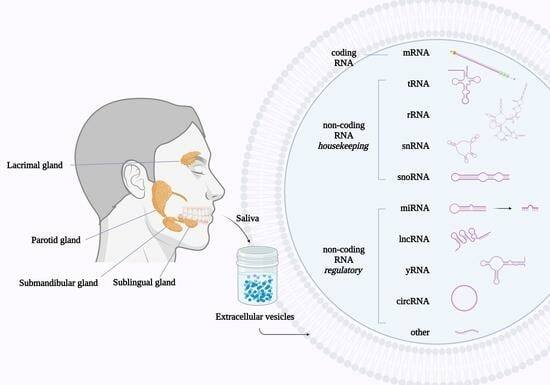
Non-Coding RNA in Salivary Extracellular Vesicles: A New Frontier in Sjögren’s Syndrome Diagnostics?
Abstract
:1. Introduction
2. Results
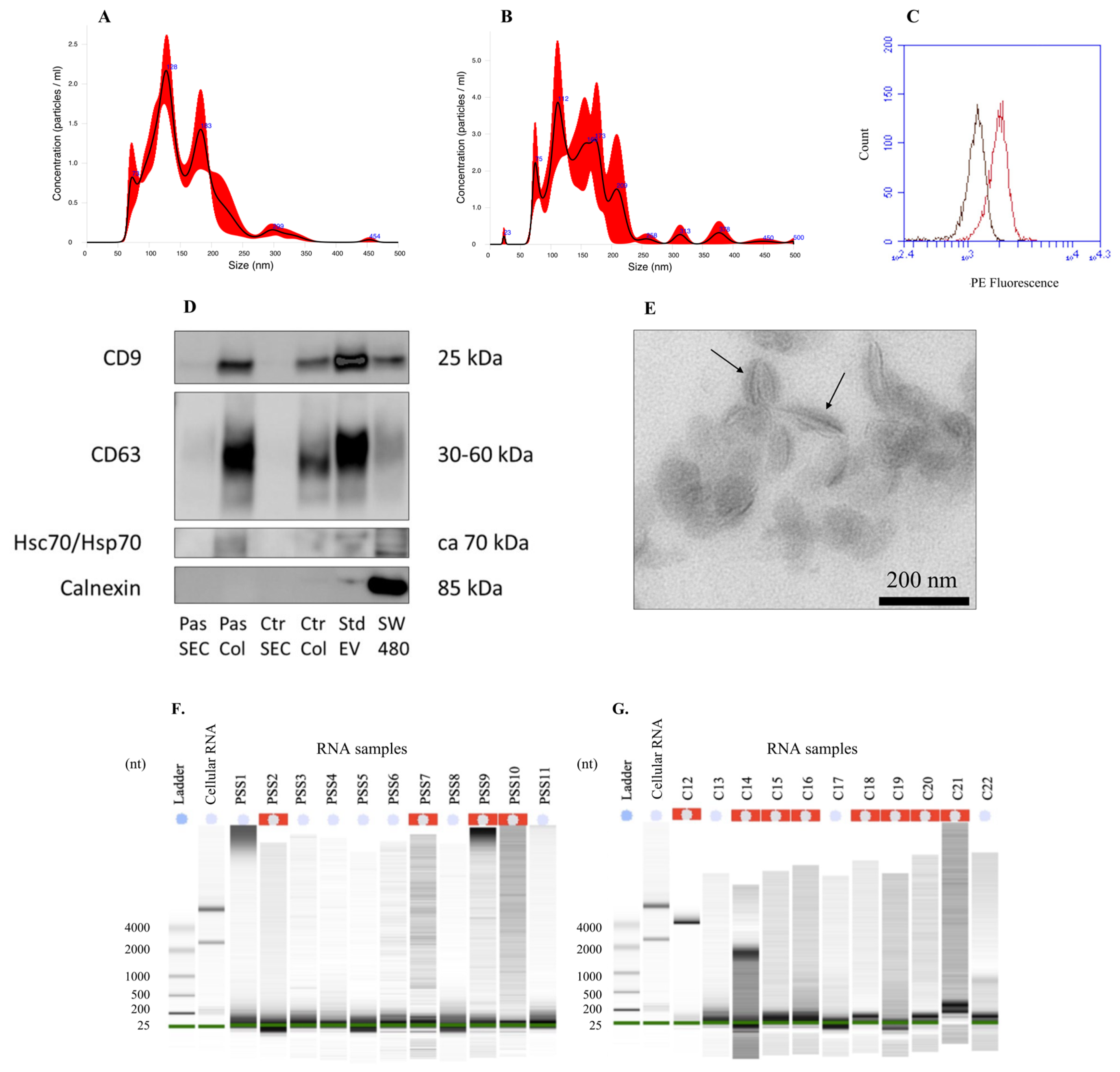
2.1. Extracellular Vesicle Characterization
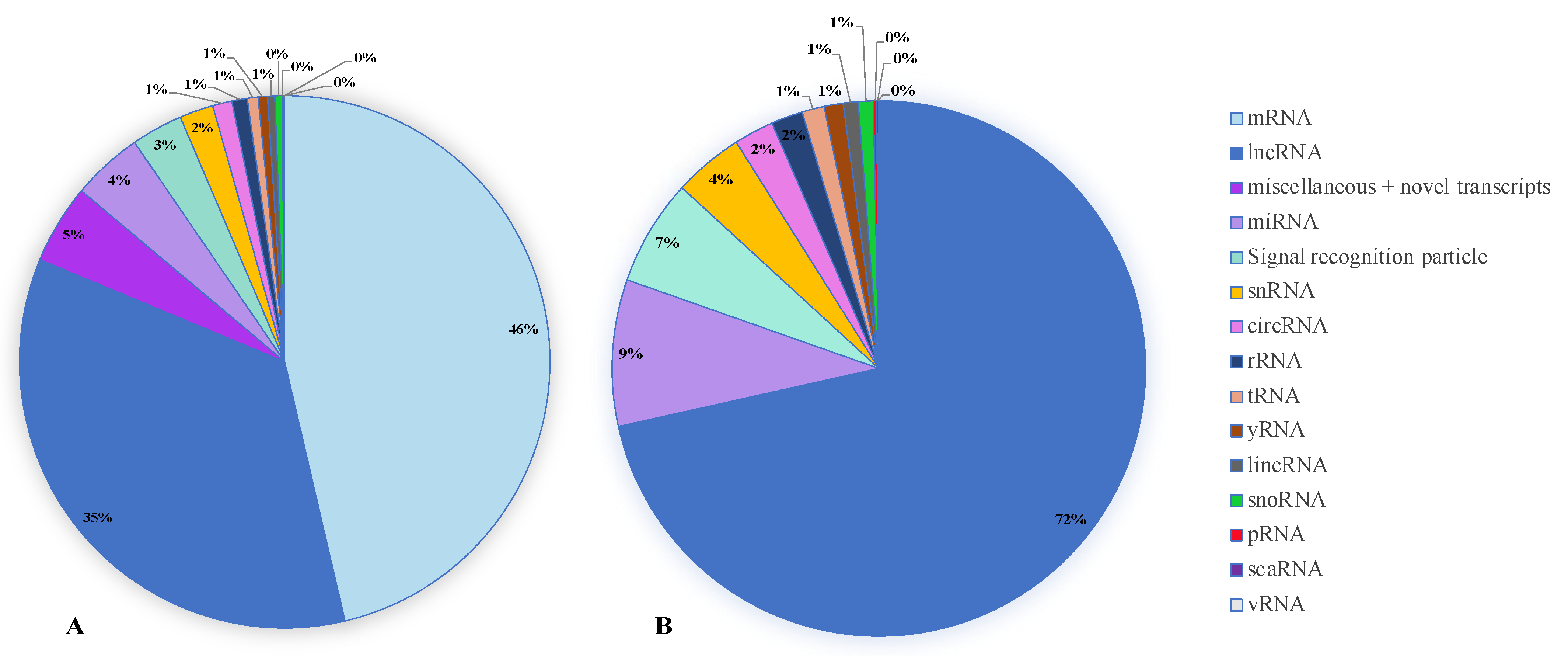
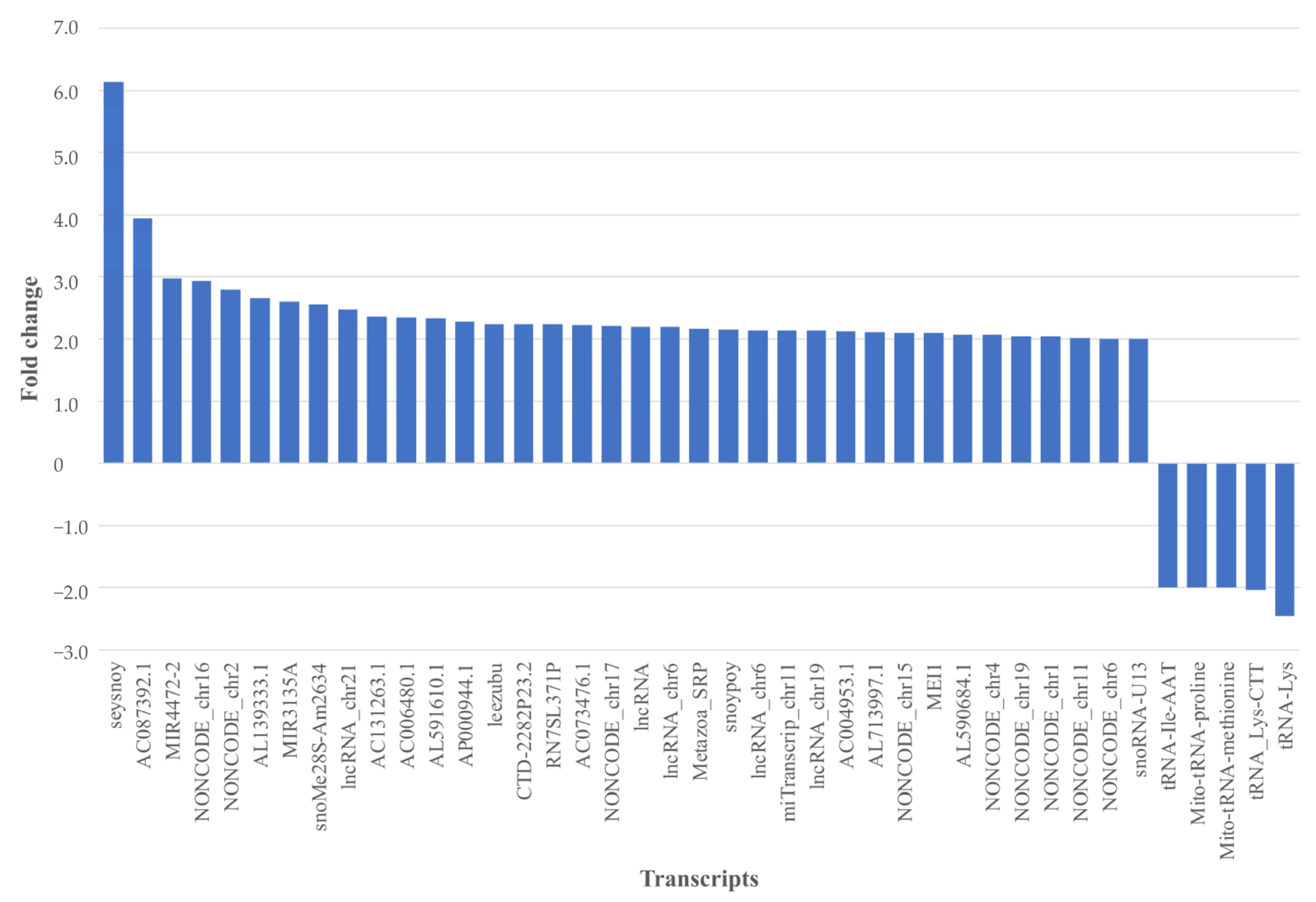
2.2. Salivary EV-RNA Analysis Using Affymetrix Clariom™ D Microarrays
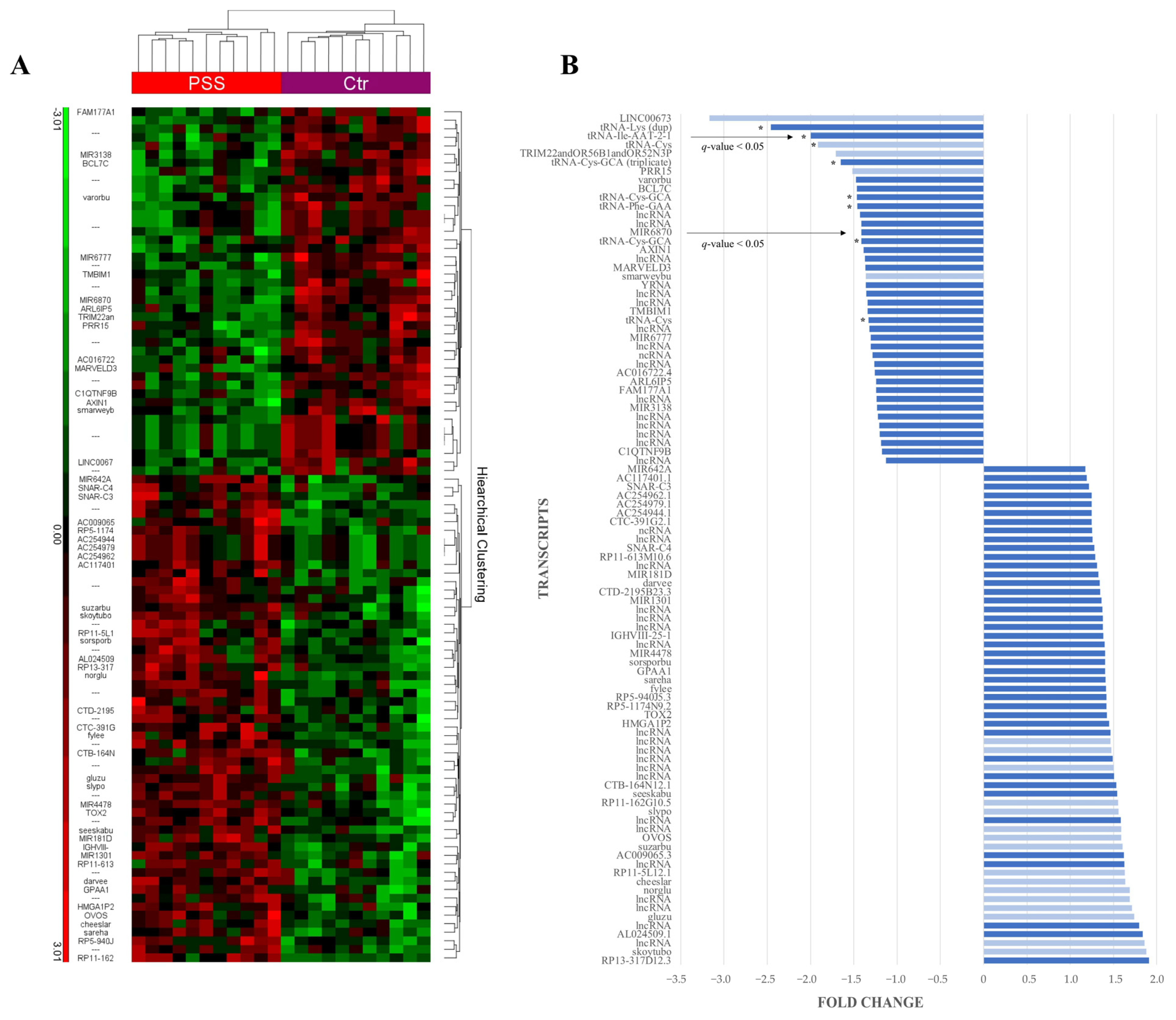
2.2.1. Comparing Affymetrix Microarray Results between pSS Patient and Control Groups
2.2.2. RNA Validation
3. Discussion
3.1. A Comparison of the mRNA Transcripts in Salivary EVs between pSS Patients and Controls
3.2. A Comparison of the miRNA Transcripts in Salivary EVs between pSS Patients and Controls
3.3. A Comparison of the tRNA Transcripts in Salivary EVs between pSS Patients and Controls
3.4. A Comparison of the yRNA Transcripts in Salivary EVs between pSS Patients and Controls
3.5. A Comparison of the lncRNA Transcripts in Salivary EVs between pSS Patients and Controls
3.6. RNA in EVs—General Considerations
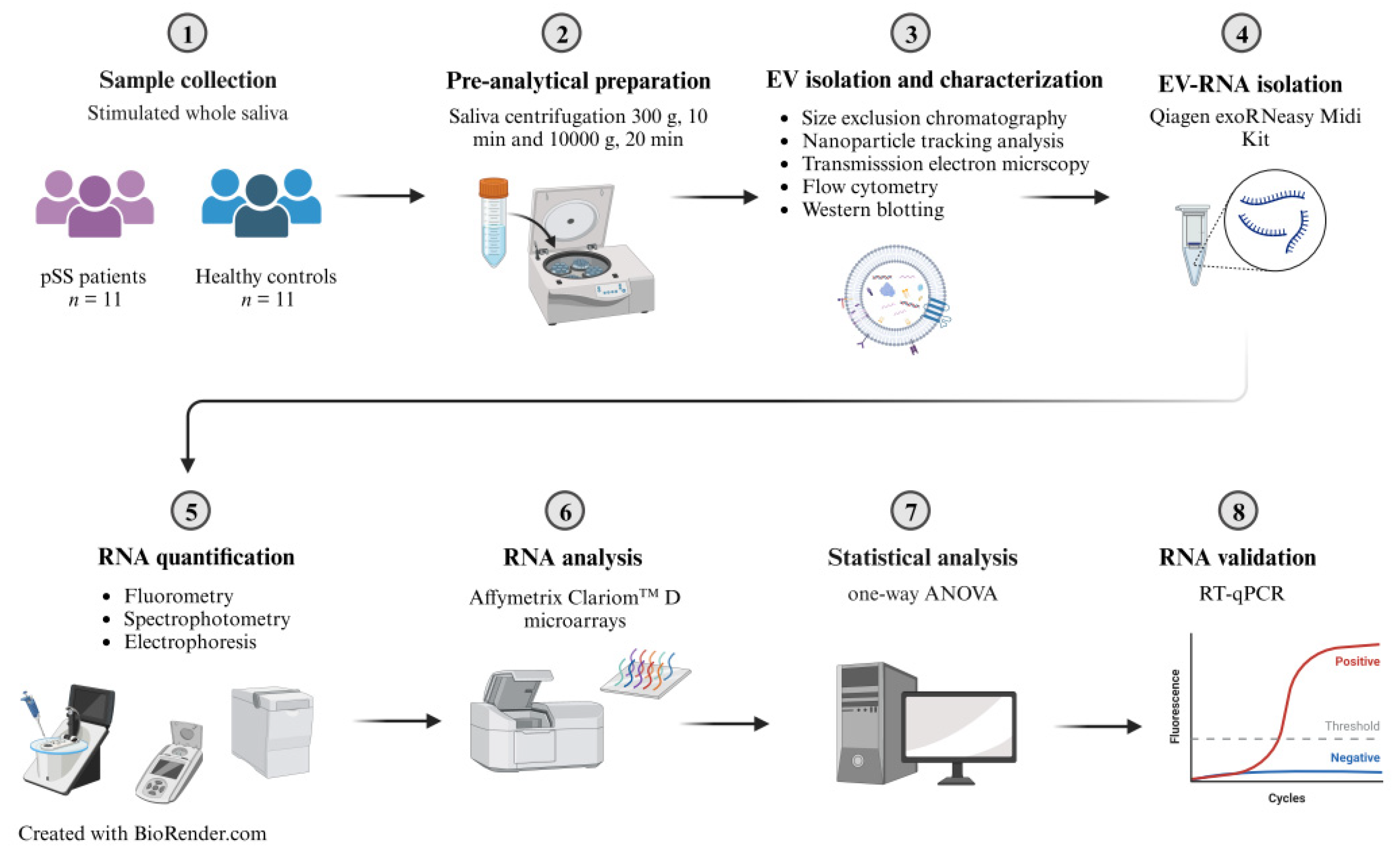
4. Materials and Methods
4.1. Study Participants and Saliva Collection
4.2. Preanalytical Sample Preparation
4.3. EV Characterization
4.4. EV-RNA Isolation Using a Qiagen exoRNeasy Midi Kit
4.5. RNA Quantification, Purity Assessment, and Characterization
4.6. RNA Analysis Using Affymetrix Clariom™ D Microarrays
4.7. RNA Validation Using RT-qPCR
4.8. Bioinformatic Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jonsson, R.; Brokstad, K.A.; Jonsson, M.V.; Delaleu, N.; Skarstein, K. Current concepts on Sjogren’s syndrome—Classification criteria and biomarkers. Eur. J. Oral. Sci. 2018, 126 (Suppl. S1), 37–48. [Google Scholar] [CrossRef]
- Negrini, S.; Emmi, G.; Greco, M.; Borro, M.; Sardanelli, F.; Murdaca, G.; Indiveri, F.; Puppo, F. Sjogren’s syndrome: A systemic autoimmune disease. Clin. Exp. Med. 2022, 22, 9–25. [Google Scholar] [CrossRef]
- Qin, B.; Wang, J.; Yang, Z.; Yang, M.; Ma, N.; Huang, F.; Zhong, R. Epidemiology of primary Sjogren’s syndrome: A systematic review and meta-analysis. Ann. Rheum. Dis. 2015, 74, 1983–1989. [Google Scholar] [CrossRef]
- Rischmueller, M.; Tieu, J.; Lester, S. Primary Sjogren’s syndrome. Best. Pract. Res. Clin. Rheumatol. 2016, 30, 189–220. [Google Scholar] [CrossRef]
- Nakamura, H.; Shimizu, T.; Kawakami, A. Role of Viral Infections in the Pathogenesis of Sjogren’s Syndrome: Different Characteristics of Epstein-Barr Virus and HTLV-1. J. Clin. Med. 2020, 9, 1459. [Google Scholar] [CrossRef]
- Shiboski, C.H.; Shiboski, S.C.; Seror, R.; Criswell, L.A.; Labetoulle, M.; Lietman, T.M.; Rasmussen, A.; Scofield, H.; Vitali, C.; Bowman, S.J.; et al. 2016 American College of Rheumatology/European League Against Rheumatism classification criteria for primary Sjogrens syndro’me: A consensus and data-driven methodology involving three international patient cohorts. Ann. Rheum. Dis. 2017, 76, 9–16. [Google Scholar] [CrossRef]
- Ramos-Casals, M.; Brito-Zeron, P.; Bombardieri, S.; Bootsma, H.; De Vita, S.; Dorner, T.; Fisher, B.A.; Gottenberg, J.E.; Hernandez-Molina, G.; Kocher, A.; et al. EULAR recommendations for the management of Sjogren’s syndrome with topical and systemic therapies. Ann. Rheum. Dis. 2020, 79, 3–18. [Google Scholar] [CrossRef]
- Jung, J.Y.; Kim, J.W.; Kim, H.A.; Suh, C.H. Salivary Biomarkers in Patients with Sjogren’s Syndrome-A Systematic Review. Int. J. Mol. Sci. 2021, 22, 12903. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- McKay, T.B.; Yeung, V.; Hutcheon, A.E.K.; Guo, X.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles in the Cornea: Insights from Other Tissues. Anal Cell. Pathol. 2021, 2021, 9983900. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microRNA biomarkers. Oral. Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef]
- Aqrawi, L.A.; Galtung, H.K.; Vestad, B.; Ovstebo, R.; Thiede, B.; Rusthen, S.; Young, A.; Guerreiro, E.M.; Utheim, T.P.; Chen, X.; et al. Identification of potential saliva and tear biomarkers in primary Sjogren’s syndrome, utilising the extraction of extracellular vesicles and proteomics analysis. Arthritis Res. Ther. 2017, 19, 14. [Google Scholar] [CrossRef]
- Zhao, J.; An, Q.; Zhu, X.; Yang, B.; Gao, X.; Niu, Y.; Zhang, L.; Xu, K.; Ma, D. Research status and future prospects of extracellular vesicles in primary Sjogren’s syndrome. Stem. Cell Res. Ther. 2022, 13, 230. [Google Scholar] [CrossRef]
- Ogawa, Y.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yanoshita, R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol. Pharm. Bull. 2008, 31, 1059–1062. [Google Scholar] [CrossRef]
- Palanisamy, V.; Sharma, S.; Deshpande, A.; Zhou, H.; Gimzewski, J.; Wong, D.T. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE 2010, 5, e8577. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef]
- Lasser, C.; Alikhani, V.S.; Ekstrom, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef]
- Eltermaa, M.; Jakobson, M.; Utt, M.; Koks, S.; Magi, R.; Starkopf, J. Genetic variants in humanin nuclear isoform gene regions show no association with coronary artery disease. BMC Res. Notes 2019, 12, 759. [Google Scholar] [CrossRef]
- Duforestel, M.; Nadaradjane, A.; Bougras-Cartron, G.; Briand, J.; Olivier, C.; Frenel, J.S.; Vallette, F.M.; Lelievre, S.A.; Cartron, P.F. Glyphosate Primes Mammary Cells for Tumorigenesis by Reprogramming the Epigenome in a TET3-Dependent Manner. Front. Genet. 2019, 10, 885. [Google Scholar] [CrossRef] [PubMed]
- Moss, D.J.H.; Pardinas, A.F.; Langbehn, D.; Lo, K.; Leavitt, B.R.; Roos, R.; Durr, A.; Mead, S.; TRACK-HD investigators; REGISTRY investigators; et al. Identification of genetic variants associated with Huntington’s disease progression: A genome-wide association study. Lancet Neurol. 2017, 16, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Karu, I.; Tahepold, P.; Ruusalepp, A.; Reimann, E.; Koks, S.; Starkopf, J. Exposure to sixty minutes of hyperoxia upregulates myocardial humanins in patients with coronary artery disease—A pilot study. J. Physiol. Pharmacol. 2015, 66, 899–906. [Google Scholar] [PubMed]
- Di Mauro, S.; Scamporrino, A.; Filippello, A.; Di Marco, M.; Di Martino, M.T.; Scionti, F.; Di Pino, A.; Scicali, R.; Malaguarnera, R.; Purrello, F.; et al. Mitochondrial RNAs as Potential Biomarkers of Functional Impairment in Diabetic Kidney Disease. Int. J. Mol. Sci. 2022, 23, 8198. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef]
- Cha, S.; Mona, M.; Lee, K.E.; Kim, D.H.; Han, K. MicroRNAs in Autoimmune Sjogren’s Syndrome. Genom. Inform. 2018, 16, e19. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Getting, S.J.; Moschos, S.A. Extracellular vesicles and their nucleic acids for biomarker discovery. Pharmacol. Ther. 2018, 192, 170–187. [Google Scholar] [CrossRef]
- Pan, C.; Stevic, I.; Muller, V.; Ni, Q.; Oliveira-Ferrer, L.; Pantel, K.; Schwarzenbach, H. Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol. Oncol. 2018, 12, 1935–1948. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Acton, R.J.; Yuan, W.; Gao, F.; Xia, Y.; Bourne, E.; Wozniak, E.; Bell, J.; Lillycrop, K.; Wang, J.; Dennison, E.; et al. The genomic loci of specific human tRNA genes exhibit ageing-related DNA hypermethylation. Nat. Commun. 2021, 12, 2655. [Google Scholar] [CrossRef]
- Liu, D.S.K.; Yang, Q.Z.C.; Asim, M.; Krell, J.; Frampton, A.E. The Clinical Significance of Transfer RNAs Present in Extracellular Vesicles. Int. J. Mol. Sci. 2022, 23, 3692. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; Santos, M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip. Rev. RNA 2017, 8, e1423. [Google Scholar] [CrossRef] [PubMed]
- Sobala, A.; Hutvagner, G. Transfer RNA-derived fragments: Origins, processing, and functions. Wiley Interdiscip. Rev. RNA 2011, 2, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Yu, J.; Zhou, P. Role of tRNA-derived fragments in cancer: Novel diagnostic and therapeutic targets tRFs in cancer. Am. J. Cancer Res. 2020, 10, 393–402. [Google Scholar]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef]
- Wang, J.; Ma, G.; Li, M.; Han, X.; Xu, J.; Liang, M.; Mao, X.; Chen, X.; Xia, T.; Liu, X.; et al. Plasma tRNA Fragments Derived from 5′ Ends as Novel Diagnostic Biomarkers for Early-Stage Breast Cancer. Mol. Ther. Nucleic Acids 2020, 21, 954–964. [Google Scholar] [CrossRef]
- Shen, Y.; Yu, X.; Zhu, L.; Li, T.; Yan, Z.; Guo, J. Transfer RNA-derived fragments and tRNA halves: Biogenesis, biological functions and their roles in diseases. J. Mol. Med. 2018, 96, 1167–1176. [Google Scholar] [CrossRef]
- Zhu, L.; Li, J.; Gong, Y.; Wu, Q.; Tan, S.; Sun, D.; Xu, X.; Zuo, Y.; Zhao, Y.; Wei, Y.Q.; et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 2019, 18, 74. [Google Scholar] [CrossRef]
- Torres, A.G.; Marti, E. Toward an Understanding of Extracellular tRNA Biology. Front. Mol. Biosci. 2021, 8, 662620. [Google Scholar] [CrossRef]
- Thompson, D.M.; Parker, R. Stressing out over tRNA cleavage. Cell 2009, 138, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Lin, Y.; Luo, Y.; Xiong, X.; Wang, L.; Durante, K.; Li, J.; Zhou, F.; Guo, Y.; Chen, S.; et al. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: A multicenter prospective study. Mol. Cancer 2022, 21, 21. [Google Scholar] [CrossRef] [PubMed]
- Driedonks, T.A.P.; Nolte-’t Hoen, E.N.M. Circulating Y-RNAs in Extracellular Vesicles and Ribonucleoprotein Complexes; Implications for the Immune System. Front. Immunol. 2018, 9, 3164. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, M.P.; Krude, T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015, 66, 20–29. [Google Scholar] [CrossRef]
- Boccitto, M.; Wolin, S.L. Ro60 and Y RNAs: Structure, functions, and roles in autoimmunity. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 133–152. [Google Scholar] [CrossRef]
- Vojtech, L.; Woo, S.; Hughes, S.; Levy, C.; Ballweber, L.; Sauteraud, R.P.; Strobl, J.; Westerberg, K.; Gottardo, R.; Tewari, M.; et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014, 42, 7290–7304. [Google Scholar] [CrossRef]
- Tosar, J.P.; Gambaro, F.; Sanguinetti, J.; Bonilla, B.; Witwer, K.W.; Cayota, A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015, 43, 5601–5616. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; ’t Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef]
- Kapsogeorgou, E.K.; Abu-Helu, R.F.; Moutsopoulos, H.M.; Manoussakis, M.N. Salivary gland epithelial cell exosomes: A source of autoantigenic ribonucleoproteins. Arthritis Rheum. 2005, 52, 1517–1521. [Google Scholar] [CrossRef]
- Greiling, T.M.; Dehner, C.; Chen, X.; Hughes, K.; Iniguez, A.J.; Boccitto, M.; Ruiz, D.Z.; Renfroe, S.C.; Vieira, S.M.; Ruff, W.E.; et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 2018, 10, eaan2306. [Google Scholar] [CrossRef]
- Wang, X.; Guo, S.; Zhou, X.; Wang, Y.; Zhang, T.; Chen, R. Exploring the Molecular Mechanism of lncRNA-miRNA-mRNA Networks in Non-Syndromic Cleft Lip with or without Cleft Palate. Int. J. Gen. Med. 2021, 14, 9931–9943. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.F.; Pegtel, D.M.; Lambertz, U.; Leonardi, T.; O’Driscoll, L.; Pluchino, S.; Ter-Ovanesyan, D.; Nolte-’t Hoen, E.N. ISEV position paper: Extracellular vesicle RNA analysis and bioinformatics. J. Extracell Vesicles 2013, 2, 22859. [Google Scholar] [CrossRef] [PubMed]
- Yeri, A.; Courtright, A.; Reiman, R.; Carlson, E.; Beecroft, T.; Janss, A.; Siniard, A.; Richholt, R.; Balak, C.; Rozowsky, J.; et al. Total Extracellular Small RNA Profiles from Plasma, Saliva, and Urine of Healthy Subjects. Sci. Rep. 2017, 7, 44061. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Aas, V.; Ovstebo, R.; Brusletto, B.S.; Aspelin, T.; Troseid, A.S.; Qureshi, S.; Eid, D.S.O.; Olstad, O.K.; Nyman, T.A.; Haug, K.B.F. Distinct microRNA and protein profiles of extracellular vesicles secreted from myotubes from morbidly obese donors with type 2 diabetes in response to electrical pulse stimulation. Front. Physiol. 2023, 14, 1143966. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cross, T.; Haug, K.B.F.; Brusletto, B.S.; Ommundsen, S.K.; Trøseid, A.-M.S.; Aspelin, T.; Olstad, O.K.; Aass, H.C.D.; Galtung, H.K.; Utheim, T.P.; et al. Non-Coding RNA in Salivary Extracellular Vesicles: A New Frontier in Sjögren’s Syndrome Diagnostics? Int. J. Mol. Sci. 2023, 24, 13409. https://doi.org/10.3390/ijms241713409
Cross T, Haug KBF, Brusletto BS, Ommundsen SK, Trøseid A-MS, Aspelin T, Olstad OK, Aass HCD, Galtung HK, Utheim TP, et al. Non-Coding RNA in Salivary Extracellular Vesicles: A New Frontier in Sjögren’s Syndrome Diagnostics? International Journal of Molecular Sciences. 2023; 24(17):13409. https://doi.org/10.3390/ijms241713409
Chicago/Turabian StyleCross, Tanya, Kari Bente Foss Haug, Berit Sletbakk Brusletto, Stine Kamilla Ommundsen, Anne-Marie Siebke Trøseid, Trude Aspelin, Ole Kristoffer Olstad, Hans Christian Dalsbotten Aass, Hilde Kanli Galtung, Tor Paaske Utheim, and et al. 2023. "Non-Coding RNA in Salivary Extracellular Vesicles: A New Frontier in Sjögren’s Syndrome Diagnostics?" International Journal of Molecular Sciences 24, no. 17: 13409. https://doi.org/10.3390/ijms241713409
APA StyleCross, T., Haug, K. B. F., Brusletto, B. S., Ommundsen, S. K., Trøseid, A.-M. S., Aspelin, T., Olstad, O. K., Aass, H. C. D., Galtung, H. K., Utheim, T. P., Jensen, J. L., & Øvstebø, R. (2023). Non-Coding RNA in Salivary Extracellular Vesicles: A New Frontier in Sjögren’s Syndrome Diagnostics? International Journal of Molecular Sciences, 24(17), 13409. https://doi.org/10.3390/ijms241713409