanism of Action and Peptide-Membrane Interactions of Aib-Rich Peptides: Multitechnique Experimental and Theoretical Analysis. ChemBioChem 2021, 22, 1656–1667. [Google Scholar] [CrossRef]
- Bobone, S.; Gerelli, Y.; De Zotti, M.; Bocchinfuso, G.; Farrotti, A.; Orioni, B.; Sebastiani, F.; Latter, E.; Penfold, J.; Senesi, R.; et al. Membrane thickness and the mechanism of action of the short peptaibol trichogin GA IV. Biochim. Biophys. Acta-Biomembr. 2013, 1828, 1013–1024. [Google Scholar] [CrossRef]
- Eid, M.; Rippa, S.; Castano, S.; Desbat, B.; Chopineau, J.; Rossi, C.; Béven, L. Exploring the membrane mechanism of the bioactive peptaibol ampullosporin a using lipid monolayers and supported biomimetic membranes. J. Biophys. 2010, 2010, 179641. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta-Biomembr. 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Salnikov, E.S.; Friedrich, H.; Li, X.; Bertani, P.; Reissmann, S.; Hertweck, C.; O’Neil, J.D.J.; Raap, J.; Bechinger, B. Structure and Alignment of the Membrane-Associated Peptaibols Ampullosporin A and Alamethicin by Oriented 15N and 31P Solid-State NMR Spectroscopy. Biophys. J. 2009, 96, 86–100. [Google Scholar] [CrossRef]
- Fox, R.O., Jr.; Richards, F.M. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-A resolution. Nature 1982, 300, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Syryamina, V.N.; De Zotti, M.; Toniolo, C.; Formaggio, F.; Dzuba, S.A. Alamethicin self-assembling in lipid membranes: Concentration dependence from pulsed EPR of spin labels. Phys. Chem. Chem. Phys. 2018, 20, 3592–3601. [Google Scholar] [CrossRef] [PubMed]
- Milov, A.D.; Tsvetkov, Y.D.; Raap, J.; De Zotti, M.; Formaggio, F.; Toniolo, C. Conformation, self-aggregation, and membrane interaction of peptaibols as studied by pulsed electron double resonance spectroscopy. Biopolymers 2016, 106, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, T.I.; Smirnov, A.I. Peptide-Membrane Interactions by Spin-Labeling EPR. Methods Enzymol. 2015, 564, 219–258. [Google Scholar] [CrossRef]
- Vicente, E.F.; Basso, L.G.M.; Cespedes, G.F.; Lorenzón, E.N.; Castro, M.S.; Mendes-Giannini, M.J.S.; Costa-Filho, A.J.; Cilli, E.M. Dynamics and Conformational Studies of TOAC Spin Labeled Analogues of Ctx(Ile21)-Ha Peptide from Hypsiboas albopunctatus. PLoS ONE 2013, 8, e60818. [Google Scholar] [CrossRef] [PubMed]
- Sammalkorpi, M.; Lazaridis, T. Modeling a Spin-Labeled Fusion Peptide in a Membrane: Implications for the Interpretation of EPR Experiments. Biophys. J. 2007, 92, 10–22. [Google Scholar] [CrossRef]
- Biondi, B.; Syryamina, V.N.; Rocchio, G.; Barbon, A.; Formaggio, F.; Toniolo, C.; Raap, J.; Dzuba, S.A. Is Cys(MTSL) the Best α-Amino Acid Residue to Electron Spin Labeling of Synthetically Accessible Peptide Molecules with Nitroxides? ACS Omega 2022, 7, 5154–5165. [Google Scholar] [CrossRef] [PubMed]
- Schreier, S.; Bozelli, J.C., Jr.; Marín, N.; Vieira, R.F.; Nakaie, C.R. The spin label amino acid TOAC and its uses in studies of peptides: Chemical, physicochemical, spectroscopic, and conformational aspects. Biophys. Rev. 2012, 4, 45–66. [Google Scholar] [CrossRef]
- Shabestari, M.H.; van Son, M.; Moretto, A.; Crisma, M.; Toniolo, C.; Huber, M. Conformation and EPR characterization of rigid, 310-helical peptides with TOAC spin labels: Models for short distances. Biopolymers 2014, 102, 244–251. [Google Scholar] [CrossRef]
- Hanson, P.; Martinez, G.; Millhauser, G.; Formaggio, F.; Crisma, M.; Toniolo, C.; Vita, C. Distinguishing Helix Conformations in Alanine-Rich Peptides Using the Unnatural Amino Acid TOAC and Electron Spin Resonance. J. Am. Chem. Soc. 1996, 118, 271–272. [Google Scholar] [CrossRef]
- Gobbo, M.; Merli, E.; Biondi, B.; Oancea, S.; Toffoletti, A.; Formaggio, F.; Toniolo, C. Synthesis and preliminary conformational analysis of TOAC spin-labeled analogues of the medium-length peptaibiotic tylopeptin B. J. Pept. Sci. 2012, 18, 37–44. [Google Scholar] [CrossRef]
- Bortolus, M.; Dalzini, A.; Formaggio, F.; Toniolo, C.; Gobbo, M.; Maniero, A.L. An EPR study of ampullosporin A, a medium-length peptaibiotic, in bicelles and vesicles. Phys. Chem. Chem. Phys. 2016, 18, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Ishiyama, D.; Satou, T.; Senda, H.; Fujimaki, T.; Honda, R.; Kanazawa, S. Heptaibin, a novel antifungal peptaibol antibiotic from Emericellopsis sp. BAUA8289. J. Antibiot. 2000, 53, 728–732. [Google Scholar] [CrossRef]
- Hou, X.; Sun, R.; Feng, Y.; Zhang, R.; Zhu, T.; Che, Q.; Zhang, G.; Li, D. Peptaibols: Diversity, bioactivity, and biosynthesis. Eng. Microbiol. 2022, 2, 100026. [Google Scholar] [CrossRef]
- Vladimir, V.; Ryakhovsky, A.; Ivanov, S. Study of intramolecular aminolysis in peptides containing N-alkylamino acids at position 2. Tetrahedron 2021, 68, 7070–7076. [Google Scholar] [CrossRef]
- Van Bohemen, A.-I.; Ruiz, N.; Zalouk-Vergnoux, A.; Michaud, A.; du Pont, T.R.; Druzhinina, I.; Atanasova, L.; Prado, S.; Bodo, B.; Meslet-Cladiere, L.; et al. Pentadecaibins I-V: 15-residue peptaibols produced by a marine-derived Trichoderma sp. of the Harzianum Clade. J. Nat. Prod. 2021, 84, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Morbiato, L.; Haneen, D.S.A.; Formaggio, F.; De Zotti, M. Total synthesis of the natural, medium-length, peptaibol pentadecaibin and study of the chemical features responsible for its membrane activity. J. Pept. Sci. 2023, 29, e3479. [Google Scholar] [CrossRef] [PubMed]
- De Zotti, M.; Biondi, B.; Peggion, C.; Park, Y.; Hahm, K.S.; Formaggio, F.; Toniolo, C. Synthesis, preferred conformation, protease stability, and membrane activity of heptaibin, a medium-length peptaibiotic. J. Pept. Sci. 2011, 17, 585–594. [Google Scholar] [CrossRef]
- Goodman, M.; Felix, A.; Moroder, L.; Toniolo, C. (Eds.) Houben–Weyl Methods of Organic Chemistry; Synthesis of Peptides and Peptidomimetics; Georg Thieme Verlag: Stuttgart, Germany, 2001; Volume E22a, ISBN 3 132 19604 5. [Google Scholar]
- Monty, O.B.C.; Simmons, N.; Chamakuri, S.; Matzuk, M.M.; Young, D.W. Solution-Phase Fmoc-Based Peptide Synthesis for DNA-Encoded Chemical Libraries: Reaction Conditions, Protecting Group Strategies, and Pitfalls. ACS Comb. Sci. 2020, 22, 833–843. [Google Scholar] [CrossRef]
- Fasman, G.D. (Ed.) Circular Dichroism and the Conformational Analysis of Biomolecules; Plenum Press: New York, NY, USA, 1996. [Google Scholar]
- Toniolo, C.; Polese, A.; Formaggio, F.; Crisma, M.; Kamphuis, J. Circular Dichroism Spectrum of a Peptide 310-Helix. J. Am. Chem. Soc. 1996, 118, 2744–2745. [Google Scholar] [CrossRef]
- Subirós-Funosas, R.; Prohens, R.; Barbas, R.; El-Faham, A.; Albericio, F. Oxyma: An Efficient Additive for Peptide Synthesis to Replace the Benzotriazole-Based HOBt and HOAt with a Lower Risk of Explosion. Chem. Eur. J. 2009, 15, 9394–9403. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Gill, E.E.; Coombs, M.R.P.; Falsafi, R.; Hancock, R.E.W.; Hoskin, D.W. MDA-MB-231 breast cancer cells resistant to pleurocidin-family lytic peptides are chemosensitive and exhibit reduced tumor-forming capacity. Biomolecules 2020, 10, 1220. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Almansour, N.M. Triple-Negative Breast Cancer: A Brief Review About Epidemiology, Risk Factors, Signaling Pathways, Treatment and Role of Artificial Intelligence. Front. Mol. Biosci. 2022, 9, 836417. [Google Scholar] [CrossRef]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer—Expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022, 19, 91–113. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Siddharth, S.; Sharma, D. Triple Negative Breast Cancer: A Mountain Yet to Be Scaled Despite the Triumphs. Cancers 2021, 13, 3697. [Google Scholar] [CrossRef] [PubMed]
- Houthaeve, G.; De Smedt, S.C.; Braeckmans, K.; De Vos, W.H. The cellular response to plasma membrane disruption for nanomaterial delivery. Nano Converg. 2022, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Wang, H.N.; Xie, S.T.; Luo, Y.; Sun, C.Y.; Chen, X.L.; Zhang, Y.Z. Antimicrobial peptaibols, novel suppressors of tumor cells, targeted calcium-mediated apoptosis and autophagy in human hepatocellular carcinoma cells. Mol. Cancer 2010, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.T.; Maruo, T.; Jansen, S.; Roble, J.; Rataiczak, R.D. Comparative Esr Studies of Solid Polycrystalline Alkali Metal Salts of Tcnq and Tcnqf4. Mol. Cryst. Liq. Cryst. 2011, 134, 21–30. [Google Scholar] [CrossRef]


| Acronym | Name | Sequence 1 |
|---|---|---|
| VX | pentadecaibin | Ac-Aib-Gly-Ala-Leu-Aib-Gln-Aib-Val-Aib-Ala-Aib-Aib-Aib-Gln-Phol |
| To1-VX | [Toac1]- pentadecaibin | Ac-Toac-Gly-Ala-Leu-Aib-Gln-Aib-Val-Aib-Ala-Aib-Aib-Aib-Gln-Phol |
| To11-VX | [Toac11]- pentadecaibin | Ac-Aib-Gly-Ala-Leu-Aib-Gln-Aib-Val-Aib-Ala-Toac-Aib-Aib-Gln-Phol |
| Tip | |
|---|---|
| 1 | For Toac-containing pentadecaibins, the use of Gln residues without side-chain protection is recommended. |
| 2 | If acid-sensitive side-chain protections are used, cleave the fully protected peptide from the resin first, then remove protections via treatment with TFA 5% and TIS 2.5% (no water) for 7 h. |
| 3 | Choose the Toac position so that it never has to act as a nucleophile onto an incoming Aib. |
| 4 | Apply TFFH with at least 20’ preactivation for Aib/Toac onto Aib couplings. Reaction time: 24 h. |
| 5 | If TFA has to be used for peptide cleavage from resin, pour the solution from the reaction vessel on fresh CH3OH to avoid the esterification of the C-terminal 1,2-aminoalcohol |
| IC50 (μM) | |||
|---|---|---|---|
| Cancer Cell Line | VX [CI 1] | To1-VX | To11-VX |
| MDA-MB-231 | 8.79 [8.38–9.24] | 7.98 [7.85–8.12] | 8.04 [7.88–8.199] |
| SK-OV-3 | 11.09 [10.25–11.95] | 9.56 [9.38–9.78] | 9.64 [9.24–9.44] |
| A549 | 9.099 [8.50–9.97] | 11.09 [8.38–14.30] | 9.70 [9.005–10.86] |
| MUG-Mel2 | 14.24 | 10.05 | 13.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morbiato, L.; Quaggia, C.; Menilli, L.; Dalla Torre, C.; Barbon, A.; De Zotti, M. Synthesis, Conformational Analysis and Antitumor Activity of the Naturally Occurring Antimicrobial Medium-Length Peptaibol Pentadecaibin and Spin-Labeled Analogs Thereof. Int. J. Mol. Sci. 2023, 24, 13396. https://doi.org/10.3390/ijms241713396
Morbiato L, Quaggia C, Menilli L, Dalla Torre C, Barbon A, De Zotti M. Synthesis, Conformational Analysis and Antitumor Activity of the Naturally Occurring Antimicrobial Medium-Length Peptaibol Pentadecaibin and Spin-Labeled Analogs Thereof. International Journal of Molecular Sciences. 2023; 24(17):13396. https://doi.org/10.3390/ijms241713396
Chicago/Turabian StyleMorbiato, Laura, Celeste Quaggia, Luca Menilli, Chiara Dalla Torre, Antonio Barbon, and Marta De Zotti. 2023. "Synthesis, Conformational Analysis and Antitumor Activity of the Naturally Occurring Antimicrobial Medium-Length Peptaibol Pentadecaibin and Spin-Labeled Analogs Thereof" International Journal of Molecular Sciences 24, no. 17: 13396. https://doi.org/10.3390/ijms241713396
APA StyleMorbiato, L., Quaggia, C., Menilli, L., Dalla Torre, C., Barbon, A., & De Zotti, M. (2023). Synthesis, Conformational Analysis and Antitumor Activity of the Naturally Occurring Antimicrobial Medium-Length Peptaibol Pentadecaibin and Spin-Labeled Analogs Thereof. International Journal of Molecular Sciences, 24(17), 13396. https://doi.org/10.3390/ijms241713396









